Abstract
Diverse optogenetic tools have allowed versatile control over neural activity. Many depolarizing and hyperpolarizing tools have now been developed in multiple laboratories and tested across different preparations, presenting opportunities but also making it difficult to draw direct comparisons. This challenge has been compounded by the dependence of performance on parameters such as vector, promoter, expression time, illumination, cell type and many other variables. As a result, it has become increasingly complicated for end users to select the optimal reagents for their experimental needs. For a rapidly growing field, critical figures of merit should be formalized both to establish a framework for further development and so that end users can readily understand how these standardized parameters translate into performance. Here we systematically compared microbial opsins under matched experimental conditions to extract essential principles and identify key parameters for the conduct, design and interpretation of experiments involving optogenetic techniques.
Optogenetics1,2 integrates genetic targeting and optical stimulation to achieve temporally precise manipulation of genetically and spatially defined cell types in intact tissue, and has influenced the study of the central nervous system and other systems across a broad range of model organisms and behaviors3,4. The adoption of optogenetics has been facilitated by the emergence of single-component (that is, with no exogenous cofactor required5), genetically targetable, microbial (type I) ‘opsin’ genes, encoding proteins that respond to illumination by certain wavelengths of light with depolarizing currents, hyperpolarizing currents or specified signal-transduction events3,4.
Since the first demonstrations that microbial opsins could be used to control action potentials6–9, the optogenetic toolbox has expanded to offer researchers an increasingly powerful and diverse selection of opsins. However, this process has also made it increasingly challenging to conclude which tool might be optimal for a given experiment. To draw these conclusions, researchers require two broad classes of information.
First, it is essential to understand the important properties of microbial opsins and how these properties vary across opsin type. It is difficult to extract a rigorous comparison from the current literature because previously published studies differ in confounding variables that contribute to differences in performance (including expression method, vector backbone, promoter, temperature, light power density and stimulation protocols). There has not been a broad empirical comparison under standardized conditions that allows isolated comparison of the tools themselves, rather than of the experimental parameters.
Second, it is important to understand how differences in these properties may result in differences in the ability to elicit or inhibit neural activity. Indeed, most papers introducing new opsin genes have also included evidence of improved functionality, often highlighting the advantage of one or several key properties. But not only do different papers highlight different aspects of performance, confounding experimental variables make it difficult to compare results from even a single performance measure across datasets. For example, the biophysical properties of the specific cell type will determine how the photocurrent is transformed into a change in membrane voltage and also how that change in membrane potential affects action-potential firing.
To begin to address these issues, we compared, in parallel, depolarizing and hyperpolarizing optogenetic tools, under conditions chosen for relevance to the mammalian nervous system. We then investigated the ability of depolarizing tools to elicit spikes in pyramidal cells and to drive fast-spiking cells at high frequencies as well as the ability of hyperpolarizing tools to inhibit action potentials in pyramidal cells. Given the complexity of the subject, this work will not answer all questions about performance under all possible circumstances, which will still inevitably be contingent on many experimental factors and will need to be addressed for each specific experimental condition. Rather, our analysis is intended to provide a starting point: to compare properties and performance under specific experimental conditions, to identify critical parameters, to develop a framework of principles that organize the currently available tools and to guide characterization of future tools.
RESULTS
Depolarizing tools and properties
Channelrhodopsins are cation channels that can give rise to neuronal depolarization when activated by light. Channelrhodopsin-2 (ChR2), isolated from the algae Chlamydomonas reinhardtii, can depolarize neurons and evoke precisely timed action potentials7–10. The subsequently developed depolarizing tools, ChR2 point mutants10–16, channelrhodopsins from other algal species identified using genomic strategies17,18 and chimeras constructed by combining channelrhodopsins11,19–21, range widely in their photo-current wavelength selectivity, kinetics and/or magnitudes. Here we compare microbial opsin genes that enable elicitation of precisely timed action potentials in response to light and exclude step-function opsins (SFOs), a distinct class of depolarizing tool that exhibits bistable photocurrents used to modulate excitability11,15.
Light-evoked spiking is a function of the size and kinetics of the photocurrent, together with the cell’s biophysical response to that photocurrent. We first quantify these photocurrent properties under voltage clamp and then compare performance in eliciting spikes under current clamp. We note some initial principles that govern the experimental setup to follow: first, to elicit spikes, these channelrhodopsins must open rapidly in response to a light pulse, conduct enough photocurrent to bring the neuron past the spiking threshold with precise timing after light onset and then close rapidly after the pulse to allow the neuron to repolarize. Second, photocurrents typically desensitize in response to sustained light; this effect can cause spike ‘failures’ if photocurrents are reduced to subthreshold amounts. To elicit consistent firing throughout prolonged pulse trains, channelrhodopsins should therefore exhibit minimal desensitization during light and/or rapid recovery from desensitization in darkness. Third, for some applications, particularly experiments in vivo, it may be important to modulate a large or distant tissue volume using low intensity light, in which case the relationship between photocurrent and light intensity becomes critical.
We first characterized, side by side, the major strongly expressing depolarizing channelrhodopsins that have been reported, which fall into three genetic classes (Fig. 1a). The first class consists of wild-type ChR2 and ChR2 mutants with several single-amino-acid substitutions: ChR2(H134R) (ChR2R10,12), ChR2(E123A) (ChETAA13), ChR2(T159C) (TC14), ChR2(E123T/ T159C) (ChETATC14) and ChR2(L132C) (CatCh16). The second class comprises hybrids formed from combining different segments of ChR1 and ChR2: ChIEF19, which has an I170V amino acid substitution relative to ChR1, channelrhodopsin fast receiver (FR20) and channelrhodopsin green receiver (GR21). The third class consists of hybrids formed by combining ChR1 and VChR1 (a ChR variant from Volvox carteri), termed C1V1, including the mutants C1V1(E162T) (C1V1T11) and C1V1(E122T/E162T) (C1V1TT11) (see Supplementary Table 1 for a summary of depolarizing tool backbones, mutations and naming convention). We compared photocurrent properties in transfected cultured hippocampal pyramidal neurons. We packaged all opsin genes identically in a lentiviral backbone, driven by the mouse excitatory neuron–specific CaMKIIoc (Camk2a) promoter and fused in frame with the gene encoding enhanced YFP (eYFP), enabling direct visualization of transfected neurons (Fig. 1b).
