Abstract
Reading and language abilities are heritable traits that are likely to share some genetic influences with each other. To identify pleiotropic genetic variants affecting these traits, we first performed a genome-wide association scan (GWAS) meta-analysis using three richly characterized datasets comprising individuals with histories of reading or language problems, and their siblings. GWAS was performed in a total of 1862 participants using the first principal component computed from several quantitative measures of reading- and language-related abilities, both before and after adjustment for performance IQ. We identified novel suggestive associations at the SNPs rs59197085 and rs5995177 (uncorrected P ≈ 10–7 for each SNP), located respectively at the CCDC136/FLNC and RBFOX2 genes. Each of these SNPs then showed evidence for effects across multiple reading and language traits in univariate association testing against the individual traits. FLNC encodes a structural protein involved in cytoskeleton remodelling, while RBFOX2 is an important regulator of alternative splicing in neurons. The CCDC136/FLNC locus showed association with a comparable reading/language measure in an independent sample of 6434 participants from the general population, although involving distinct alleles of the associated SNP. Our datasets will form an important part of on-going international efforts to identify genes contributing to reading and language skills.
Keywords: pleiotropic variants, CLDRC, developmental dyslexia, GWAS, language, meta-analysis, reading, reading disability, SLIC, specific language impairment
Reading disability (RD, also known as developmental dyslexia) refers to a significant difficulty in reading that cannot be explained by obvious causes, such as sensory impairments or lack of educational opportunity (Shaywitz et al. 1990). Specific language impairment (SLI) is diagnosed as an unexpected difficulty or delay in acquiring spoken language abilities, despite normal hearing and intelligence, and in absence of overt neurological deficits (Bishop 1994). RD and SLI are among the most prevalent neurocognitive disorders of school-aged children, with prevalence ≈5–8% in many populations (Shaywitz et al. 1990; Tomblin et al. 1997). Both are complex disorders with moderate to high heritabilities (30–70%) as assessed by studies of families and twins (Barry et al. 2007; Fisher & DeFries 2002).
RD and SLI display high comorbidity: 43% of SLI children are later diagnosed with RD and up to 55% of dyslexic children meet criteria for SLI (McArthur et al. 2000; Snowling et al. 2000). Moreover, RD and SLI show comorbidity with other neurodevelopmental traits including attention deficit hyperactivity disorder (ADHD) (Pennington 2006; Willcutt et al. 2010) and speech sound disorders (Newbury & Monaco 2010; Pennington & Bishop 2009). It is likely that these disorders arise due to some shared genetic/neurobiological mechanisms, as well as non-shared causal factors (Newbury et al. 2011; Paracchini 2011). A study of twins by Harlaar et al. (2008) indicated that an association between early language and later reading is underpinned by common environmental and genetic influences, and a family study by Logan et al. (2011) also found significant genetic correlations of reading and language measures.
Variants of several genes have previously been associated with RD, most notably DYX1C1 (15q21, Taipale et al. 2003), KIAA0319 and DCDC2 (6p22, Cope et al. 2005; Francks et al. 2004; Meng et al. 2005), MRPL19/C2ORF3 (2p12, Anthoni et al. 2007) and ROBO1 (3p12, Bates et al. 2011; Hannula-Jouppi et al. 2005). Similarly, some loci have been implicated in SLI; variants in genes, such as CNTNAP2 (7q36, Vernes et al. 2008) and CMIP and ATP2C2 (16q23-24, Newbury et al. 2009) show associations with quantitative traits in children with typical SLI, while rare mutations of FOXP2 (7q31, Fisher & Scharff 2009) cause a monogenic speech and language disorder. These genes were mostly identified through linkage analysis followed by either positional cloning or else targeted association mapping. Functional analyses suggest that some of these genes mediate important processes in central nervous system (CNS) development, such as neuronal migration, axonal guidance and neurite outgrowth (Carrion-Castillo et al. 2013; Vernes et al. 2011). A subset of the candidate genes may contribute to both RD and SLI, again indicating a partial genetic overlap for these traits (Bates et al. 2011; Newbury et al. 2011; Scerri et al. 2011b). Crucially, an overwhelming majority of the heritable variance in reading and language skills is unexplained, and the molecular mechanisms that contribute to RD and SLI remain largely unknown (Newbury & Monaco 2010; Peterson & Pennington 2012).
Some of the genetic variation contributing to RD and SLI is also likely to impact on reading/language skills in the general population (Bates et al. 2011; Luciano et al. 2007; Paracchini et al. 2008, 2011; Scerri et al. 2011b; Whitehouse et al. 2011). To detect previously undiscovered associations of common genetic variants with reading and language skills, it is therefore appropriate to sample broad ranges of the trait distributions in study datasets, while screening over the entire genome.
In recent years, a small number of studies have tried to identify genes involved in reading and/or language through genome-wide association scanning (GWAS). An early GWAS for reading ability used DNA pooling of low vs. high reading ability groups in ∼1500 7-year-old children, and a relatively low density SNP microarray with ∼107,000 SNPs (Meaburn et al. 2008). The SNPs showing the largest allele frequency differences between low and high ability groups were further genotyped and tested in an additional sample of 4,258 children, with 10 SNPs finally showing nominally significant association with continuous variation in reading ability (Meaburn et al. 2008). A GWAS on mismatch negativity, which is a potential endophenotype of dyslexia derived from electroencephalography, has also been reported based on 386 dyslexic children, and showed replicable association of the SNP rs4234898 on 4q32 along with the haplotype rs4234898-rs11100040 (Roeske et al. 2011). These were shown to affect mRNA expression levels of SLC2A3 (12p13), which codes for a neuronal glucose transporter, suggesting a possible role of glucose levels in memory performance necessary for speech perception in dyslexia (Roeske et al. 2011). More recently, a genome-wide linkage and association scan using ∼133,000 SNPs, in 718 subjects from 101 dyslexia-affected families, reported a borderline significant association with dyslexia status at rs9313548, near FGF18 (5q35.1), which is a gene involved in laminar positioning of cortical neurons during development (Field et al. 2013).
Two GWAS studies have directly attempted to identify shared genetic contributions to reading and language abilities. Luciano et al. (2013), in a GWAS on quantitative reading and language traits in two population datasets (N ∼ 6500), found the strongest association between rs2192161, in the ABCC13 pseudogene (21q11.2), and a nonword repetition measure (p ∼ 7 × 10–8), while rs4807927 (DAZAP1, 19p13.3) showed association with both word reading and a composite reading–spelling factor score (p ∼ 10–6 for both traits). In the same study, CDC2L1, CDC2L2, LOC728661 (1p36.33) and RCAN3 (1p36.11) showed significant gene-based associations with the reading–spelling factor (Luciano et al. 2013). A case-control GWAS using a relatively small number of RD (N = 353), language impairment (N = 163) and comorbid cases (N = 174), in comparison to general population controls (N = 4117), identified nominally significant associations for the comorbid cases at rs12636438 and rs1679255 in ZNF385D (3p24.3) (Eicher et al. 2013). These SNPs also showed associations with a vocabulary measure and white matter volumes of brain fibre tracts previously implicated in language, in an independent dataset (Eicher et al. 2013).
In the present study, we carried out a GWAS meta-analysis for genetic variants influencing reading and language abilities. We included three long-established datasets comprising children with reading or language problems, along with their siblings. This approach complemented other recent GWAS studies of reading/language performance (Eicher et al. 2013; Luciano et al. 2013) because it included continuous trait variance across a broad range of reading and language abilities, but also involved a pronounced enrichment for poor performance while not applying an arbitrary dichotomy between RD/SLI cases and controls.
Within each dataset, we tested single nucleotide polymorphisms (SNPs), along with single base insertions/deletions (indels), for association with the first Principal Component (PC) derived from a range of reading- and language-related quantitative traits. We then meta-analysed the GWAS results from the separate datasets, followed by gene- and pathway-level analysis, and we checked the most significant associations arising from our analysis within the GWAS results generated by Luciano et al. (2013).
Although we used PC-based analysis as a form of data reduction for the purposes of GWAS, we also investigated the two most significant SNP associations arising from our meta-analysis by using multivariate association modelling in each dataset, and by testing of these SNPs against the individual measures separately. This approach would help to understand the cross-phenotypic effects involved. In other words, the PC-based GWAS was used to identify potential genetic effects on shared variance between multiple reading and language measures, and then pleiotropy was investigated in more detail through univariate analysis and multivariate modelling, for individual SNPs implicated by the PC-based GWAS meta-analysis. In addition, in order to more closely match the trait measurement across all datasets, we repeated the GWAS and meta-analysis using the first PC of only single word reading and spelling ability, because these were the only two measures available in all datasets.
Some genetic effects on reading and language may be pleiotropic for IQ, whereas other effects may be largely or wholly independent of IQ (Bishop & Snowling 2004; Pennington & Bishop 2009). To detect the latter type of effect it is advantageous to remove the shared variance with IQ that is present in measures of reading and language, prior to association testing. We therefore performed our GWAS analyses both with and without IQ-adjustment of the reading and language measures. In addition, Luciano et al. (2013) analysed only IQ-adjusted data, so that for cross-comparing of results an IQ-adjustment was desirable to include in this study.
Subjects and methods
Datasets
UK Reading Disability (UK-RD)
This dataset comprised children diagnosed with RD, and their siblings, collected at the Dyslexia Research Centre clinics in Oxford and Reading, or the Aston Dyslexia and Development Clinic in Birmingham, UK. Ethical approval was acquired from the Oxfordshire Psychiatric Research Ethics Committee (OPREC O01.02) and written informed consent of the participants (or their parents) was obtained. The total number of participants was 983, mean age 11.7 years, age range 5–31, from 608 independent nuclear families. All children, regardless of diagnosis, were administered psychometric tests of reading- and language-related abilities, as well as assessments of verbal and non-verbal IQ (details further below). A subset of this dataset has been analysed in previous studies on reading (Becker et al. 2013) and handedness traits (Brandler et al. 2013; Scerri et al. 2011a), but no GWAS of reading-/language-related traits has previously been reported.
SLI Consortium (SLIC)
The SLI Consortium dataset comprised children affected by SLI, along with their siblings, recruited from five centres across the UK; The Newcomen Centre at Guy's Hospital, London (now called Evelina Children's Hospital); the Cambridge Language and Speech Project (CLASP); the Child Life and Health Department at the University of Edinburgh; the Department of Child Health at the University of Aberdeen and the Manchester Language Study, as described in previous reports by the SLI Consortium (Falcaro et al. 2008; Newbury et al. 2009; The SLI Consortium 2002, 2004). This sample included 49 families from the Guy's Hospital, London cohort which had not been included in previous SLI Consortium studies. Ethical agreement was given by local ethics committees of the hospitals involved in the consortium, and all subjects provided informed consent. All children in this sample were assessed for a number of reading- and language-related traits (see below) regardless of their language ability. For this study, we obtained genome-wide genotype data for affected probands and their available siblings, for a total of 548 participants, mean age 10 years, age range 5–19, from 288 independent nuclear families. The SLIC dataset has been used for prior linkage studies (Falcaro et al. 2008; The SLI Consortium 2002, 2004), and targeted candidate gene analyses (Newbury et al. 2009; Vernes et al. 2008). More recently, it has been used for investigating copy number variants (Ceroni et al. 2014), identification of chromosomal abnormalities (Simpson et al. 2014) and in a genome-wide search for parent-of-origin effects on SLI (Nudel et al. 2014). However, no GWAS for continuous language and reading scores has yet been reported for this (or any other) SLI sample.
Colorado Learning Disabilities Research Centre (CLDRC)
The Colorado Learning Disabilities Research Centre (CLDRC) dataset was derived from an ongoing study on the aetiology of learning disabilities run in 27 school districts in Colorado, USA (DeFries et al. 1997; Willcutt et al. 2005). Pairs of twins were initially recruited based on a school report of RD, ADHD or other learning disabilities in one or both of the twins; they were then administered a number of psychometric tests for several learning-related skills, along with their additional co-siblings, and DNA was collected for genetic studies. The Institutional Review Boards of the University of Nebraska Medical Center and of the University of Colorado at Boulder had approved the protocol, and written informed consent of the participants (or their parents) was obtained.
For this study, for MZ twin pairs, we selected one child per pair based on the maximum availability of reading- and language-related trait data, or otherwise randomly. The sample of twins and siblings available for this study comprised 749 participants in total, mean age 11.7 years, age range 8–19, from 343 unrelated twinships/sibships. Of these, 266 of the twinships/sibships (a total of 585 participants) were originally recruited via a proband with a history of RD, and 77 of the twinships/sibships (164 participants in total) were originally recruited via a proband with a history of ADHD. We analysed these two subsets separately for GWAS before meta-analysing the results together with those from the other datasets listed above. The two subsets are indicated hereafter as CLDRC-RD and CLDRC-ADHD. As for the other datasets, no prior GWAS has been reported.
Genotype data generation, quality control (QC) and imputation
DNA was extracted from whole blood or buccal swab samples and prepared for genotyping using standard protocols. Genome-wide genotype data were generated for each dataset using Illumina® SNP arrays. These were the HumanHap 550k for a first genotyping wave of 200 subjects from UK-RD, and the Human OmniExpress (730k SNPs) for SLIC, CLDRC and the remaining UK-RD samples. Data were processed using Illumina's BeadStudio®/GenomeStudio® software, following the manufacturer's guidelines. All datasets then underwent a first round of quality control, using functions in the software PLINK v1.07 (Purcell et al. 2007; http://pngu.mgh.harvard.edu/∼purcell/plink/), in which all SNPs deviating from Hardy–Weinberg Equilibrium (HWE, P < 1 × 10–6), with Minor Allele Frequency (MAF) < 1% and call frequency < 99%, were filtered out. In addition, samples were excluded if they showed inconsistencies in genome-wide identity-by-descent sharing with their siblings and unrelated individuals, or sex mismatches, or call rates <98%. Multi-dimensional scaling (MDS) analysis of genome-wide genotype data was used to identify any subjects that did not cluster together with the majority of the dataset, and these were discarded, as were any outliers for genome-wide homozygosity. These QC steps were followed by genotype phasing using MACH v1.0 (Liu et al. 2010; http://www.sph.umich.edu/csg/abecasis/MACH/index.html) and imputation of SNPs and single-base indels using Minimac (Howie et al. 2012; http://genome.sph.umich.edu/wiki/Minimac), with the 1000 Genomes Project reference dataset (GIANT all populations panel, Phase 1, v3; The 1000 Genomes Project Consortium, 2012; http://www.1000genomes.org). We excluded poorly imputed polymorphisms (with r2 < 0.3), and deleted individual genotypes with imputation quality scores <0.9. A final quality control procedure was then run on the imputed data, using PLINK, in which we discarded SNPs with HWE P < 5 × 10–6, MAF < 1%, and call frequency <95%. Key features of the QC are shown in Table1. Further details are reported in Appendix S4.
Table 1.
Genotype quality control (QC) filters used and number of samples/markers discarded at each step (see Methods and Appendix S4 for details)
| QC step | CLDRC (749)† | UK-RD (200 + 818)‡ | SLIC (548) |
|---|---|---|---|
| HWE P < 1 × 10–6 (SNPs) | 57 | 12,631§; 191 | 54 |
| MAF < 1% (SNPs) | 74,770 | 23,467; 77,342 | 1,718 |
| Call freq < 99% (SNPs) | 0 ¶ | 82,052; 0¶ | 72,043 |
| Call rate < 98% (samples) | 0 ¶ | 3; 0 ¶ | 9 |
| IBD sharing (samples) | 11 | 1; 7 | 17 |
| Sex mismatch (samples) | 3 | 0; 8** | 13†† |
| Homozygosity outlier (samples) | 6 | 1; 3 | 2 |
| MDS outlier (samples) | 0 | 0; 2 | 5 |
| HWE P < 5 × 10–6 (SNPs)* | 2,166 | 2,779 | 2,096 |
| MAF < 1% (SNPs)* | 3,640,742 | 1,980,500 | 3,260,639 |
| Call freq <95% (SNPs)* | 1,729,493 | 1,704,412 | 1,766,376 |
| Call rate < 95%, MDS outliers, IBD sharing (samples)* | 0 | 0 | 0 |
| Passing QC | 729 (6,427,200) | 959 (6,190,549) | 502 (6,240,842) |
Final number of samples (and SNPs in brackets) passing the genotype QC are reported in the bottom row. Note that these numbers do not also account for QC of the trait scores.
After imputation QC. Before this step, imputed SNPs with r2 < 0.3 were filtered out, and all the genotypes with quality score <0.9 were set to missing.
As CLDRC-RD and CLDRC-ADHD were processed together and drawn from the same population, we treated them as a single dataset in the genotype QC.
As UK-RD samples had been genotyped on two different Illumina® platforms (see Methods) the subsets were analysed separately before imputation, and pre-imputation QC details are therefore reported for both the subsets (first genotyping wave with HumanHap 550k and second genotyping wave with Human OmniExpress). Note that 35 samples were genotyped on both of the arrays, and one of these samples showed inconsistent genotyping and was therefore discarded in both subsets.
The high number of SNPs discarded at this stage was due to the fact that no quality filter had been applied on this subset during genotype call process (see Appendix S4).
In these cases, SNPs with call frequency <99% and samples with call rate <98% had already been discarded during genotype call process (see Appendix S4).
Includes three sex chromosome abnormalities carriers.
Includes nine samples with sex chromosome abnormalities and one with X chromosome call rate <95%.
At the end of the genotype QC process, we had data for 959 participants and 6,190,549 polymorphisms in UK-RD, 729 participants and 6,427,000 polymorphisms in CLDRC, and 502 participants and 6,240,842 polymorphisms in SLIC, with 5,518,496 polymorphisms shared across all three datasets.
Reading and language measures
Table2 lists the reading- and language-related traits that were assessed in the different datasets, as detailed in prior publications (Compton et al. 2001; Francks et al. 2004; Friend & Olson 2010; The SLI Consortium 2002, 2004). Further information on these measures is given in Tables5. To remove outliers, trait scores were excluded when they were more than three standard deviations from the relevant sample mean. Subjects with three or more such outliers were excluded from the dataset (one participant in UK-RD and one in CLDRC-RD). Reading/language traits had been previously age-adjusted according to normative data (Compton et al. 2001; Francks et al. 2004; Friend & Olson 2010; The SLI Consortium 2002, 2004). When a measure differed significantly from normality, we performed a within-dataset rank-normalization to attain normality and improve the suitability for principal components analysis (see Appendix S4 for details). We also excluded subjects showing full scale IQ < 70 (one participant from CLDRC-RD, and four participants from SLIC). This left 564 subjects in CLDRC-RD, 958 in UK-RD, 498 in SLIC and 163 in CLDRC-ADHD, which were used for the computation of the First Principal Component. Pairwise trait correlations within each dataset were calculated as the median correlation over 100 repeat random samplings of one individual from each independent sibship (see Appendix S4).
Table 2.
Phenotypic traits available (when labelled by ‘x’) and measures used for PC1 derivation within each dataset (labelled with relative loadings on PC1 in parentheses)
| Trait | Description (ability assessed) | CLDRC-RD (564) | UK-RD (958) | SLIC (498) | CLDRC-ADHD (163) |
|---|---|---|---|---|---|
| WRead | Reading real words | x (0.918) | x (0.918) | x (0.902) | x (0.871) |
| WSpell | Spelling real words | x (0.813) | x (0.852) | x (0.862) | x (0.764) |
| PD | Ability to convert letter strings into sounds, according to given phonetic rules | x (0.895, 0.861)* | x (0.809) | x (0.821, 0.729)* | |
| PA | Ability to recognize and manipulate speech sounds (phonemes) | x (0.801) | x† | x (0.744) | |
| OC | Ability to recognize a word as an orthographic unit and to retrieve the corresponding phonological form | x (0.764) | x (0.888) | x (0.644) | |
| NWR | Ability to repeat nonsense words orally presented | x (0.493) | x (0.665) | x (0.355) | |
| ELS | Sentence recalling and production (expressive domain of language) | x (0.856) | |||
| RLS | Listening and auditory comprehension (receptive domain of language) | x (0.837) | |||
| VIQ | Verbal reasoning | x | x | x | x |
| PIQ | Logical reasoning | x | x | x | x |
| PC1 | Common variance in reading and language skills | 544 | 914 | 245 | 159 |
| IQ-adjusted PC1 | Common variance in reading and language skills, not shared with general cognitive abilities | 544 | 878 | 245 | 159 |
Sample sizes of the datasets (after genotype and phenotype QC) are reported in the header row. Sample sizes involved in the PC1 meta-analysis are reported at the bottom of the table (since we excluded participants with at least one missing measure among the traits involved in principal component analysis).
WRead, word reading; WSpell, word spelling; PD, phonological decoding; PA, phoneme awareness; OC, orthographic coding; NWR, nonword repetition; ELS/RLS, expressive/receptive language score; VIQ/PIQ, verbal/performance IQ.
Loadings of nonword reading and phonological choice (respectively) on PC1s.
Trait excluded from the PCA due to the low number of measures available.
Table 5.