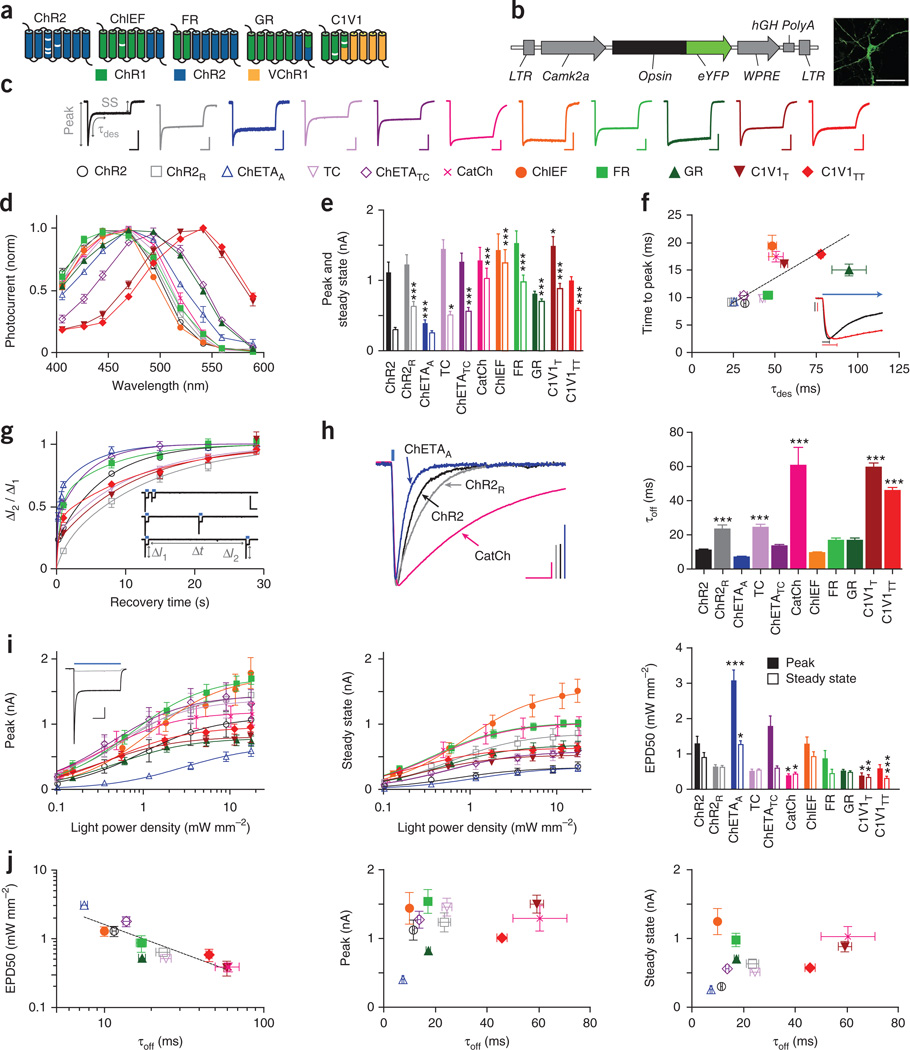
Figure 1.
Properties of depolarizing optogenetic tools. (a) Depolarizing tool classes. White bars indicate mutations. (b) Construct design and representative image for in vitro characterization. Scale bar, 50 µm. (c) Normalized representative photocurrents. Scale bars, 400 pA, 200 ms. Horizontal scale bar applies to all traces. Color and shape legend applies throughout the figure. (d) Action spectra (n = 5–11). (e) Peak (filled bars) and steady-state (hollow bars) photocurrents to 1 s light (n = 8–27). (f) Time to peak (n = 8–27) versus τdes (n = 8–50). Traces show normalized representative ChR2 (black) and C1V1TT (red) onset photocurrents. Vertical scale bars represent 200 pA, bars indicate time to peak, and blue arrow indicates ongoing light pulse. (g) Recovery from desensitization (n = 5–20). Vertical and horizontal scale bars represent 1 nA and 2 s. (h) Normalized representative traces and summary plots of τoff (n = 8–53). Scale bars, 200 pA and 25 ms. (i) Peak and steady-state photocurrents across light intensities. Inset, representative ChR2 photocurrents at low (light gray) versus high (dark gray) light intensity. Scale bars, 250 pA and 250 ms. EPD50 for peak (filled bars) and steady state (hollow bars) (n = 5–15). (j) τoff versus EPD50 and peak and steady-state photocurrents. All population data are plotted as mean ± s.e.m. *P < 0.05, **P < 0.01 and *** P < 0.001. Unless otherwise indicated, C1V1T and C1V1TT were activated with 560-nm light, and all other tools were activated with 470-nm light at ∼5 mW mm−2.
We tested these depolarizing tool variants under voltage-clamp conditions, analyzing factors that contribute to photocurrent amplitude. Because many photocurrent properties are dependent on light intensity, we matched light power density across experiments, at an irradiance (5 mW mm−2) chosen to mimic typical effective and safe experimental conditions at target neurons in vivo4. Although most uses of these depolarizing tools involve pulsed light, the response to constant light (Fig. 1c and Supplementary Fig. 1a) provides crucial insight into the same channel dynamics that underlie the response to any pattern of light stimulation, with increasing relevance for longer and higher frequency stimulation patterns which, in the limit, approach the constant-light condition. The macroscopic photocurrent is a result of the aggregate activity of the membrane-resident channel population, giving rise to a peak photocurrent followed by a smaller steady-state photocurrent owing to a proportion of the population shuttling to the desensitized state22,23. Based on the action spectra (Fig. 1d), we performed subsequent characterizations using 560-nm light for C1V1T and C1V1TT, and blue light (470 nm) for all other channelrhodopsins, although we noted a moderate red-shift in ChETATC, as previously observed14.
Compared to ChR2, most depolarizing tools had similar mean peak photocurrents (1–1.5 nA), although ChETAA photocurrents were significantly smaller (<0.5 nA) and C1V1T photocurrents were significantly larger than ChR2 (P < 0.05; Fig. 1e and Supplementary Fig. 1b). However, pointing to one of the most important challenges faced when aiming to achieve performance consistency, we observed that ChR2 displayed the most desensitization or the lowest steady-state/peak ratio (0.30 ± 0.01 (values throughout are mean ± s.e.m.), Supplementary Fig. 1c), resulting in significantly smaller steady-state photocurrents than all other tools except for ChETAA (P < 0.001–0.05). In contrast, CatCh, ChIEF and GR had the highest steady-state/peak ratios (>0.75), demonstrating capability for stable photocurrent responses across prolonged illumination. Although cells expressing different opsins differed significantly in the mean total fluorescence from these eYFP fusion proteins, with ChR2R, ChETAA and ChETATC being the brightest (P < 0.001), this measure was not a good predictor of group differences in photocurrents among opsins; there was also significant variation in photocurrent/fluorescence ratio, with CatCh, ChIEF, C1V1T and C1V1TT showing the highest ratios (P < 0.001–0.01; Supplementary Fig. 1d,e).
Because photocurrent peak timing results from the balance between activation and desensitization of the proteins (Fig. 1f), we next examined the kinetics of those processes most relevant to the temporal precision and stability of the photocurrent response. ChR2 had the fastest time to peak, and CatCh, ChIEF, GR, C1V1T and C1V1TT were significantly slower (>15 ms, P < 0.001, Supplementary Fig. 1f). As activation and desensitization occur simultaneously, photocurrents are expected to peak more rapidly if desensitization rate is faster. We measured desensitization kinetics (τdes; Supplementary Fig. 1g) with a monoexponential fit of the photocurrent transition from peak to steady state, and indeed the desensitization kinetics were highly correlated with time to peak (R2 = 0.53, Spearman correlation coefficient R = 0.77, P < 0.01; Fig. 1f).