Language-/reading-related traits available in the CLDRC dataset
| Trait | Test | Test description* | Statistical elaboration† |
|---|---|---|---|
| WRead | Peabody Individual Achievement Test (PIAT)1 | Reading aloud in sequence single real words increasing in semantic and phonetic difficulty, until errors are made in five out of any seven consecutive items (untimed) | C, A, S, R |
| Timed oral reading2,3 | Reading aloud a series of single real words within 2 seconds of their presentation, until errors are made in 10 out of any 20 consecutive items | ||
| WSpell | PIAT1 | Choosing the correct spelling of a series of real words (of increasing difficulty) orally presented, among four orthographically and often phonologically similar alternatives printed on a card (for each word), until errors are made in five out of seven consecutive responses | A, S |
| PD | Oral Nonword Reading Task2,3 | Reading aloud a series of single-syllable nonsense words (structure ranging from vcv to cccvcv) | C, A, S, R |
| Reading aloud a series of two-syllables nonsense words | |||
| Phonological Choice (Silent Nonword Reading Task)2,3 | Choosing which of three nonsense words would sound like a real word if read aloud (for n triplets of nonwords) | A, S, R | |
| PA | Phoneme Segmentation and Transposition Task3 | Taking the first phoneme of a word, putting it at the end and add the sound/ay/(for n words, e.g. rope → ope-ray) | C, A, S, R |
| Phoneme Deletion Task3 | Repeating nonwords within 2 seconds of their oral presentation, then removing a specified phoneme and pronouncing the resulting words within another 4 seconds (e.g. ‘say prot..now say prot without the/r/’ ‘pot’) | ||
| OC | Word-Pseudohomophone Choice2,4 | Speeded forced-choice to distinguish a real word from a phonologically similar nonword (for n pairs of words-nonwords; e.g. rane vs. rain) | C, A, S, R |
| Homophone Choice*,4 | Selecting which of two homophones visually presented answers a question asked orally by the tester (for n pairs of words, e.g. ‘Which is a flower?’ rose rows) | ||
| NWR | Gathercole & Baddeley5 | Repeating tape-recorded nonsense words of increasing length and complexity | A, S, R |
| vIQ | WISC-R/WAIS-R6 | Comprehension (explaining situations, actions, or activities that the examinee is expected to be familiar with) | None |
| Information (general cultural knowledge test) | |||
| Similarities (explaining how two words are alike/similar) | |||
| Vocabulary (defining a provided word) | |||
| pIQ | WISC-R/WAIS-R6 | Block design (arranging blocks to duplicate a given image/design) | None |
| Object assembly (correctly assembling the parts that an object is divided into, like a puzzle) | |||
| Picture arrangement (arranging a number of given pictures from left to right to tell the intended story) | |||
| Picture completion (identifying the missing part in a series of pictures representing common objects) |
Superscript numbers after each test indicate the initial reference for it, where further details on the test can be found:
Dunn & Markwardt 1970;
Olson et al. 1989;
Olson et al. 1994a;
Olson et al. 1994b;
Gathercole et al. 1994;
Wechsler 1974.
Where more than one battery is administered, the total score is computed as a sum of the raw scores from each subtest (IQ measures), as an average of z-scores derived from accuracy scores (% of correct responses) and median correct reaction times of the two subtests (nonword reading), or as the arithmetic average of the raw scores from each subtest (all the other measures).
Legend of statistical elaborations: C, composite score; A, age-adjusted (score regressed against age and age2); S, standardized against the normative mean of a control population; R, further rank-normalized (using Blom's formula), because the trait distribution after standardization differed from normality (Shapiro–Wilk test P value < 0.05).
Table 3.
Language-/reading-related traits available in the UK-RD dataset
| Trait | Test | Test description* | Statistical elaboration† |
|---|---|---|---|
| WRead | British Ability Scale (BAS)/Wide Range Achievement Test-Revised (WRAT-R)1,2 | Reading aloud a series of real words presented on a card | A, S, R |
| WSpell | BAS/WRAT-R1,2 | Writing words that are dictated by the test administrator | A, S, R |
| PD | Castles & Coltheart (C&C)3,4 | Reading aloud nonsense words of increasing difficulty, according to English grapheme-phoneme conversion rules | A, S, R |
| Nonword reading | |||
| PA | Spoonerism test5,6 | Simple phoneme deletion and substitution (e.g. replace the first sound in dog with \l\ to make log) | A, S, R |
| Complex phoneme deletion and substitution | |||
| Spoonerism (swapping the first sounds of two words, e.g. from spoon, dog to doon, spog) | |||
| OC | C&C3,4 | Reading aloud irregular words of increasing difficulty (i.e. words whose pronunciation does not follow the English grapheme-phoneme conversion rules, e.g. yacht) | A, S, R |
| Irregular word reading | |||
| vIQ | BAS/Wechsler Adult Intelligence Scale – Revised (WAIS-R)7 | Similarities subtest only (explaining how two/three words are similar or go together) | A, S, R |
| pIQ | BAS1 | Matrices subtest only (predicting missing components of increasingly complex matrices containing abstract symbols) | A, S |
Superscript numbers after each test indicate the initial reference for it, where further details on the test can be found.
Elliot et al. 1979;
Jastak & Wilkinson 1984;
Castles & Coltheart 1993;
Coltheart & Leahy 1996;
Gallagher & Frederickson 1995;
Frederickson 1995;
Wechsler 1981.
Where more than one battery is administered, the total score is usually computed as a sum of the raw scores from each subtest.
Legend of trait adjustments: A, age-adjusted; S, standardized against the normative mean of the population of reference; R, further rank-normalized (using Blom's formula), because the trait distribution after standardization differed from normality (Shapiro–Wilk test P < 0.05).
Table 4.
Language-/reading-related traits available in the SLIC dataset
| Trait | Test | Test description* | Statistical elaboration† |
|---|---|---|---|
| WRead | Wechsler Objectives of Reading Dimensions (WORD)1 | Reading single real words of increasing difficulty | A, S, R |
| WSpell | WORD1 | Spelling of single real words | A |
| NWR | Gathercole & Baddeley2 | Repeating tape-recorded nonsense words of increasing length and complexity | A, S, R |
| ELS | Clinical Evaluation of Language Fundamentals Revised (CELF-R)3 | Formulating sentences (formulating sentences about visual stimuli using a targeted word or phrase) | A, S, R |
| Recalling sentences (imitating sentences presented by the examiner) | |||
| Sentence assembly (producing two semantically/grammatically correct sentences from visually and orally presented words/groups of words) | |||
| RLS | CELF-R3 | Oral directions (pointing to pictured objects in response to oral directions) | A, S, R |
| Semantic relations (listening to a sentence and selecting the two choices that answer a target question, out of four possible answers) | |||
| Word classes (choosing two related words and describing their relationship) | |||
| vIQ | Wechsler Intelligence Scale for Children (WISC)/WAIS4 | Arithmetic (solving orally administered arithmetic word problems) | A |
| Comprehension (explaining situations, actions, or activities that the examinee is expected to be familiar with) | |||
| Digit span (reciting a sequence of digits presented by the examiner by recalling them in the same/reverse order) | |||
| Information (general cultural knowledge test) | |||
| Similarities (explaining how two words are alike/similar) | |||
| Vocabulary (defining a provided word) | |||
| pIQ | WISC/WAIS4 | Block design (arranging blocks to duplicate a given image/design) | A, S, R |
| Coding (marking rows of shapes with different lines/transcribing symbols under digits, according to a given code) | |||
| Object assembly (correctly assembling the parts that an object is divided into, like a puzzle) | |||
| Picture arrangement (arranging a number of given pictures from left to right to tell the intended story) | |||
| Picture completion (identifying the missing part in a series of pictures representing common objects) |
Superscript numbers after each test indicate the initial reference for it, where further details on the test can be found:
Rust et al. 1993;
Gathercole et al. 1994;
Semel et al. 1992;
Wechsler et al. 1992.
Where more than one battery is administered, the total score is usually computed as a sum of the raw scores from each subtest.
Legend of statistical elaborations: A, age-adjusted; S, standardized against the normative mean of the population of study, when required (Shapiro–Wilk test P < 0.05); R, further rank-normalized (using Blom's formula), because the trait distribution after standardization differed from normality (Shapiro–Wilk test P < 0.05).
First Principal Component score computation
The First Principal Component from all of the language- and reading-related traits available (PC1, Table2) was derived in each dataset, through the SPSS® 20.0 Factor Analysis (Principal Component extraction method, hereafter called PCA). This reduced our correlated measures into a smaller set of latent variables (factors or principal components) that can explain the maximum amount of shared variance (Field 2005). In each dataset, only linear components with Eigenvalue >1 were extracted, allowing for correlation among the components (oblique rotation, direct oblim method) and excluding subjects with any missing measure (missing listwise option). A Kaiser–Meyer–Olkin measure of sampling adequacy and a Bartlett's test of sphericity were run in all the PCAs. These tests revealed a high common variance (KMO = 0.8–0.9) and a significant interdependence (Bartlett's test P value < 0.05) among the variables examined in each dataset, justifying the PCAs.
The proportion of total variance explained by PC1 was 75.3% in UK-RD, 68.6% in SLIC, 64.5% in CLDRC-RD and 52.0% in CLDRC-ADHD. In all the datasets PC2 explained no more than 13% of the total variance. All of the PC1s showed a broad pattern of loadings across the traits (Table2). The total number of participants for which we finally obtained genotype and PC1 data (i.e. all datasets combined) was 1862. We also obtained residuals from regressing PC1 against performance IQ (which had not been included in PC1 computation), again separately within each dataset. A measure of performance IQ was not available for 36 of the 1862 participants, and therefore the total sample size for IQ-adjusted PC1 was 1826.
We also derived a first principal component score within each dataset from only word reading and spelling, because these were the only measures available in all datasets and therefore provided a possibility to match traits as closely as possible across datasets. The first PC derived from word reading and spelling is referred to as PC1read hereafter. The proportion of variance in word reading and spelling explained by PC1read was 86.9% in UK-RD, 88% in CLDRC-RD, 93.4% in SLIC and 80.1% in CLDRC-ADHD. As only two measures were used to construct PC1read then the measures loaded equally onto this component, and the loadings were high in all datasets (≥0.9). PC1read was therefore a highly comparable construct across datasets (see Appendix S3). Moreover, the correlation between PC1 and PC1read was high in each dataset (Pearson's r = 0.925 in CLDRC-RD, 0.947 in UK-RD, 0.914 in SLIC and 0.917 in CLDRC-ADHD), so that PC1 itself could also be regarded as highly comparable across datasets. Note that these correlations were based on repeat random sampling of one member from each unrelated sibship (as for all pairwise trait correlations; see above). The total number of subjects across all datasets for PC1read was 1913, and for IQ-adjusted PC1read it was 1875. We primarily focused on PC1 for our subsequent genetic analysis (below), because this would maximize the chance of identifying SNPs that affect variance shared between both reading and language measures. However, we also repeated GWAS meta-analysis using PC1read to provide a comparable analysis that would be minimally affected by the heterogeneity of available measures across datasets.
Genetic association analyses
Sibling-pair GWAS
Sibling-based genome-wide association analyses were conducted using PC1 and PC1read scores separately within each dataset, both before and after IQ-adjustment, and using the ‘total’ association option of the QFAM function implemented in PLINK v1.07 (http://pngu.mgh.harvard.edu/∼purcell/plink/; Purcell et al. 2007). This method tests for association at each SNP by regressing trait scores on genotypes in an additive linear model. To correct for non-independence of siblings, permutations were run (i.e. label-swapping of phenotypes/genotypes) to obtain empirical significance levels (further details in Appendix S4).
GWAS meta-analysis (GWASMA)
The results from GWAS in the separate datasets were then meta-analysed together. This was implemented in the programme METAL (http://www.sph.umich.edu/csg/abecasis/Metal/index.html; Willer et al. 2010). We chose an approach that does not assume equivalence of allelic effect sizes between datasets, which was appropriate given the heterogeneity of study recruitment and assessment. Put briefly, the GWAS meta-analysis tested each SNP for a genetic effect, across the contributing datasets, computing an overall z-score for that SNP determined by the P value, the direction of the allelic effect on the quantitative trait, and the sample size of each study involved in the meta-analysis.