We next explored the kinetics of recovery from desensitization in darkness, another important determinant of photocurrent stability in response to pulsed light. We delivered two 1-s light pulses separated by varying intervals of darkness (Δt); for each light pulse, we quantified ΔI, the difference between the peak and steady-state photocurrent magnitude (Fig. 1g). We divided ΔI from the second pulse (ΔI2) by ΔI from the first pulse (ΔI1) and plotted the ratio against Δt (Fig. 1g). We excluded CatCh, ChIEF and GR from this analysis because they had high (>75%) steady-state/peak ratios (very little desensitization), which prevented accurate measurement of this parameter and rendered measurement of this parameter less functionally relevant. The data were well described by a bi-exponential fit (R2 of 0.77–0.98), from which we quantified time required for 50% recovery from desensitization (Supplementary Fig. 1h). ChETAA had significantly faster recovery (P < 0.05), and ChETATC and FR both trended in that direction; ChR2R and C1V1T recovery was significantly slower (P < 0.01).
Finally, we measured off kinetics (τoff), the rate of channel closure at the end of the light pulse. Fast off kinetics are important for avoiding sustained depolarization between light pulses. We calculated τoff from a monoexponential fit of the decay of the photocurrent after light offset. As τoff has been shown to depend on parameters including pulse duration9, we used a short (3-ms) light pulse, a typical duration for eliciting spikes in vivo (Fig. 1h). The proteins had widely varied off kinetics: compared to ChR2 (11.6 ± 0.4 ms), under these conditions ChETAA trended toward more rapid deactivation (7.5 ± 0.4 ms), ChETATC, ChIEF, FR and GR had similar kinetics (10–17 ms), and the remaining tools were slower; in particular CatCh and C1V1T were significantly slower (both ∼60 ms, P < 0.001).
An additional critical consideration for in vivo use is activation at low light, for modulation of large volumes of tissue or minimally invasive stimulation of cells far from the light source11. We therefore quantified photocurrent responses across a range of light power densities, ∼0.1–20 mW mm-2. Although we focus this analysis on peak and steady-state photocurrent magnitudes, we note that light power density greatly affects not only the photocurrent size but also many other features characterized above, including steady-state/peak ratio, time to peak (quantified for ChR2 in Supplementary Fig. 1i) and desensitization kinetics, as seen in the representative ChR2 photocurrent responses (Fig. 1i).
We measured both peak and steady-state photocurrents across the range of light power densities and fit the data with a one-site specific binding curve (R2 = 0.34–0.82; Fig. 1i) as has been used previously19. For safety reasons we did not push to saturating light intensities, so we did not constrain the peak value of the fit; we therefore extrapolated the maximal peak and steady-state photocurrents achievable with unbounded light intensity, ChIEF showing the largest predicted peak (1,740 ± 140 pA) and steady state (1,540 ± 110 pA, Supplementary Fig. 1j). Both peak and steady-state photocurrents increased with light power density, but steady-state photocurrents saturated more quickly, resulting in a lower steady-state/peak ratio at higher light power densities.
Absolute (non-normalized) photocurrents help define the ‘operational’ light sensitivity of the different opsins, providing insight into practical cell performance at different light power densities. ChIEF, for example, has the highest photocurrent peak and steady state at higher light power densities, whereas ChR2 shows peak photocurrents that are moderate and steady-state photocurrents that are among the smallest. However, these absolute photocurrent values are highly subject to protein expression levels under our specific experimental conditions (transfected cultured neurons, CaMKIIα promoter and so on). To focus on differences independent of expression, we normalized photocurrents in each cell (Supplementary Fig. 1k,l) and used the curve fit to calculate the light power density required to achieve half-maximal activation (EPD50 or effective power density for 50% activation, analogous to an EC50; Fig. 1i), a measure of sensitivity independent of protein expression. Compared to ChR2 (EPD50 for the peak was 1.3 ± 0.2 mW mm−2), the ChETAA peak photocurrent was less sensitive (3.1 ± 0.3 mW mm−2, P < 0.001), whereas those for CatCh and C1V1T were more sensitive (∼0.4 ± 0.1 mW mm−2, P < 0.05).
Differences in EPD50 (population light sensitivity) may arise from differences in intrinsic protein light sensitivity (the efficiency with which individual molecules are activated by light24) or from differences in off kinetics. At lower light intensities a smaller fraction of the pool is recruited in the beginning of the pulse, so there is a larger pool of molecules in the closed state, which can continue to be recruited as the light pulse continues; moreover, with slow off kinetics, members of the population of opsins on a cell will accumulate in the open state as the light pulse continues. In fact, τoff and EPD50 were highly correlated (R2 = 0.84, Spearman correlation coefficient R = −0.91, P < 0.001; Fig. 1j), suggesting that population light sensitivity is dominated by off kinetics, although intrinsic light sensitivity may account for deviations from the curve. This correlation supports a theoretical tradeoff between off kinetics (precision of spiking) and sensitivity (volume of activation).
To summarize experimentally important characteristics of the depolarizing tools, we plotted τoff against peak and steady-state photocurrents (Fig. 1j), both calculated from 1-s pulses of ∼5 mW mm-2 light. These summary plots can be used as a starting point to guide opsin choice for experiments with different stimulation paradigm timing and patterns (which will help determine the relevance of the peak versus the steady state), and with different requirements for photocurrent size and kinetics.
Evoking spiking in pyramidal cells in vitro
We next compared the same depolarizing tools under current clamp to investigate evoked spiking in pyramidal cells. As ChETAA had small photocurrents in this preparation (not consistently larger than the rheobase of pyramidal cells25), and the GR activation spectrum limits combinatorial experiments relative to the C1V1 tools without providing a photocurrent advantage, we excluded those two opsins from this analysis, focusing on ChR2, ChR2R, TC, ChETATC, CatCh, ChIEF, FR, C1V1T and C1V1TT. Although subsets of these depolarizing tools have previously been compared14,16,19,26, to our knowledge no empirical study to date has attempted the broader comparison.
Because we performed these experiments separately from those in Figure 1, we first verified that steady-state photocurrent values were comparable (Supplementary Fig. 2a). Although we could not directly measure peak photocurrents owing to escaped spikes (we collected previous data in the presence of sodium-channel blockers, which could not be used in these experiments), we also calculated expected peak values based on the steady-state/peak ratios from the previous dataset (Supplementary Fig. 2b). We first compared the health of opsin-expressing cells; relative to eYFP-transfected controls, all opsin-expressing cells trended toward larger mean holding currents, with significant differences reached for ChIEF (P < 0.01), C1V1T (P < 0.01) and C1V1TT (P < 0.05). ChIEF-expressing cells also had significantly lower input resistance and membrane capacitance (P < 0.05 for both; Supplementary Fig. 2c–e). As these lower input resistances could impede spiking in response to a given input, we compared spiking fidelity of opsin- and eYFP-expressing cells in response to trains of short (5-ms) 400-pA current injections over a range of frequencies (Supplementary Fig. 2f). eYFP-expressing cells trended toward more successful spiking at higher pulse frequencies, and two-way ANOVA revealed a significant interaction between cell population and spiking success across frequencies (F54,738 = 1.83; P = 0.0004). However, Bonferroni post-tests of spiking performance relative to eYFP were not significant for any opsin at any frequency.
We stimulated cells with trains of 40 pulses (2-ms pulse width) ranging from 5 Hz to 100 Hz, at 20 mW mm−2, 6 mW mm−2 and 2 mW mm−2 (Fig. 2a). The performance of some tools, such as ChR2, deteriorated markedly with decreasing light power densities, whereas that of others remained stable or even (in the case of CatCh) improved. Bonferroni post-tests between ChR2 and CatCh showed that, although CatCh performed significantly worse than ChR2 at high light intensities (P < 0.01 at 10 and 20 Hz), it performed better than ChR2 at low light intensities (P < 0.05 at 5 and 10 Hz), consistent with previous reports16. A likely explanation is the difference in light sensitivity, with ChR2 (which has a higher EPD50) creating insufficient photocurrent to generate reliable spikes at lower light power densities.