Gene-based analysis
The results of the GWASMA on PC1 were used as input for gene-based association analyses using VEGAS v0.8.27 (http://gump.qimr.edu.au/VEGAS/; Liu et al. 2010). This software performs association tests for ∼18,000 autosomal genes, by assigning multiple SNPs to each individual gene according to their genomic locations, and then combining the evidence for association across all SNPs assigned to a given gene, while taking into account the linkage disequilibrium (LD) structure between SNPs. Each tested gene also included potentially regulatory regions located up to 50 kb beyond the 5′- and 3′-untranslated regions (UTRs). A Bonferroni-corrected significance threshold was set at P < 2.8 × 10−6 to account for the number of genes tested (see Appendix S4 for details).
Pathway-based analysis
Finally, a pathway/network-based association analysis was run using the PC1 GWASMA results, with the programme INRICH v1.0 (http://atgu.mgh.harvard.edu/inrich/started.html; Lee et al. 2012). This tool tests for an enrichment of association within predefined gene sets, through a permutation-based approach. We defined associated genomic intervals as those containing an individual association P < 0.001 in the GWASMA results. Gene boundaries were again defined as extending 50 kb beyond the 5′- and 3′-UTRs. Three candidate gene lists, based on the gene sets of the Gene Ontology Database (http://www.geneontology.org/), were tested for an enrichment of association. These represented three distinct neurobiological hypotheses on the aetiology of reading and language disabilities: axon guidance (including all the GO sets containing the term ‘axon guidance’), neuronal migration (including all the GO sets containing the term ‘neuron migration’) and steroid sex hormone biology (including all the GO sets containing the terms ‘steroid’, ‘androgen’, ‘oestrogen’, ‘progesterone’ and ‘testosterone’). Further details on the analysis can be found in Appendix S4.
Further analysis of top association signals
Effect sizes on different traits
We repeated the regressions of PC1 and IQ-adjusted PC1 on the genotypes of our two most significantly associated SNPs from GWAS meta-analysis, in an additive linear model, in order to conveniently obtain the regression r 2 as indicative measures of effect sizes. To generate measures unbiased by sample relatedness, regression r 2 were calculated in R (R Core Team 2013, http://www.r-project.org/) as the median r 2 over 100 repeat random samplings of one individual from each independent sibship, separately in each dataset.
We further investigated each of our top two association signals by running QFAM univariate association tests in PLINK v1.07 (Purcell et al. 2007) for each individual trait that was used in constructing PC1, and separately in each dataset. This analysis provided an initial assessment of pleiotropy for these loci. We also performed multivariate association analysis for these two loci, in PLINK Multivariate v1.06 (https://genepi.qimr.edu.au/staff/manuelF/multivariate/main.html; Ferreira & Purcell 2009), again separately in each dataset and using each of the reading/language traits that were used in constructing PC1. PLINK multivariate extracts the linear combination of traits that explains the largest possible amount of covariance between the SNP and all of the traits. The loading produced for each trait represent its contribution to the multivariate association. MQFAM ‘total’ association was run, with adaptive permutations to adjust for sample relatedness (see Appendix S4 for details).
Assessment of top association signals in two additional datasets
Our two most significant association signals from PC1 meta-analysis were checked against published and unpublished results from the recent GWASMA of reading and language abilities reported by Luciano et al. (2013). This prior study analysed two population datasets, the Brisbane Adolescent Twin Sample (BATS) and the Avon Longitudinal Study of Parents and their Children (ALSPAC). BATS is a cohort of twins and their non-twin siblings recruited from ongoing studies of melanoma risk factors and cognition in an Australian population-based sample (Wright et al. 2001). Subjects had been administered psychometric tests assessing regular-, irregular-, and nonword reading, and spelling, together with the Schonell graded word reading test, and nonword repetition (see Luciano et al. 2013). ALSPAC is a longitudinal, population-based sample recruited from the county of Avon, UK (Boyd et al. 2013). The study website contains details of all the data available through a fully searchable data dictionary (http://www.bris.ac.uk/alspac/researchers/data-access/data-dictionary). Ethical approval was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees. Participants (all free of neurological/psychiatric conditions) had been tested for word reading, nonword reading, spelling and nonword repetition (see Luciano et al. 2013). BATS and ALSPAC had been genotyped using Illumina® 610k Quad Bead and HumanHap 550k Quad chips, respectively, and imputed using the HapMap Phase II CEU reference panel (NCBI build 36) (The International HapMap 3 Consortium, 2010). A total of 6434 subjects (962 from BATS and 5472 from ALSPAC) were meta-analysed by Luciano et al. (2013), for three different traits: word reading, nonword repetition and a composite/component score of reading and spelling (called hereafter the reading–spelling factor).
Results
GWAS meta-analysis
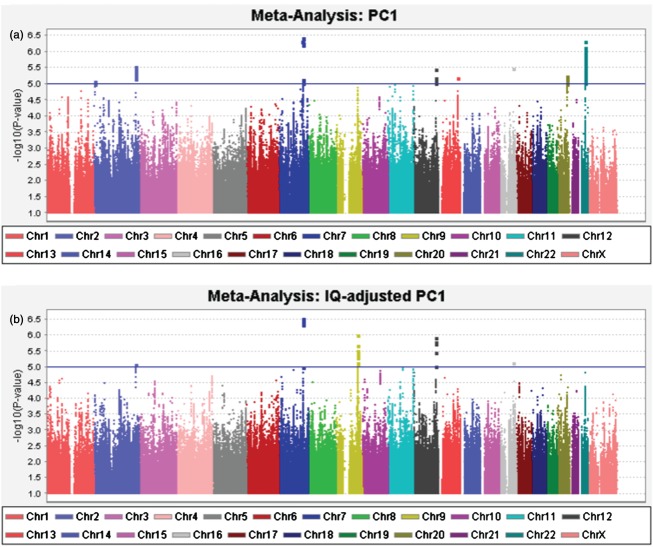
Table6 describes the most significant associations from the meta-analyses on PC1 (N = 1862) and IQ-adjusted PC1 (N = 1826). Figure 1 shows genome-wide Manhattan Plots. QQ-plots revealed no evidence of population stratification affecting the meta-analysis results, or of genome-wide significant associations [Fig. S1a,b (Appendix S1)]. The most significant association was observed for rs59197085 in PC1 and IQ-adjusted PC1 meta-analyses (P = 3.86 × 10–7 for PC1, and P = 3.01 × 10–7 for IQ-adjusted PC1; A/G, MAF ∼ 8%). This SNP is located at 7q32.1, within CCDC136 (coiled-coil domain containing 136, or NAG6) and ∼10 kb upstream of FLNC [filamin C; Fig. S1c (Appendix S1)]. The second most significantly associated region, before IQ-adjustment, was located on 22q12.3, SNP rs5995177 (P = 5.01 × 10–7, A/G, MAF ∼ 8%), within RBFOX2 [also known as RNA-binding motif protein 9, or RBM9; Fig. S1d (Appendix S1)]. The association was less significant after IQ-adjustment of PC1 (P = 1.5 × 10–5), and this difference was not merely due to the loss of 36 subjects in the IQ-adjusted analysis (investigated by performing a repeat PC1 analysis in the same reduced set of subjects as were available for IQ-adjusted PC1, data not shown).
Table 6.
Top association signals (P < 1 × 10–6) in the PC1 and IQ-adjusted PC1 meta-analyses
| Chr | SNP* | Position (hg19) | Allele1 | Allele2 | Freq Allele1 (%) | P value | Direction† | Gene (distance)‡ | Variant type |
|---|---|---|---|---|---|---|---|---|---|
| PC1 | |||||||||
| 7 | rs59197085 | 128460756 | a | g | 7.971 | 3.86 × 10–7 | – – – – | FLNC(–9.726)|CCDC136(0) | Intronic |
| 7 | rs58845495 | 128462847 | t | c | 92.029 | 4.09 × 10–7 | + + + + | FLNC(–7.635)|CCDC136(+0.664) | |
| 7 | 7:128439695:I | 128439695 | i | r | 7.94 | 4.99 × 10–7 | – – – – | CCDC136(0) | Intronic |
| 22 | rs5995177 | 36309553 | a | g | 8.049 | 5.01 × 10–7 | – – – – | RBFOX2(0) | Intronic |
| 7 | rs3734972 | 128470838 | t | c | 7.983 | 5.66 × 10–7 | – – – – | FLNC(0)|CCDC136(+8.655) | Exonic,synonymous |
| 7 | rs3800560 | 128461094 | t | c | 7.971 | 6.25 × 10–7 | – – – – | FLNC(–9.388)|CCDC136(0) | Intronic |
| 22 | rs12158565 | 36316843 | c | g | 87.23 | 7.57 × 10–7 | + + + + | RBFOX2(0) | Intronic |
| 22 | rs5755979 | 36290707 | t | c | 12.77 | 9.05 × 10–7 | – – – – | RBFOX2(0) | Intronic |
| 22 | rs5750202 | 36339542 | t | c | 12.77 | 9.06 × 10–7 | – – – – | RBFOX2(0) | Intronic |
| 22 | rs5750203 | 36339998 | a | t | 87.23 | 9.72 × 10–7 | + + + + | RBFOX2(0) | Intronic |
| IQ-adjusted PC1 | |||||||||
| 7 | rs59197085 | 128460756 | a | g | 7.971 | 3.01 × 10–7 | – – + – | FLNC(–9.726)|CCDC136(0) | Intronic |
| 7 | rs58845495 | 128462847 | t | c | 92.029 | 3.23 × 10–7 | + + – + | FLNC(–7.635)|CCDC136(+0.664) | |
| 7 | rs3800560 | 128461094 | t | c | 7.971 | 3.95 × 10–7 | – – + – | FLNC(–9.388)|CCDC136(0) | Intronic |
| 7 | 7:128439695:I | 128439695 | i | r | 7.94 | 4.48 × 10–7 | – – + – | CCDC136(0) | Intronic |
| 7 | rs3734972 | 128470838 | t | c | 7.983 | 4.68 × 10–7 | – – + – | FLNC(0)|CCDC136(+8.655) | Exonic,synonymous |
Single-base indels were not filtered out from the imputed polymorphisms since they were reliably called in the imputation reference (1000 Genomes, Phase I v3), and were tested for association as they could represent coding frameshift variants of biological interest.
The direction of effect of Allele1 is reported for datasets in the following order: CLDRC-RD, UK-RD, SLIC, CLDRC-ADHD.
Physical distance (kb) from closest genes (in a ±10 kb range from each marker) is indicated, along with orientation based on the direction of transcription (‘–’, upstream of 5'-UTR; ‘+’, downstream of 3′-UTR).
Figure 1.
Manhattan plots of the meta-analyses of the first principal component scores. (a) PC1; (b) IQ-adjusted PC1. The blue line represents the nominal suggestive significance threshold (P = 1 × 10–5).
Table S2a,b (Appendix S2) shows all SNPs with association P < 1 × 10–5 in GWAS meta-analysis of PC1 or IQ-adjusted PC1. No genome-wide significant associations were observed in the GWAS in the individual datasets (data not shown).
The results of our complementary PC1read meta-analysis (Appendix S3) were consistent with the PC1 meta-analysis, with rs59197085 and rs5995177 among the top suggestive associations (P ∼ 10–6). This was expected given the high correlations between PC1 and PC1read in each dataset (all correlations >0.9, see above).