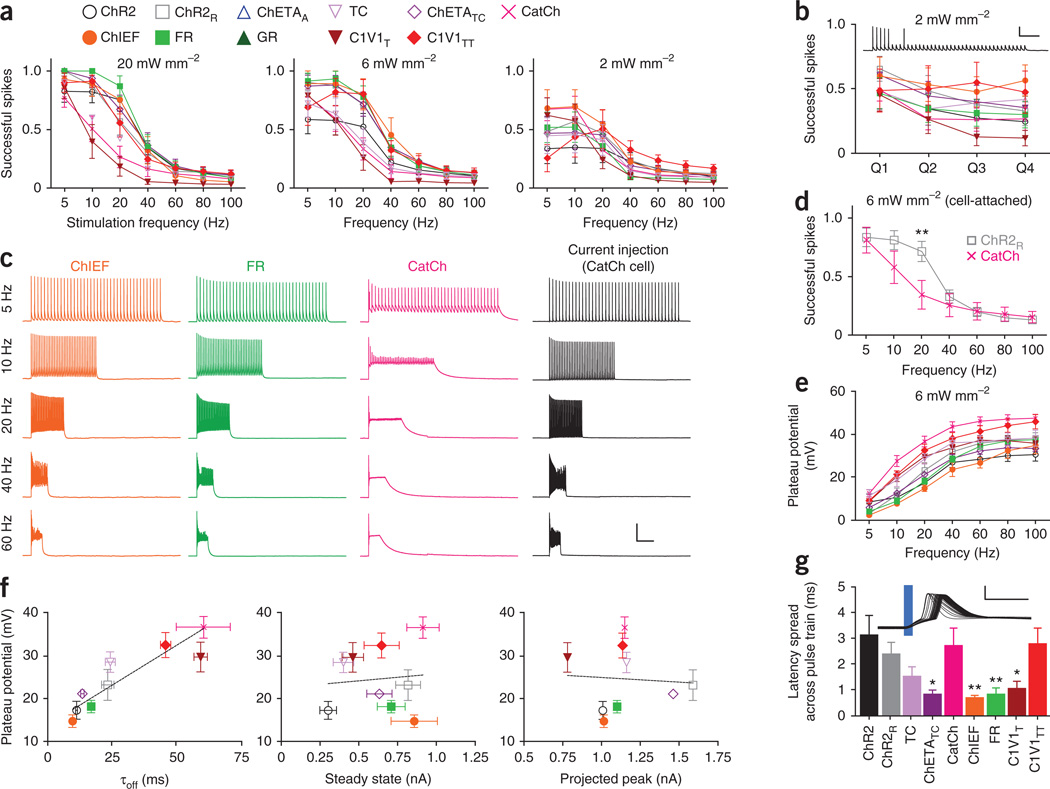
Figure 2.
Performance of depolarizing tools. (a) Proportion of successfully evoked spikes (of 40 pulses; 5–100 Hz) at different light intensities (n = 8–18). Colors and shapes apply throughout the figure. (b) Temporal stationarity at 20 Hz, 2 mW mm−2(n = 8–18), based on the proportion of successful spikes in each quartile of pulses. Vertical and horizontal scale bars represent 40 mV and 1 s, respectively. (c) Representative evoked spiking across stimulation frequencies for ChIEF, FR and CatCh with closely matched ∼1.5 nA steady-state photocurrents at 6 mW mm−2. Vertical and horizontal scale bars represent 40 mV and 1 s, respectively. (d) Comparison of spiking performance between ChR2R (n = 19) and CatCh (n = 12) in cell-attached mode at 6 mW mm−2. (e) Plateau potential across pulse frequencies at 6 mW mm−2(n = 5–17). (f) Mean plateau potential for each opsin plotted against τoff, steady-state photocurrents and projected peak photocurrents. All values taken from the 6 mW mm−2 condition. (g) Latency spread across a pulse train, illustrated by representative traces of 40 consecutive ChR2 spikes in a train, aligned to the light pulse and overlaid. Vertical and horizontal scale bars represent 40 mV and 10 ms, respectively. All population data are plotted as mean ± s.e.m. *P < 0.05 and **P < 0.01. C1V1T and C1V1TT were activated with 560-nm light, and all other opsins were activated with 470-nm light.
As commonly observed, we also noted that spiking failures tended to occur later in the pulse train for most of these optogenetic tools, as seen in a representative ChR2 trace (Fig. 2b). To quantify spiking stability over time, to which we refer as ‘temporal stationarity’, we divided the pulse train into quartiles and calculated the number of successful spikes in each quartile (Fig. 2b and Supplementary Fig. 2g). One possible explanation for these late failures is insufficient photocurrent toward the end of the pulse train, owing to photocurrent desensitization. However, slow off kinetics could also cause spike failures owing to interactions with the host cell repolarization and spike-firing mechanisms, particularly at higher frequencies. To explore this factor, we chose photocurrent-matched cells expressing the faster tools ChIEF and FR, and the slower tool CatCh, and examined spike performance across frequencies (Fig. 2c). CatCh elicited spiking at lower frequencies but suffered from more spike failures at high frequencies, though even the same CatCh-expressing cell could spike at high frequency in response to injection of current pulses.
It has been hypothesized that the CatCh photocurrent activates calcium-dependent BK channels that could help repolarize the cell16; pipette solutions could mask such an effect through altered calcium buffering. We therefore repeated these experiments for both CatCh and ChR2R, comparing evoked spiking in whole-cell versus cell-attached modes under identical stimulation conditions (Fig. 2d and Supplementary Fig. 3). We observed no significant differences in performance between those two methods when expressing either opsin, and ChR2R outperformed CatCh at the two higher light power densities (Bonferroni post-tests: P < 0.01 at 10 Hz, 20 mW mm−2 and P < 0.01 at 20 Hz, 6 mW mm−2; Fig. 2d and Supplementary Fig. 3).
CatCh-expressing cells displayed a larger sustained depolarization (plateau potential), which we quantified across all opsins and conditions (Fig. 2e and Supplementary Fig. 4). This is an important issue to address because plateau potentials can lead to spike failures by impairing the voltage-dependent de-inactivation of host-cell sodium channels; ChETA mutations have previously been shown to virtually eliminate the plateau potential in fast-spiking interneurons13, but left unclear the best strategy to use for pyramidal neurons because those ChETAs did not give rise to strong photocurrents in cultured pyramidal cells (Fig. 1). We found that among strongly expressing opsins in pyramidal cells, mean plateau potentials were highly correlated with off kinetics (R2 = 0.81, Spearman’s correlation coefficient R = 0.97, P < 0.001) but not with either steady-state (R2 = 0.03, Spearman’s correlation coefficient R = 0.13, P = 0.66) or projected peak photocurrent magnitudes (R2 = 0.02, Spearman’s correlation coefficient R = 0.23, P = 0.74; Fig. 2f). For example, CatCh plateau potentials were significantly larger than for ChR2 at all frequencies above 5 Hz (P < 0.001, Bonferroni post-test; Supplementary Fig. 4); ChIEF and TC appeared to most effectively normalize the plateau potential in these cells19,26, but all of these tools displayed a pronounced frequency-dependent emergence of missed spikes compared with current injection (Supplementary Fig. 5), consistent with the idea that finite off kinetics can lead to difficulty driving pyramidal neurons at high frequency owing to accumulation of plateau potentials. Indeed, this is a cell type–specific phenomenon: fast-spiking cells will continue to spike in the presence of a sustained depolarizing current, whether electrically or optically induced, whereas pyramidal cells are more susceptible to depolarization block (Supplementary Fig. 6).