Effect sizes and profiles of top associations
rs59197085 (CCDC136/FLNC) explained 3% of PC1 variance and 3.2% of IQ-adjusted PC1 variance in our largest GWAS dataset (UK-RD), and 1.3% of PC1 variance and 1.5% of IQ-adjusted PC1 variance in the next largest dataset (CLDRC-RD). The estimated effect sizes in the smaller datasets were ≤0.2%. Estimated effect sizes for rs5995177 (RBFOX2) were more consistent across datasets. This SNP explained 1.2% of PC1 and IQ-adjusted PC1 variance in UK-RD, and 1.8% of PC1 variance and 1.2% of IQ-adjusted PC1 variance in CLDRC-RD, while estimated effect sizes in the smaller datasets were between 0.6% and 1.6% of variance.
Both rs59197085 and rs5995177 showed broad profiles of association across the measures that were used to construct PC1, as assessed from the PLINK multivariate loadings and corresponding QFAM univariate association P values shown in Table7. These findings suggest pleiotropic effects of the two SNPs on reading and language.
Table 7.
Effect of the top association signals rs59197085 (7q32.1) and rs5995177 (22q12.3) on the single reading and language traits used in constructing PC1
| Trait | CLDRC-RD | UK-RD | SLIC | CLDRC-ADHD |
|---|---|---|---|---|
| rs59197085 | ||||
| WRead | –0.66 (0.024) | –0.87 (5.3 × 10–5) | –0.29 (0.626) | –0.5 (0.427) |
| WSpell | –0.89 (3.8 × 10–3) | –0.75 (1.1 × 10–3) | 0.08 (0.862) | –0.1 (0.871) |
| PD |
|
–0.86 (1.6 × 10–5) |
|
|
| PA | –0.65 (0.029) | –0.49 (0.018)† | 0.35 (0.588) | |
| OC | –0.64 (0.036) | –0.89 (3 × 10–6) | –0.04 (0.95) | |
| NWR | –0.34 (0.269) | –0.57 (0.32) | –0.28 (0.686) | |
| ELS | –0.25 (0.807) | |||
| RLS | 0.08 (0.821) | |||
| rs5995177 | ||||
| WRead | –0.66 (0.027) | –0.81 (2 × 10–3) | –0.71 (0.116) | 0.01 (0.98) |
| WSpell | –0.81 (6.9 × 10–3) | –0.82 (1.1 × 10–3) | –0.52 (0.262) | –0.33 (0.359) |
| PD |
|
–0.77 (1.8 × 10–3) |
|
|
| PA | –0.72 (0.023) | –0.72 (2.5 × 10–3)† | –0.65 (0.046) | |
| OC | –0.68 (0.026) | –0.57 (0.017) | –0.02 (0.968) | |
| NWR | –0.04 (0.922) | –0.23 (0.674) | 0.06 (0.876) | |
| ELS | –0.82 (0.057) | |||
| RLS | –0.61 (0.206) | |||
These were computed for each trait as PLINK Multivariate MQFAM loadings and PLINK univariate QFAM association P values (in brackets) and refer to the minor alleles (A for both SNPs).
WRead, word reading; WSpell, word spelling; PD, phonological decoding; PA, phoneme awareness; OC, orthographic coding; NWR, nonword repetition; ELS/RLS, expressive/receptive language score.
Loading on nonword reading and phonological choice (respectively).
Although PA had been excluded from the PCA in UK-RD (due to the low number of measures available), it was tested in this case to have a term of comparison to the other datasets.
Gene-based meta-analysis
The strongest gene-based associations inferred from the PC1 and IQ-adjusted PC1 meta-analyses are reported in Table S2c,d (Appendix S2). While no gene exceeded the appropriate genome-wide significance threshold for this analysis (P < 2.8 × 10−6), CCDC136, FLNC and RBFOX2 were among the most significantly associated genes, with the latter approaching the significance threshold in the PC1 analysis (P = 5 × 10–6). However, after conditioning on the most significant association signal within each gene, no other SNP within each of these genes showed significant evidence for having an independent residual effect, after correction for multiple testing [Table S2e,f (Appendix S2)]. For this analysis the gene boundaries were defined in the same way as for gene-based analysis (see above).
Pathway-based meta-analysis
We assessed evidence for an excess of association signals within the genes of three neurobiological pathways that are prominent in prior literature on reading and language: axon guidance, neuronal migration and steroid sex hormone biology (see Discussion for the relevant citations). None of the three tested sets showed significant associations with PC1 or IQ-adjusted PC1 [Table S2g,h (Appendix S2)], although the association between PC1 and the steroid-related pathway approached significance (P = 0.051).
Assessment of top associations within previous GWAS results
We assessed our most significant associations from PC1 meta-analyses within published and unpublished results from the previous GWAS study of the BATS/ALSPAC datasets, for which the reading and language measures were IQ-adjusted (Luciano et al. 2013). FLNC and CCDC136 showed nominally significant associations in gene-based (VEGAS) analyses of reading-related traits in BATS/ALSPAC (CCDC136 P = 0.034 for reading-spelling factor and P = 0.003 for word-reading; FLNC P = 0.009 for word- reading; see Table S3 of Luciano et al. 2013). The reading–spelling factor in the BATS/ALSPAC datasets was the most comparable trait to the IQ-adjusted PC1 score of this study. As the study of Luciano et al. 2013 had used the HapMap2 reference dataset for genotype imputation, it was not possible to directly investigate the most highly associated SNPs from this study in the BATS/ALSPAC datasets. We therefore investigated association for two HapMap2 SNPs that were closest to our top hits on 7q32 and 22q12.3. rs3734972 (PC1 P = 5.66 × 10–7, IQ-adjusted PC1 P = 4.68 × 10–7; T/C, minor allele T, MAF ≈ 8%) lies ∼10 kb away from rs59197085 on 7q32 and is in high LD with it [R 2 = 0.89, see local association plot, Fig. S1c (Appendix S1)]. rs3734972 showed a P value of 0.032 with the IQ-adjusted reading-spelling factor in BATS/ALSPAC. The allelic trend was in the opposite direction to that observed in the UK-RD/SLIC/CLDRC datasets, with the T allele having a positive effect on the trait score in the BATS/ALSPAC cohorts. rs12158565 (PC1 P = 7.57 × 10–7, IQ-adjusted PC1 P = 4.65 × 10–5; C/G, minor allele G, MAF ≈ 13%) was the second most significant association in 22q12.3, mapping ∼7 kb from the top SNP at this locus rs5995177, and in low LD with it (R 2 = 0.083), as are all the other suggestively associated SNPs in 22q12.3 [see local association plot, Fig. S1d (Appendix S1)]. rs12158565 showed no evidence of association in BATS/ALSPAC (P = 0.81).
Discussion
This study aimed to identify pleiotropic variants having effects on reading and language abilities by analysing continuous traits in multiple datasets. Our study is complementary to two recently published GWAS: one using a similar approach but in general population samples (Luciano et al. 2013), and another contrasting a relatively small number of categorically defined RD-SLI comorbid cases with unaffected controls (Eicher et al. 2013).
Our study is novel and distinct for several reasons:
First, we analysed continuous variation in reading and language skills while also having an enrichment of participants with low abilities (i.e. through analysing poor performing probands together with their siblings), and without applying a dichotomous classification into cases and controls that necessarily involves arbitrary thresholding. Our design was therefore suited to detect genetic effects on susceptibility to RD and SLI that also act across the entire distribution of reading and language skills.
Second, we specifically focused on shared neurobiological mechanisms underlying language and reading, by analysing the first principal component of all of the reading- and language-related measures available in each dataset, followed by investigating the cross-phenotypic effects of the resulting top GWAS hits through univariate association analysis using each individual measure. We additionally followed this with a confirmatory analysis focused only on word reading and spelling, because these measures provided the closest matching possibility across our datasets. The first principal component (PC1) of all available measures extracted a large proportion of shared trait variance across the two domains of reading and language, and was highly correlated with the component derived from only reading and spelling (PC1read).
Third, we performed GWAS both before and after IQ-adjustment of PC1. This was done in order to identify both genetic variants having effects broadly across reading, language and general cognitive abilities, and variants having effects on reading and language but independently of general cognitive ability. This approach also facilitated a comparison of our top results with those from datasets investigated in Luciano et al. (2013).
We checked within our GWASMA results 18 specific SNPs that had been highlighted to show the most promising candidate associations by the authors of previous GWAS studies of reading and/or language (Eicher et al. 2013; Field et al. 2013; Luciano et al. 2013; Meaburn et al. 2008; Roeske et al. 2011). Seventeen of these SNPS showed no nominally significant association within our GWASMA results (data not shown). Only rs10485609 (Meaburn et al. 2008) showed a nominally significant association (P = 0.013 for PC1, P = 0.015 for IQ-adjusted PC1; allele A was associated with lower performance, which was a consistent allelic direction of effect with that reported by Meaburn et al. 2008), but this was not significant after multiple testing correction for 18 tests.
Like the other recently published GWAS efforts in this field, our study did not find any individual associations that achieved genome-wide significance (threshold P = 5 × 10–8). However, we did identify two novel, suggestive results of particular interest, on 7q32.1 and 22q12.3, with the most significant associations at rs59197085 and rs5995177, respectively. As shown in Table7, both SNPs displayed a broad pattern of association across multiple reading and language traits, consistent with effects on neurobiological processes shared between reading and language cognition. In the regression model these SNPs explained a notable proportion (up to 3.2%) of variance in PC1 and IQ-adjusted PC1 scores, particularly in the largest datasets (CLDRC-RD and UK-RD), although these effect sizes are likely to be overestimated since this is the first report of these associations (Ioannidis 2008). Gene based-tests were consistent with the results of the SNP-based analysis for FLNC, CCDC136 and RBFOX2, and the gene-based P values were found to be largely or wholly reflective of the individual top associations within each of these genes.
rs5995177 is an intronic variant localized within RBFOX2 (RNA-binding protein, fox-1 homologue 2, also known as RBM9), a protein that regulates alternative splicing and is active in neurons. RBFOX2 is highly expressed in the foetal brain and has important roles in CNS development (Gehman et al. 2012). The homologous gene RBFOX1 has been implicated in several neurodevelopmental disorders, including Rolandic Epilepsy (Lal et al. 2013) and Autism Spectrum Disorder (Voineagu et al. 2011), and is a downstream target of FOXP2, a transcription factor implicated in monogenic speech and language disorders (Ayub et al. 2013). The high comorbidity between Rolandic Epilepsy and RD (Clarke et al. 2007) and the presence of a FOXP2 binding site ∼5 kb from rs5995177 (The ENCODE Project Consortium, 2012), further support a link of RBFOX2 with reading and language abilities. Thus convergent evidence from multiple lines of research makes RBFOX2 an intriguing candidate gene for future studies. There was no evidence of association of this locus with reading and language measures in the results of the population-based study of Luciano et al. (2013).
rs59197085 is located in CCDC136 (coiled-coil domain containing 136, or NAG6) and ∼10 kb upstream of FLNC (filamin C). This SNP, along with the nearby SNPs rs3800560, rs58845495 and rs3734972, forms roughly 10-kb haplotypes spanning the region between CCDC136 and FLNC and partially overlapping these genes [see local association plot, Fig. S1c (Appendix S1)]. CCDC136 encodes a poorly characterized tumour suppressor which has been found to be downregulated in gastric carcinoma (Zhang et al. 2004) and is highly expressed in the cerebellum and in the occipital cortex (Allen Human Brain Atlas, Hawrylycz et al. 2012; http://human.brain-map.org). Filamin C (or filamin gamma) is a structural protein that crosslinks actin filaments into orthogonal networks in the cortical cytoplasm and participates in cytoskeleton re-modelling, suggesting a possible role in cell motility and migration. Functions of FLNC have been demonstrated in muscle tissues, where mutations are responsible for several forms of myopathies (Duff et al. 2011). However, its pattern of expression includes spinal cord, cerebellum, corpus callosum, basal ganglia and some localized areas in the frontal, temporal and occipital cortex (Allen Human Brain Atlas, Hawrylycz et al. 2012). Its homologue FLNA (filamin A) is involved in neuronal migration and is implicated in an X-linked dominant form of periventricular heterotopia, a neurological disorder that sometimes involves reading and spelling problems (Robertson 2005).
Associations within the 7q32 region are particularly interesting in light of data from two previous independent studies that have each reported evidence for linkage between a microsatellite marker in this region (D7S530, located ∼650 kb from our peaks of association) and RD status (Kaminen et al. 2003) or else nonword spelling and irregular word reading (Bates et al. 2007). There was also evidence of association, at the gene level, with reading and language measures for FLNC, and CCDC136 in the BATS/ALSPAC datasets studied by Luciano et al. (2013). At the SNP level, one of our most significantly associated SNPs from GWASMA, rs3734972 also showed association with an IQ-adjusted reading–spelling score in the BATS/ALSPAC datasets. However, the allelic directions of effect on the traits in this study and the study by Luciano et al. were opposite.
We sought to detect an excess of association signals within genes belonging to each of three candidate gene sets based on different biological functions: axon guidance, neuronal migration and steroid hormone biology. Axon guidance and neuronal migration are functions linked to some of the previously identified candidate genes in RD and SLI; ROBO1 (Hannula-Jouppi et al. 2005), DCDC2 (Meng et al. 2005), KIAA0319 (Peschansky et al. 2010), DYX1C1 (Tammimies et al. 2013) and FOXP2 (Vernes et al. 2011). A potential involvement of neuronal migration deficits in RD aetiology represents a longstanding hypothesis of the field (see Galaburda & Cestnick 2003). The steroid hypothesis was motivated by literature suggesting links between sex hormone biology, language performance and the brain architecture that subserves reading and language (Good et al. 2001; Lombardo et al. 2012; Shapleske et al. 1999; Whitehouse et al. 2012); and by evidence of interaction between Oestrogen Receptors and DYX1C1, both at the gene (Tammimies et al. 2012) and at the protein level (Massinen et al. 2009). None of the three gene sets showed a significant excess of association signals, although the steroid hormone biology set approached significance in this analysis.
In carrying out GWASMA studies of complex cognitive traits across multiple datasets collected by different research teams, an obvious limitation is that the specific trait measurements that are available may be quite diverse. Even when tests are similar, and hypothesized to measure corresponding cognitive processes, they may still create a substantial source of heterogeneity for a meta-analysis effort. In this study we sought to overcome this limitation by focusing on a principal component (PC1) capturing a majority of the shared variance between reading- and language-related traits. In spite of the phenotypic heterogeneity of our datasets, this measure can be considered comparable across datasets for a number of reasons. First, the loadings of the individual traits on PC1 scores were similar across the datasets. Second, dropping one or more traits from our PC1 computation did not substantially affect the resulting PC1 scores (data not shown). Third, the First Principal Component derived only from word reading and spelling (PC1read) was strongly correlated with PC1. Word reading and spelling were the only two measures available in all of the datasets and provided the closest phenotype matching possible across datasets. Not surprisingly, given the high correlations between PC1 and PC1read in all datasets, the association meta-analysis using PC1read (Appendix S3) produced results consistent with PC1-based meta-analysis. We therefore conclude that PC1 was a sufficiently well matched construct across datasets to support GWASMA, in which we nonetheless allowed for heterogeneity of effect sizes across datasets to avoid assuming a perfect matching. It is interesting that a single PC can capture comparable variation across a diverse range of reading and language traits and in the presence of heterogeneity of measurement across datasets. This indicates a robust unifying dimension to much of this variation, and supports a genetic approach framed around pleiotropy.
The use of a principal component can lead to some loss of information, both in terms of detecting trait-specific genetic effects, and of reducing the sample size (because individuals with one or more missing trait values were excluded from the analysis). However, as we aimed to identify shared genetic effects on reading and language, the use of PC1 scores, followed by investigating cross-phenotypic associations of the top SNPs at the level of individual traits, was an appropriate approach to analysing these multivariate datasets. There is now a need for a larger international meta-analysis effort that incorporates further datasets. This would improve the power to detect pleiotropic variants affecting reading and language.
Acknowledgments
The authors declare no conflicts of interest. This work was supported by the Max Planck Society, the University of St Andrews, the EU (Neurodys, 018696), and the US National Institutes of Health (Grant ref: P50 HD027802). Genotyping at the Wellcome Trust Centre for Human Genetics was supported by the Wellcome Trust (090532/Z/09/Z) and a Medical Research Council Hub Grant (G0900747 91070). Silvia Paracchini is a Royal Society University Research Fellow. Dianne Newbury is an MRC Career Development Fellow and a Junior Research Fellow at St John's College, University of Oxford. We thank Dr Manuel Ferreira and Dr Phil H Lee for useful advice regarding the use of PLINK Multivariate and INRICH tools. Dr Margaret Wright and Prof Nicholas Martin are the principal investigators for the BATS IQ data collection and genotyping.
Members of the SLI Consortium: Wellcome Trust Centre for Human Genetics, Oxford: D. F. Newbury, N. H. Simpson, R. Nudel, A. P. Monaco; Max Planck Institute for Psycholinguistics, Nijmegen: S. E. Fisher, C. Francks; Newcomen Centre, Guy's Hospital, London: G. Baird, V. Slonims, K Dworzynski; Child and Adolescent Psychiatry Department and Medical Research Council Centre for Social, Developmental, and Genetic Psychiatry, Institute of Psychiatry, London: P. F. Bolton; Medical Research Council Centre for Social, Developmental, and Genetic Psychiatry Institute of Psychiatry, London: E. Simonoff; Department of Reproductive and Developmental Sciences, University of Edinburgh: A. O'Hare; Molecular Medicine Centre, University of Edinburgh: J. Seckl; Department of Speech and Language Therapy, Royal Hospital for Sick Children, Edinburgh: H. Cowie; Speech and Hearing Sciences, Queen Margaret University College: A. Clark and J. Watson; Department of Educational and Professional Studies, University of Strathclyde: W. Cohen; Department of Child Health, the University of Aberdeen: A. Everitt, E. R. Hennessy, D. Shaw, P. J. Helms; Audiology and Deafness, School of Psychological Sciences, University of Manchester: Z. Simkin, G. Conti-Ramsden; Department of Experimental Psychology, University of Oxford: D. V. M. Bishop; Biostatistics Department, Institute of Psychiatry, London: A. Pickles.
ALSPAC: We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The UK Medical Research Council and the Wellcome Trust (Grant ref: 092731) and the University of Bristol provide core support for ALSPAC. This publication is the work of the authors and DME will serve as guarantor for the contents of this paper that are related to ALSPAC.
Supporting Information
Appendix S1: QQ plots and association plots of the top association signals from analysis based on PC1 and IQ-adjusted PC1. Contribution of each dataset to the strength of the association in the PC1 and IQ-adjusted PC1 meta-analysis, for the top association signals.
Appendix S2: SNP-, gene- and pathway-based meta-analysis results from analysis based on PC1 and IQ-adjusted PC1. Test for independent associations within the top associated regions, after conditioning on the most significant local association signal.
Appendix S3: Analyses based on PC1read and IQ-adjusted PC1read (trait loadings on PC1read, Manhattan plots, QQ plots and top associations)
Appendix S4: Genotype calls and QC protocols; correlation patterns of reading and language measures; statistical analyses, commands and parameters.