As we performed this analysis at room temperature (20–22 °C) and because channelrhodopsin kinetics accelerate markedly with temperature, the difference between current-evoked and light-evoked spiking may be smaller in vivo or in warmed solution. Another recent comparison of the functional properties of four depolarizing tools (ChR2, ChR2R, TC and ChETATC) across a similar range of frequencies and light power densities also reported large plateau potentials at high frequencies and light intensities, correlated with poor spiking performance14. In general higher-fidelity spiking had been seen in that study14 at higher frequencies; this effect may be attributable to the higher recording temperature (29–31 °C), highlighting the importance of direct experimental comparison under matched conditions.
Finally, we analyzed parameters of spiking precision: the proportion of light pulses that evoked multiple spikes, the mean spike latency (the time from light onset to spike threshold) and the mean latency spread across a pulse train (Fig. 2g and Supplementary Fig. 7a–c). In pyramidal neurons, ChIEF, ChETATC and FR consistently displayed the lowest latency, and latency spread across trains, over different stimulation conditions (Supplementary Fig. 7).
Ultrafast depolarizing tools and properties
Next we aimed to identify the most promising optogenetic tools for evoking precise and very-high-frequency spikes in fast-spiking cells. ChR2(E123T/H134R) (ChETATR) is a standard tool currently used for eliciting high-frequency stimulation and has already been shown to out-perform ChR2 in driving stimulation up to 200 Hz13. However, it is not clear how the properties of ChETATR compare with those of the single-mutant ChR2(E123T) (ChETAT) or with the additional ChETA mutants ChR2(E123A) (ChETAA) and ChR2(E123A/H134R) (ChETAAR), which have not yet been tested in neurons. Furthermore, no ChETA has ever been tested directly against ChIEF in any functional context. Based on ChIEF’s photocurrent properties (fast kinetics, low desensitization in the face of sustained light) and its ability to elicit high-fidelity spikes in pyramidal cells at the high end of their frequency capacity, ChIEF is another promising candidate for eliciting reliable, precise spiking at even higher frequencies. We therefore sequentially (i) compared the properties of the ChETA mutants to identify the most promising, (ii) tested the performance of those ChETA mutants in eliciting high-frequency spikes in fast-spiking neurons in Pvalb ::cre mice27,28 and (iii) tested the performance of the best-performing ChETA mutant against ChIEF.
We analyzed ChETAA, ChETAAR, ChETAT and ChETATR, with ChR2 included as a benchmark (Fig. 3a). As in the experiments described in Figure 1, we analyzed photocurrent properties in response to 1-s light stimulation under voltage clamp (Fig. 3a). Because these ChETA variants respond to blue light, we used 470-nm light for this comparison; but we noted that ChETAT has a moderate red-shift (Fig. 3b), as described previously13 and as seen for ChETATC (Fig. 1d). All ChETAs had smaller peak photo-currents than ChR2 (< 0. 5 nA versus > 1 nA, P < 0.001; Fig. 3c and Supplementary Fig. 8a) but in light of the small steady-state/ peak ratio of ChR2 (Supplementary Fig. 8b) displayed steady-state photocurrents comparable to those of ChR2 (∼0.3 nA, P > 0.05). Relative to the double mutants, ChETA single mutants exhibited both faster time to peak and faster desensitization kinetics, consistent with the correlation between the two parameters (Supplementary Fig. 8c–e). The single-mutation ChETA variants also had faster recovery from desensitization (Fig. 3d and Supplementary Fig. 8f) and faster off kinetics: under these matched conditions of illumination and temperature, ChETAA and ChETAT had τoff values of 7.5 ± 0.4 ms and 6.8 ± 0.2 ms, respectively, whereas the double mutants and ChR2 had τoff values several milliseconds slower (P < 0.001).
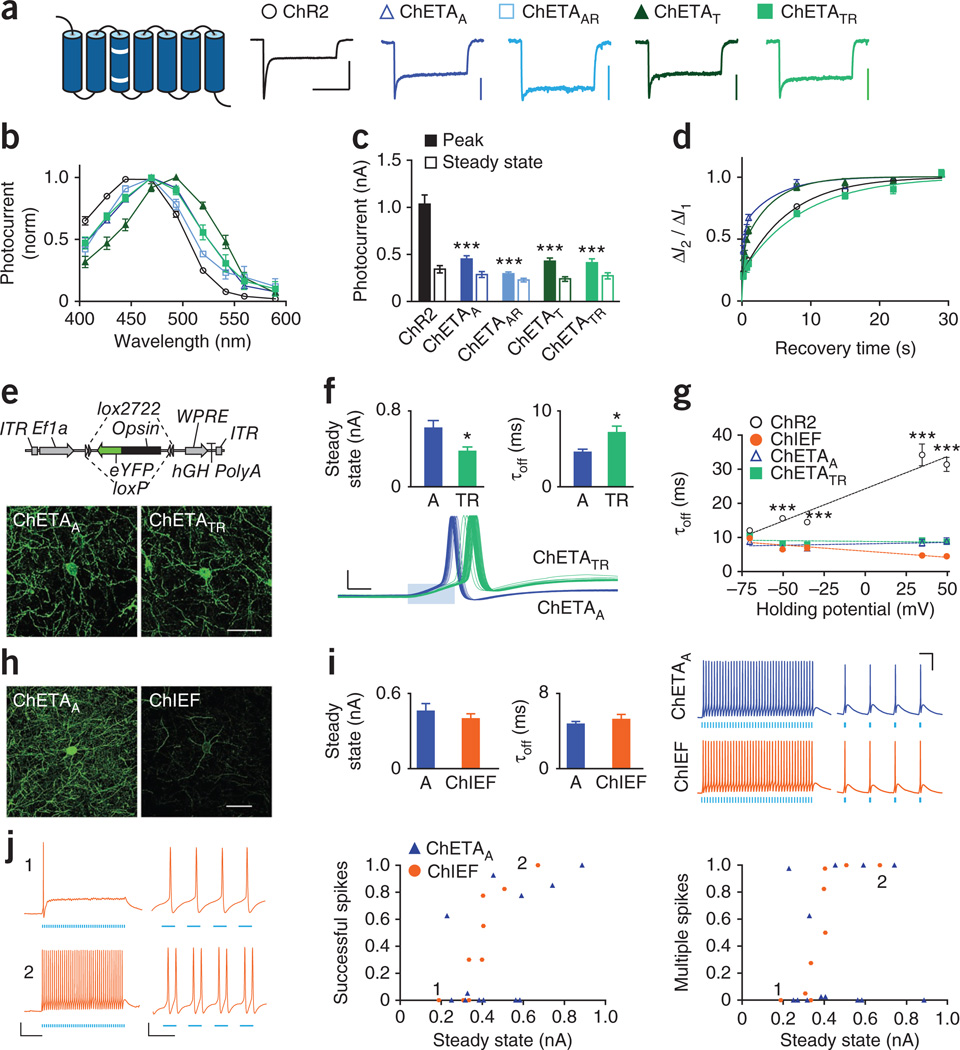
Figure 3.
Properties and performance of ultrafast depolarizing tools. (a) Schemata and normalized photocurrents for ChETAs and ChR2. White bars indicate mutations. Colors and shapes apply throughout the figure. Scale bars, 500 pA and 500 ms. Horizontal scale bar applies to all traces. (b) Action spectra (n = 5–12). (c) Peak (filled bars) and steady-state (hollow bars) photocurrents (n = 9–35). (d) Recovery from desensitization (n = 8–20). (e) ChETAA and ChETATR expression in fast-spiking neurons using a Cre recombinase-dependent strategy. Scale bar, 50 µm. (f) Steady-state photocurrents (n = 9),τoff n = 7), and consecutively evoked spikes for ChETAA and ChETATR (5 Hz, 2-ms light pulses). Scale bars, 20 mV and 1 ms. (g) τoff at −70 mV to +50 mV (n = 7–12). (h) ChETAA and ChIEF expression (scale bar, 50 µm). (i) Steady-state photocurrents (n = 9–13),τoff (n = 7), and evoked high and low frequency firing (200 Hz and 20 Hz). Scale bars, 25 mV and 25 ms. (j) ChIEF-expressing neurons with small (190 pA) or large (510 pA) photocurrents, under stringent or permissive conditions (1 ms or 5 ms pulse width). Vertical scale bar, 20 mV. Horizontal scale bars, 50 ms (left) and 10 ms (right). Spiking performance and multiple spike likelihood (under those same conditions) for all cells. All population data is plotted as mean ± s.e.m. *P < 0.05 and *** P < 0.001. Cells were illuminated with 470-nm light at ∼5 mW mm−2, unless otherwise specified.
We completed our characterization of ChETA properties by examining sensitivity to light. ChR2 had the largest peak photo-currents across light power densities, followed by ChETATR, but steady-state photocurrents were more similar (Supplementary Fig. 8g,h). A comparison of the normalized curves, to remove differences in expression or trafficking, revealed that cells expressing either of the single-mutant ChETAs showed significantly higher peak EPD50 values (>3 mW mm−2), ∼ 2–3 times greater (less sensitive) than those expressing ChR2 or the double mutants (P < 0.001, Supplementary Fig. 8i,j).
Evoking high-frequency spiking in parvalbumin neurons
As single-mutant ChETA variants had faster kinetics and double mutants (in particular ChETATR) had larger photocurrents in vitro, we compared the functional performance of ChETATR and ChETAA. (Although ChETAT was similar overall to ChETAA, it had a slightly reduced steady-state/peak ratio, which would impair spike fidelity over sustained pulse trains, as well as a spectral red-shift disadvantageous for combinatorial experiments.) We targeted fast-spiking parvalbumin cells capable of firing up to and above 200 Hz so that tool performance itself, rather than the biophysics of the cell, would be the limiting factor in achieving high-frequency spike fidelity. To achieve this, we used opsins in a Cre-dependent (double-floxed inverted open reading frame (DIO)) configuration28–30 under the control of the ubiquitous promoter EF1α (Ef1a; Fig. 3e and Online Methods). We packaged constructs in an adeno-associated viral vector (AAV serotype 2/5) and stereotactically injected into the prefrontal cortex of transgenic Pvalb ::cre mice to obtain whole-cell recordings from fast-spiking parvalbumin interneurons in an acute slice preparation.
Both ChETAA and ChETATR expressed well in vivo (Fig. 3e). Expressing cells had no difference in cell health measures of input resistance or resting potential (Supplementary Fig. 9a). In contrast to the results in culture, ChETAA actually had larger steady-state photocurrents than ChETATR (610 ± 80 pA versus 370 ± 40 pA, P = 0.02; Fig. 3f). Although both tools showed faster off kinetics (the expected result because we performed this experiment at 32 °C rather than room temperature), the relationship seen in culture was maintained, with ChETAA having faster off kinetics (4.5 ± 0.4 ms versus 7.1 ± 0.9 ms, P = 0.02; Fig. 3f). To test the performance of the two ChETAs, we stimulated cells with trains of 40 light pulses with 1 ms, 2 ms and 5 ms pulse widths of 5–200 Hz (100 Hz for 5-ms pulse width) with the cell resting at either −60 or −70 mV membrane potential. Overall, the two ChETAs performed very similarly across conditions, showing no detectable difference in temporal stationarity, plateau potential, successfully evoked spikes or multiple evoked spikes (Supplementary Figs. 9b – d and 10a). However, ChETAA triggered spikes with significantly shorter latency and reduced latency spread across the pulse train (Fig. 3f and Supplementary Fig. 10b,c) in several of the more stringent conditions (for 1-ms pulses at −60 mV, F1,15 = 5.81, P = 0.03 for latency; F1,15 = 8.28, P = 0.01 for latency spread). The lower latency of ChETAA may be attributable to a combination of faster time to peak and larger peak photocurrent, whereas its lower latency spread across the pulse train may be related to faster recovery from desensitization. These results identify ChETAA as perhaps best-suited among these ChETAs for eliciting high-frequency spikes with high precision in fast-spiking cells.
We next directly compared ChETAs and ChIEF. First, we considered the fact that our τoff measurements were obtained from neurons held at −70 mV, whereas spiking neurons are (over time) substantially more depolarized. Previous studies have shown that ChR2 τoff varies with membrane potential31 and that the E123T mutation can abolish that voltage dependence14. However, no studies to date have investigated the voltage dependence of kinetics in ChETAA or ChIEF. We therefore quantified τoff for ChR2, ChETAA, ChETATR and ChIEF, while applying holding potentials of −70 mV to +50 m V. Kinetics responded very differently to voltage (F12,144 = 27.81, P < 0.0001), with ChR2 slowing with depolarization (P < 0.001), both ChETAs staying constant and ChIEF appearing to accelerate (P < 0.001; Fig. 3g). Although ChR2 only trended slower than the others at −70 mV, ChR2 was highly significantly (P < 0.001) slower at all more depolarized voltages (Bonferroni post-test).
We investigated kinetic properties of ChR2, ChETAA and ChIEF by comparing τoff in response to the first light pulse on the cell (dark-adapted state) versus τoff in response to a subsequent light pulse after 1 s of illumination (light-adapted state). Whereas ChR2 and ChETAA showed no significant change in kinetics between those conditions, ChIEF kinetics slightly accelerated (Supplementary Fig. 11a). Note that we chose this faster, light-adapted τoff as the more relevant measure to report for ChIEF in our initial characterization (Fig. 1h), as most optogenetics experiments involve multiple light pulses. The ChIEF off kinetics (particularly under the conditions described above) along with its large, stable photocurrents (Fig. 1) suggest that it may also be highly suited for driving high-frequency firing, but no studies to date have demonstrated ChIEF functionality in driving action potentials at greater than 75 Hz26. We therefore tested ChIEF against ChETAA in fast-spiking parvalbumin neurons in acute slices.