References
- Anthoni H, Zucchelli M, Matsson H, Muller-Myhsok B, Fransson I, Schumacher J, Massinen S, Onkamo P, Warnke A, Griesemann H, Hoffmann P, Nopola-Hemmi J, Lyytinen H, Schulte-Korne G, Kere J, Nothen MM. Peyrard-Janvid M. A locus on 2p12 containing the co-regulated MRPL19 and C2ORF3 genes is associated to dyslexia. Hum Mol Genet. 2007;16:667–677. doi: 10.1093/hmg/ddm009. [DOI] [PubMed] [Google Scholar]
- Ayub Q, Yngvadottir B, Chen Y, Xue Y, Hu M, Vernes SC, Fisher SE. Tyler-Smith C. FOXP2 targets show evidence of positive selection in European populations. Am J Hum Genet. 2013;92:696–706. doi: 10.1016/j.ajhg.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry JG, Yasin I. Bishop DV. Heritable risk factors associated with language impairments. Genes Brain Behav. 2007;6:66–76. doi: 10.1111/j.1601-183X.2006.00232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TC, Luciano M, Castles A, Coltheart M, Wright MJ. Martin NG. Replication of reported linkages for dyslexia and spelling and suggestive evidence for novel regions on chromosomes 4 and 17. Eur J Hum Genet. 2007;15:194–203. doi: 10.1038/sj.ejhg.5201739. [DOI] [PubMed] [Google Scholar]
- Bates TC, Luciano M, Medland SE, Montgomery GW, Wright MJ. Martin NG. Genetic variance in a component of the language acquisition device: ROBO1 polymorphisms associated with phonological buffer deficits. Behav Genet. 2011;41:50–57. doi: 10.1007/s10519-010-9402-9. [DOI] [PubMed] [Google Scholar]
- Becker J, Czamara D, Scerri TS, et al. Genetic analysis of dyslexia candidate genes in the European cross-linguistic NeuroDys cohort. Eur J Hum Genet. 2013;22:675–680. doi: 10.1038/ejhg.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DV. Is specific language impairment a valid diagnostic category? Genetic and psycholinguistic evidence. Philos Trans R Soc Lond B Biol Sci. 1994;346:105–111. doi: 10.1098/rstb.1994.0134. [DOI] [PubMed] [Google Scholar]
- Bishop DVM. Snowling MJ. Developmental dyslexia and specific language impairment: same or different? Psychol Bull. 2004;130:858–886. doi: 10.1037/0033-2909.130.6.858. [DOI] [PubMed] [Google Scholar]
- Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, Molloy L, Ness A, Ring S. Davey Smith G. Cohort profile: the ‘Children of the 90s’—the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2013;42:111–127. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandler WM, Morris AP, Evans DM, Scerri TS, Kemp JP, Timpson NJ, St Pourcain B, Smith GD, Ring SM, Stein J, Monaco AP, Talcott JB, Fisher SE, Webber C. Paracchini S. Common variants in left/right asymmetry genes and pathways are associated with relative hand skill. PLoS Genet. 2013;9:e1003751. doi: 10.1371/journal.pgen.1003751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrion-Castillo A, Franke B. Fisher SE. Molecular genetics of dyslexia: an overview. Dyslexia. 2013;19:214–240. doi: 10.1002/dys.1464. [DOI] [PubMed] [Google Scholar]
- Castles A. Coltheart M. Varieties of developmental dyslexia. Cognition. 1993;47:149–180. doi: 10.1016/0010-0277(93)90003-e. [DOI] [PubMed] [Google Scholar]
- Ceroni F, Simpson NH, Francks C, Baird G, Conti-Ramsden G, Clark A, Bolton PF, Hennessy ER, Donnelly P, Bentley DR, Martin H, Parr J, Pagnamenta AT, Maestrini E, Bacchelli E, Fisher SE. Newbury DF. Homozygous microdeletion of exon 5 in ZNF277 in a girl with specific language impairment. Eur J Hum Genet. 2014 doi: 10.1038/ejhg.2014.4. DOI: 10.1038/ejhg.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke T, Strug LJ, Murphy PL, Bali B, Carvalho J, Foster S, Tremont G, Gagnon BR, Dorta N. Pal DK. High risk of reading disability and speech sound disorder in Rolandic epilepsy families: case–control study. Epilepsia. 2007;48:2258–2265. doi: 10.1111/j.1528-1167.2007.01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coltheart M. Leahy J. Assessment of lexical and non-lexical reading abilities in children: some normative data. Aust J Psychol. 1996;48:136–140. [Google Scholar]
- Compton DL, DeFries JC. Olson RK. Are RAN- and phonological awareness-deficits additive in children with reading disabilities? Dyslexia. 2001;7:125–149. doi: 10.1002/dys.198. [DOI] [PubMed] [Google Scholar]
- Cope N, Harold D, Hill G, Moskvina V, Stevenson J, Holmans P, Owen MJ, O'Donovan MC. Williams J. Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am J Hum Genet. 2005;76:581–591. doi: 10.1086/429131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFries JC, Filipek PA, Fulker DW, Olson RK, Pennington BF, Smith SD. Wise BW. Colorado Learning Disabilities Research Center. Learn Disabil Multidisciplinary J. 1997;8:7–19. [Google Scholar]
- Duff RM, Tay V, Hackman P, et al. Mutations in the N-terminal actin-binding domain of filamin C cause a distal myopathy. Am J Hum Genet. 2011;88:729–740. doi: 10.1016/j.ajhg.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn LM. Markwardt FC. Examiner's Manual: Peabody Individual Achievement Test. Circle Pines, MN: American Guidance Service; 1970. [Google Scholar]
- Eicher JD, Powers NR, Miller LL, et al. Genome-wide association study of shared components of reading disability and language impairment. Genes Brain Behav. 2013;8:792–801. doi: 10.1111/gbb.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot CD, Murray DJ. Pearson LS. The British Ability Scales. Slough, UK: NFER; 1979. [Google Scholar]
- Falcaro M, Pickles A, Newbury DF, Addis L, Banfield E, Fisher SE, Monaco AP, Simkin Z. Conti-Ramsden G. Genetic and phenotypic effects of phonological short-term memory and grammatical morphology in specific language impairment. Genes Brain Behav. 2008;7:393–402. doi: 10.1111/j.1601-183X.2007.00364.x. [DOI] [PubMed] [Google Scholar]
- Ferreira MA. Purcell SM. A multivariate test of association. Bioinformatics. 2009;25:132–133. doi: 10.1093/bioinformatics/btn563. [DOI] [PubMed] [Google Scholar]
- Field A. Discovering Statistics Using SPSS. London, UK: SAGE; 2005. [Google Scholar]
- Field LL, Shumansky K, Ryan J, Truong D, Swiergala E. Kaplan BJ. Dense-map genome scan for dyslexia supports loci at 4q13, 16p12, 17q22; suggests novel locus at 7q36. Genes Brain Behav. 2013;12:56–69. doi: 10.1111/gbb.12003. [DOI] [PubMed] [Google Scholar]
- Fisher SE. DeFries JC. Developmental dyslexia: genetic dissection of a complex cognitive trait. Nat Rev Neurosci. 2002;3:767–780. doi: 10.1038/nrn936. [DOI] [PubMed] [Google Scholar]
- Fisher SE. Scharff C. FOXP2 as a molecular window into speech and language. Trends Genet. 2009;25:166–177. doi: 10.1016/j.tig.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Francks C, Paracchini S, Smith SD, Richardson AJ, Scerri TS, Cardon LR, Marlow AJ, MacPhie IL, Walter J, Pennington BF, Fisher SE, Olson RK, DeFries JC, Stein JF. Monaco AP. A 77-kilobase region of chromosome 6p22.2 is associated with dyslexia in families from the United Kingdom and from the United States. Am J Hum Genet. 2004;75:1046–1058. doi: 10.1086/426404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederickson N. Phonological Assessment Battery. London, UK: Educational Psychology Publishing; 1995. [Google Scholar]
- Friend A. Olson RK. Phonological spelling and reading deficits in children with spelling disabilities. Sci Stud Read. 2010;12:90–105. doi: 10.1080/10888430701773876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaburda AM. Cestnick L. Developmental dyslexia. Rev Neurol. 2003;36(Suppl 1):3–9. [PubMed] [Google Scholar]
- Gallagher A. Frederickson N. The phonological assessment battery (PhAB): an initial assessment of its theoretical and practical utility. Educ Child Psychol. 1995;12:53–67. [Google Scholar]
- Gathercole SE, Willis CS, Baddeley AD. Emslie H. The children's test of nonword repetition: a test of phonological working memory. Memory. 1994;2:103–127. doi: 10.1080/09658219408258940. [DOI] [PubMed] [Google Scholar]
- Gehman LT, Meera P, Stoilov P, Shiue L, O'Brien JE, Meisler MH, Ares M, Otis TS. Black DL. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 2012;26:445–460. doi: 10.1101/gad.182477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good CD, Johnsrude I, Ashburner J, Henson RN, Friston KJ. Frackowiak RS. Cerebral asymmetry and the effects of sex and handedness on brain structure: a voxel-based morphometric analysis of 465 normal adult human brains. Neuroimage. 2001;14:685–700. doi: 10.1006/nimg.2001.0857. [DOI] [PubMed] [Google Scholar]
- Hannula-Jouppi K, Kaminen-Ahola N, Taipale M, Eklund R, Nopola-Hemmi J, Kääriäinen H. Kere J. The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLoS Genet. 2005;1:e50. doi: 10.1371/journal.pgen.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlaar N, Hayiou-Thomas ME, Dale PS. Plomin R. Why do preschool language abilities correlate with later reading? A twin study. J Speech Lang Hear Res. 2008;51:688–705. doi: 10.1044/1092-4388(2008/049). [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J. Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JPA. Why most discovered true associations are inflated. Epidemiology. 2008;19:640–648. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
- Jastak S. Wilkinson GS. Wide Range Achievement Test – Revised (WRAT-R) San Antonio, TX: The Psychological Corporation; 1984. [Google Scholar]
- Kaminen N, Hannula-Jouppi K, Kestilä M, Lahermo P, Muller K, Kaaranen M, Myllyluoma B, Voutilainen A, Lyytinen H, Nopola-Hemmi J. Kere J. A genome scan for developmental dyslexia confirms linkage to chromosome 2p11 and suggests a new locus on 7q32. J Med Genet. 2003;40:340–345. doi: 10.1136/jmg.40.5.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal D, Reinthaler EM, Altmüller J, Toliat MR, Thiele H, Nürnberg P, Lerche H, Hahn A, Møller RS, Muhle H, Sander T, Zimprich F. Neubauer BA. RBFOX1 and RBFOX3 mutations in Rolandic epilepsy. PLoS One. 2013;8:e73323. doi: 10.1371/journal.pone.0073323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PH, O'Dushlaine C, Thomas B. Purcell SM. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28:1797–1799. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG. Macgregor S. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan J, Petrill S, Flax J, Justice L, Hou L, Bassett A, Tallal P, Brzustowicz L. Bartlett C. Genetic covariation underlying reading, language and related measures in a sample selected for specific language impairment. Behav Genet. 2011;41:651–659. doi: 10.1007/s10519-010-9435-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo MV, Ashwin E, Auyeung B, Chakrabarti B, Taylor K, Hackett G, Bullmore ET. Baron-Cohen S. Fetal testosterone influences sexually dimorphic gray matter in the human brain. J Neurosci. 2012;32:674–680. doi: 10.1523/JNEUROSCI.4389-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano M, Lind PA, Duffy DL, Castles A, Wright MJ, Montgomery GW, Martin NG. Bates TC. A haplotype spanning KIAA0319 and TTRAP is associated with normal variation in reading and spelling ability. Biol Psychiatry. 2007;62:811–817. doi: 10.1016/j.biopsych.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Luciano M, Evans DM, Hansell NK, Medland SE, Montgomery GW, Martin NG, Wright MJ. Bates TC. A genome-wide association study for reading and language abilities in two population cohorts. Genes Brain Behav. 2013;12:645–652. doi: 10.1111/gbb.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massinen S, Tammimies K, Tapia-Páez I, Matsson H, Hokkanen M-E, Söderberg O, Landegren U, Castrén E, Gustafsson J-Å, Treuter E. Kere J. Functional interaction of DYX1C1 with estrogen receptors suggests involvement of hormonal pathways in dyslexia. Hum Mol Genet. 2009;18:2802–2812. doi: 10.1093/hmg/ddp215. [DOI] [PubMed] [Google Scholar]
- McArthur GM, Hogben JH, Edwards VT, Heath SM. Mengler ED. On the “specifics” of specific reading disability and specific language impairment. J Child Psychol Psychiatry. 2000;41:869–874. [PubMed] [Google Scholar]
- Meaburn EL, Harlaar N, Craig IW, Schalkwyk LC. Plomin R. Quantitative trait locus association scan of early reading disability and ability using pooled DNA and 100 K SNP microarrays in a sample of 5760 children. Mol Psychiatry. 2008;13:729–740. doi: 10.1038/sj.mp.4002063. [DOI] [PubMed] [Google Scholar]