Under matched conditions, ChETAA expressed much more strongly in vivo than ChIEF by fluorescence (P < 0.001; Fig. 3h and Supplementary Fig. 11b). However, ChIEF expression was qualitatively more membrane-localized, as seen in the higher-magnification image and as reported previously26. Consistent with this, ChIEF had significantly (P = 0.037) more photocurrent per fluorescence unit (Supplementary Fig. 11b), such that steady-state photocurrent magnitudes were comparable between the two (460 ± 50 pA for ChETAA versus 390 ± 40 pA for ChIEF; P = 0.27; Fig. 3i). ChETAA photocurrents were significantly larger in slice than in culture (P < 0.05 and P < 0.001 when expressed in vivo for 4 weeks and 6 weeks, respectively; Supplementary Fig. 11c), whereas ChIEF expressed more strongly in culture than in slice (P < 0.01 when expressed in vivo for 4 weeks; not examined at later time points), highlighting the importance of using matched cell preparations for comparison of performance. Despite the difference in opsin expression, neurons expressing both ChETAA and ChIEF had no difference in average input resistance or resting potential (Supplementary Fig. 11d). ChETAA off kinetics trended slightly faster (4.7 ± 0.3 ms versus 5.2 ± 0.5 ms; P = 0.17; Fig. 3i).
To test the performance of ChETAA and ChIEF, we stimulated cells under the same conditions described above. ChETAA and ChIEF elicited 200-Hz firing and generated precise, single action potentials in response to single light pulses (Fig. 3i and Supplementary Fig. 12a,b). Statistically, ChETAA and ChIEF showed no differences in plateau potential (P = 0.23–0.71; Supplementary Fig. 12a). Consistent with the fact that these fast-spiking cells are less subject to depolarization block, we observed that cells with larger photocurrents tended to elicit more successful spikes, as exemplified by two representative ChIEF cells with small (190 pA) and large (510 pA) photocurrents (Fig. 3j and Supplementary Fig. 12c); we saw the same trend in an analysis of ChR2-expressing neurons with larger photocurrents (Supplementary Fig. 12d). Despite not showing larger photocurrents than ChETAA, ChIEF trended toward more successful spiking (with significance obtained under some select conditions such as 2 ms pulses at −70 mV: F121 = 8.46, P = 0.01; Supplementary Fig. 12b), but ChIEF also trended toward more extra (spurious) spikes (Supplementary Fig. 13). Finally, ChETAA and ChIEF showed no consistent difference in temporal stationarity (Supplementary Fig. 13a), and had similarly fast latency and small latency spread across pulse train, for most conditions (Supplementary Fig. 14a,b).
Trade-off between kinetics and light sensitivity
Although our analysis excluded the SFOs, which have been compared elsewhere11,15 and by virtue of their orders-of-magnitude slower off kinetics are more appropriate for modulating excitability than for eliciting precisely timed spikes, we hypothesized that the same principles that govern enhanced population light sensitivity in SFOs might also explain variations in light sensitivity among the faster channelrhodopsins. Although the molecular perspective on light sensitivity considers only the efficiency by which an individual protein is activated by light (intrinsic sensitivity), cells expressing optogenetic protein populations with slower off kinetics will effectively appear more light-sensitive in the face of prolonged light stimulation, owing to greater integration of photons over time by the population at low (nonsaturating) light powers.
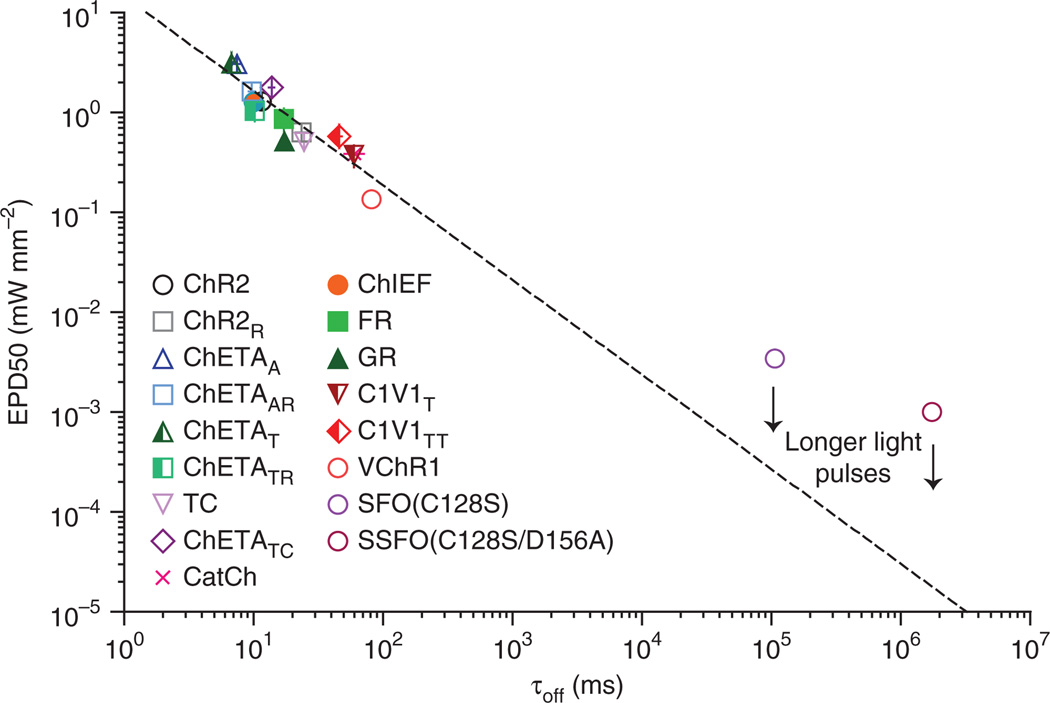
To test the universality of this principle, we plotted τoff versus EPD50 for all tools characterized in Figures 1 and 3 as well as VChR1 (ref. 17), ChR2(C128S) (an SFO15), and the recently described ChR2(C128S/D156A) stable SFO (SSFO11; Fig. 4). Indeed, the ChETAs, with generally faster off kinetics and larger EPD50s, fit well with the inverse correlation seen in Figure 1j. With the addition of the ChETAs, the combined dataset was highly correlated (Spearman correlation coefficient R = −0.92, P < 0.0001). The SFOs, which have much slower off kinetics, were also close to the established relationship. Notably, EPD50 for these SFOs will be highly dependent upon the duration of the light pulse, with longer stimulation enabling more integration of photons and higher population sensitivity (lower EPD50). Whereas the light pulse duration (1 s) used to measure EPD50 for all non-SFOs was sufficient to reach saturation, even the longer light pulses used for SFO and SSFO (5 s and 20 s, respectively) did not result in photocurrent saturation; the resulting EPD50 values were therefore overestimated and longer light pulses would be expected to bring measured values toward the consistent relationship shown. Together, these data highlight a useful and fundamental tradeoff between kinetics and population light sensitivity.
Figure 4.
Relationship between off kinetics and light sensitivity of optogenetic tools. Summary plot (on a log-log scale) of the relationship between τoff versus EPD50 for all depolarizing tools from Figures 1 and 3, plus VChR1, SFO(C128S) and SSFO(C128S/D156A). Dashed line represents best fit regression with R2 = 0.83; Spearman correlation coefficient R = −0.93, P < 0.001. Values for SFO and SSFO were estimated from previous publications11,15 and did not contribute to the regression or correlation calculations.