- Meng H, Smith SD, Hager K, Held M, Liu J, Olson RK, Pennington BF, DeFries JC, Gelernter J, O'Reilly-Pol T, Somlo S, Skudlarski P, Shaywitz SE, Shaywitz BA, Marchione K, Wang Y, Paramasivam M, LoTurco JJ, Page GP. Gruen JR. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci U S A. 2005;102:17053–17058. doi: 10.1073/pnas.0508591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury DF. Monaco AP. Genetic advances in the study of speech and language disorders. Neuron. 2010;68:309–320. doi: 10.1016/j.neuron.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury DF, Winchester L, Addis L, et al. CMIP and ATP2C2 modulate phonological short-term memory in language impairment. Am J Hum Genet. 2009;85:264–272. doi: 10.1016/j.ajhg.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury DF, Paracchini S, Scerri TS, Winchester L, Addis L, Richardson AJ, Walter J, Stein JF, Talcott JB. Monaco AP. Investigation of dyslexia and SLI risk variants in reading- and language-impaired subjects. Behav Genet. 2011;41:90–104. doi: 10.1007/s10519-010-9424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudel R, Simpson NH, Baird G, O'Hare A, Conti-Ramsden G, Bolton PF, Hennessy ER the SLIC. Ring SM, Smith GD, Francks C, Paracchini S, Monaco AP, Fisher SE. Newbury DF. Genome-wide association analyses of child genotype effects and parent-of-origin effects in specific language impairment. Genes Brain Behav. 2014;13:418–429. doi: 10.1111/gbb.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson R, Wise B, Conners F, Rack J. Fulker D. Specific deficits in component reading and language skills: genetic and environmental influences. J Learn Disabil. 1989;22:339–348. doi: 10.1177/002221948902200604. [DOI] [PubMed] [Google Scholar]
- Olson R, Forsberg H, Wise B. Rack J. Measurement of word recognition, orthographic, and phonological skills. In: Lyon GR, editor; Frames of Reference for the Assessment of Learning Disabilities: New Views on Measurement Issues. Baltimore, MD: Paul H. Brookes; 1994a. pp. 243–278. [Google Scholar]
- Olson R, Forsberg H. Wise B. Genes, environment, and the development of orthographic skills. In: Berninger VW, editor; The Varieties of Orthographic Knowledge I: Theoretical and Developmental Issues. Dordrecht, The Netherlands: Kluwer Academics; 1994b. pp. 27–71. [Google Scholar]
- Paracchini S. Dissection of genetic associations with language-related traits in population-based cohorts. J Neurodev Disord. 2011;3:365–373. doi: 10.1007/s11689-011-9091-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paracchini S, Steer MSC, Buckingham BSL-L, Morris PDA, Ring PDS, Scerri DPT, Stein FRCPJ, Pembrey MDM, Ragoussis PDJ, Golding PDJ. Monaco PDA. Association of the KIAA0319 dyslexia susceptibility gene with reading skills in the general population. Am J Psychiatry. 2008;165:1576–1584. doi: 10.1176/appi.ajp.2008.07121872. [DOI] [PubMed] [Google Scholar]
- Paracchini S, Ang QW, Stanley FJ, Monaco AP, Pennell CE. Whitehouse AJ. Analysis of dyslexia candidate genes in the Raine cohort representing the general Australian population. Genes Brain Behav. 2011;10:158–165. doi: 10.1111/j.1601-183X.2010.00651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington BF. From single to multiple deficit models of developmental disorders. Cognition. 2006;101:385–413. doi: 10.1016/j.cognition.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Pennington BF. Bishop DV. Relations among speech, language, and reading disorders. Annu Rev Psychol. 2009;60:283–306. doi: 10.1146/annurev.psych.60.110707.163548. [DOI] [PubMed] [Google Scholar]
- Peschansky VJ, Burbridge TJ, Volz AJ, Fiondella C, Wissner-Gross Z, Galaburda AM, Turco JJL. Rosen GD. The effect of variation in expression of the candidate dyslexia susceptibility gene homolog Kiaa0319 on neuronal migration and dendritic morphology in the rat. Cereb Cortex. 2010;20:884–897. doi: 10.1093/cercor/bhp154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RL. Pennington BF. Developmental dyslexia. Lancet. 2012;379:1997–2007. doi: 10.1016/S0140-6736(12)60198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ. Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- Robertson SP. Filamin A: phenotypic diversity. Curr Opin Genet Dev. 2005;15:301–307. doi: 10.1016/j.gde.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Roeske D, Ludwig KU, Neuhoff N, Becker J, Bartling J, Bruder J, Brockschmidt FF, Warnke A, Remschmidt H, Hoffmann P, Muller-Myhsok B, Nothen MM. Schulte-Korne G. First genome-wide association scan on neurophysiological endophenotypes points to trans-regulation effects on SLC2A3 in dyslexic children. Mol Psychiatry. 2011;16:97–107. doi: 10.1038/mp.2009.102. [DOI] [PubMed] [Google Scholar]
- Rust J, Golombok S. Trickey G. Wechsler Objective Reading Dimensions. Sidcup, UK: Psychological Corporation; 1993. [Google Scholar]
- Scerri TS, Brandler WM, Paracchini S, Morris AP, Ring SM, Richardson AJ, Talcott JB, Stein J. Monaco AP. PCSK6 is associated with handedness in individuals with dyslexia. Hum Mol Genet. 2011a;20:608–614. doi: 10.1093/hmg/ddq475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scerri TS, Morris AP, Buckingham L-L, Newbury DF, Miller LL, Monaco AP, Bishop DVM. Paracchini S. DCDC2, KIAA0319 and CMIP are associated with reading-related traits. Biol Psychiatry. 2011b;70:237–245. doi: 10.1016/j.biopsych.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semel EM, Wiig EH. Secord W. Clinical Evaluation of Language Fundamentals – Revised. San Antonio, TX: Psychological Corporation; 1992. [Google Scholar]
- Shapleske J, Rossell SL, Woodruff PWR. David AS. The planum temporale: a systematic, quantitative review of its structural, functional and clinical significance. Brain Res Rev. 1999;29:26–49. doi: 10.1016/s0165-0173(98)00047-2. [DOI] [PubMed] [Google Scholar]
- Shaywitz SE, Shaywitz BA, Fletcher JM. Escobar MD. Prevalence of reading disability in boys and girls. Results of the Connecticut Longitudinal Study. JAMA. 1990;264:998–1002. [PubMed] [Google Scholar]
- Simpson NH, Addis L, Brandler WM, Slonims V, Clark A, Watson J, Scerri TS, Hennessy ER, Bolton PF, Conti-Ramsden G, Fairfax BP, Knight JC, Stein J, Talcott JB, O'Hare A, Baird G, Paracchini S, Fisher SE, Newbury DF Consortium, S.L.I. Increased prevalence of sex chromosome aneuploidies in specific language impairment and dyslexia. Dev Med Child Neurol. 2014;4:346–353. doi: 10.1111/dmcn.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowling M, Bishop DV. Stothard SE. Is preschool language impairment a risk factor for dyslexia in adolescence? J Child Psychol Psychiatry. 2000;41:587–600. doi: 10.1111/1469-7610.00651. [DOI] [PubMed] [Google Scholar]
- Taipale M, Kaminen N, Nopola-Hemmi J, Haltia T, Myllyluoma B, Lyytinen H, Muller K, Kaaranen M, Lindsberg PJ, Hannula-Jouppi K. Kere J. A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain. Proc Natl Acad Sci U S A. 2003;100:11553–11558. doi: 10.1073/pnas.1833911100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammimies K, Tapia-Páez I, Rüegg J, Rosin G, Kere J, Gustafsson J-Å. Nalvarte I. The rs3743205 SNP is important for the regulation of the dyslexia candidate gene DYX1C1 by estrogen receptor β and DNA methylation. Mol Endocrinol. 2012;26:619–629. doi: 10.1210/me.2011-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammimies K, Vitezic M, Matsson H, Le Guyader S, Burglin TR, Ohman T, Stromblad S, Daub CO, Nyman TA, Kere J. Tapia-Paez I. Molecular networks of DYX1C1 gene show connection to neuronal migration genes and cytoskeletal proteins. Biol Psychiatry. 2013;73:583–590. doi: 10.1016/j.biopsych.2012.08.012. [DOI] [PubMed] [Google Scholar]
- The International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The SLI Consortium. A genomewide scan identifies two novel loci involved in specific language impairment. Am J Hum Genet. 2002;70:384–398. doi: 10.1086/338649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The SLI Consortium. Highly significant linkage to the SLI1 locus in an expanded sample of individuals affected by specific language impairment. Am J Hum Genet. 2004;74:1225–1238. doi: 10.1086/421529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomblin JB, Records NL, Buckwalter P, Zhang X, Smith E. O'Brien M. Prevalence of specific language impairment in kindergarten children. J Speech Lang Hear Res. 1997;40:1245–1260. doi: 10.1044/jslhr.4006.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, Alarcon M, Oliver PL, Davies KE, Geschwind DH, Monaco AP. Fisher SE. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, Oliver PL, Spiteri E, Lockstone HE, Puliyadi R, Taylor JM, Ho J, Mombereau C, Brewer A, Lowy E, Nicod J, Groszer M, Baban D, Sahgal N, Cazier J-B, Ragoussis J, Davies KE, Geschwind DH. Fisher SE. Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genet. 2011;7:e1002145. doi: 10.1371/journal.pgen.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ. Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D. Manual for the Wechsler Intelligence Scale for Children – Revised. New York, NY: The Psychological Corporation; 1974. [Google Scholar]
- Wechsler D. Manual for the Wechsler Adult Intelligence Scale – Revised. New York, NY: Psychological Corporation; 1981. [Google Scholar]
- Wechsler D, Golombok S. Rust J. WISC-IIIUK: Wechsler Intelligence Scale for Children: UK manual. 3rd edn. Sidcup, UK: The Psychological Corporation; 1992. [Google Scholar]
- Whitehouse AJ, Bishop DV, Ang QW, Pennell CE. Fisher SE. CNTNAP2 variants affect early language development in the general population. Genes Brain Behav. 2011;10:451–456. doi: 10.1111/j.1601-183X.2011.00684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse AJ, Mattes E, Maybery MT, Sawyer MG, Jacoby P, Keelan JA. Hickey M. Sex-specific associations between umbilical cord blood testosterone levels and language delay in early childhood. J Child Psychol Psychiatry. 2012;53:726–734. doi: 10.1111/j.1469-7610.2011.02523.x. [DOI] [PubMed] [Google Scholar]
- Willcutt EG, Pennington BF, Olson RK, Chhabildas N. Hulslander J. Neuropsychological analyses of comorbidity between reading disability and attention deficit hyperactivity disorder: in search of the common deficit. Dev Neuropsychol. 2005;27:35–78. doi: 10.1207/s15326942dn2701_3. [DOI] [PubMed] [Google Scholar]
- Willcutt EG, Betjemann RS, McGrath LM, Chhabildas NA, Olson RK, DeFries JC. Pennington BF. Etiology and neuropsychology of comorbidity between RD and ADHD: the case for multiple-deficit models. Cortex. 2010;46:1345–1361. doi: 10.1016/j.cortex.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y. Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M, De Geus E, Ando J, Luciano M, Posthuma D, Ono Y, Hansell N, Van Baal C, Hiraishi K, Hasegawa T, Smith G, Geffen G, Geffen L, Kanba S, Miyake A, Martin N. Boomsma D. Genetics of cognition: outline of a collaborative twin study. Twin Res. 2001;4:48–56. doi: 10.1375/1369052012146. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Sheng SR, Wang XY, Bin LH, Wang JR. Li GY. Expression of tumor related gene NAG6 in gastric cancer and restriction fragment length polymorphism analysis. World J Gastroenterol. 2004;10:1361–1364. doi: 10.3748/wjg.v10.i9.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: QQ plots and association plots of the top association signals from analysis based on PC1 and IQ-adjusted PC1. Contribution of each dataset to the strength of the association in the PC1 and IQ-adjusted PC1 meta-analysis, for the top association signals.
Appendix S2: SNP-, gene- and pathway-based meta-analysis results from analysis based on PC1 and IQ-adjusted PC1. Test for independent associations within the top associated regions, after conditioning on the most significant local association signal.
Appendix S3: Analyses based on PC1read and IQ-adjusted PC1read (trait loadings on PC1read, Manhattan plots, QQ plots and top associations)
Appendix S4: Genotype calls and QC protocols; correlation patterns of reading and language measures; statistical analyses, commands and parameters.