Hyperpolarizing tools and properties
We next compared hyperpolarizing optogenetic tools head to head. Although each experiment will have unique requirements for hyperpolarizing photocurrent properties, some common guiding principles initially seem clear. (i) In most experimental applications, hyperpolarizing photocurrents will need to be sufficiently large to robustly and safely inhibit spiking even in the presence of excitatory inputs. (ii) As with excitatory tools, higher light sensitivity will likely enable modulation of larger volumes of tissue, the use of lower light powers and/or less invasive light delivery. (iii) Precise, time-locked inhibition will presumably require photocurrents with rapid onset and offset, whereas longer-term inhibition will require photocurrents that are stable, with minimal desensitization. (iv) Finally the nature of the action spectrum will dictate feasibility of combining with other light-activated reagents in the same preparation32–34.
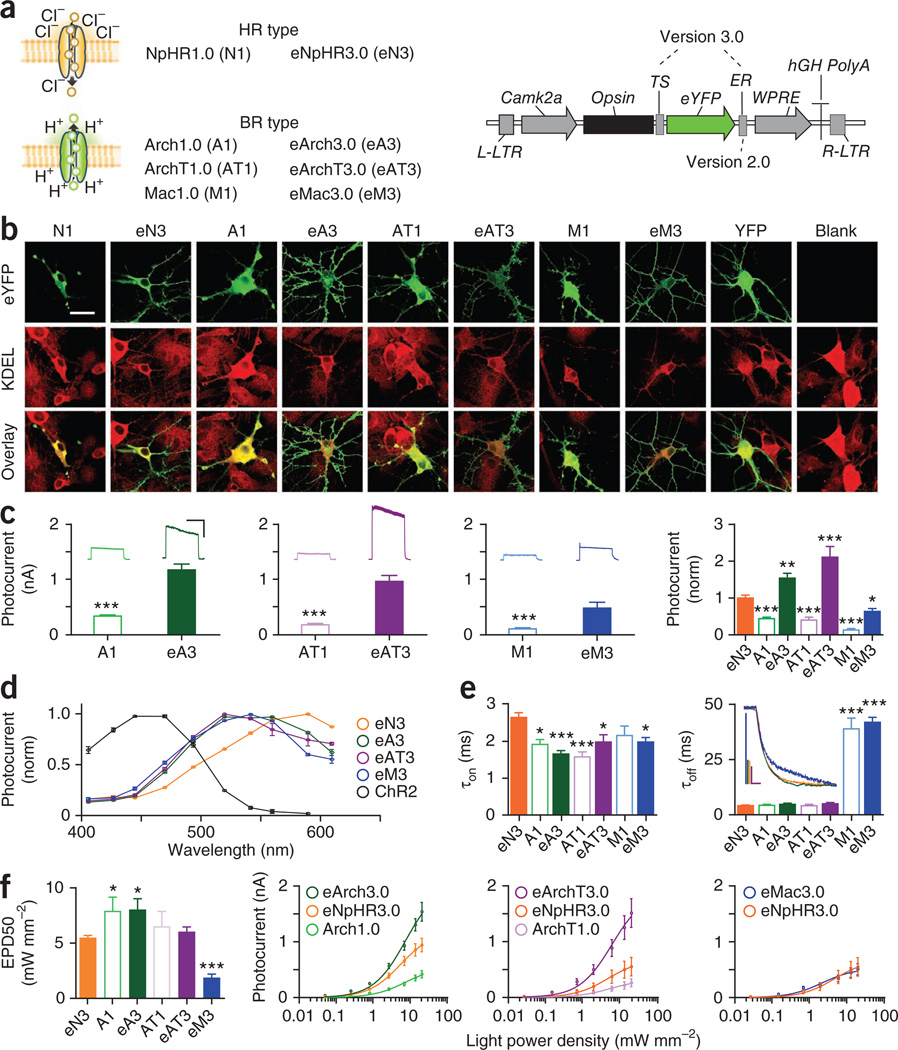
The first hyperpolarizing tool shown to be efficacious in neurons was the Natromonas pharaonis halorhodopsin (NpHR), a yellow light-activated chloride pump that has now been used in preparations ranging across mammalian brain slice32, freely moving worms32, cultured neurons32,34 and behaving mammals35–38. Two versions modified for enhanced membrane targeting in mammalian neurons, termed eNpHR2.0 (ref. 39) and eNpHR3.0 (ref. 33) have been reported. The outward proton pumps Arch40 (from Halorubrum sodomense), ArchT41 (from Halorubrum strain TP009), eBR33 (from Halobacterium) and Mac40 (from Leptosphaeria maculans) have also recently been shown to achieve neuronal inhibition. eNpHR3.0 has larger photocurrents than eNpHR2.0 (ref. 33), and Arch has larger photocurrents than eNpHR2.0 (ref. 40), but no direct comparison between eNpHR3.0 and Arch or any of the proton pumps has yet been reported to our knowledge. Below we present a direct comparison of the most potent hyperpolarizing opsins (Fig. 5a), including new membrane trafficking–enhanced versions of proton pumps resulting in the highest expression and largest inhibitory photocurrents yet described. We first characterize properties in vitro and then test the functional performance of two of the most promising candidates in acute slice.
Figure 5.
Properties of hyperpolarizing tools. (a) NpHR is an inward chloride pump (halorhodopsin type; HR), whereas Arch, ArchT, and Mac are outward proton pumps (bacteriorhodopsin type; BR). The 3.0 versions include the endoplasmic reticulum export sequence (ER) after the fluorophore (which constitutes the 2.0 version) as well as a trafficking sequence (TS) between opsin and fluorophore. (b) Confocal images of 1.0 (the originally described version of the molecule) and 3.0 versions (green) expressed in culture and immunolabeled with an ER marker (KDEL; red). Scale bar, 25 µm. (c) Representative traces and raw photocurrents in response to 1 s light for 1.0 (open bars) versus 3.0 versions (closed bars) for Arch (n = 15–19), ArchT (n = 14–16) and Mac (n = 8–12). Vertical and horizontal scale bars represent 500 pA and 500 ms, respectively. Photocurrents were normalized to eNpHR3.0 values from within the same experiment to enable direct comparisons across opsins (n = 8–35). (d) Action spectra for 3.0 versions (n = 7–20) alongside ChR2 (black). (e) τon and τoff (n = 7–35). Vertical and horizontal scale bars represent 200 pA and 5 ms, respectively. (f) EPD50 values for all hyperpolarizing opsins (n = 5–14). Raw photocurrent versus light power density plotted alongside within-experiment eNpHR3.0 (n = 5–14). Population data are plotted as mean ± s.e.m. *P < 0.05, **P < 0.01 and ***P < 0.001. Unless otherwise indicated, eNpHR3.0 was activated with 590-nm light, and all other tools were activated with 560-nm light, both at ∼5 mW mm−2.
We first fused each hyperpolarizing tool in-frame with enhanced YFP (eYFP), cloned the opsins into an identical lentiviral backbone with the excitatory CaMKIIα promoter and expressed the opsins in cultured neurons (Fig. 5a,b). eNpHR3.0 was well-targeted to the membrane, but unmodified (1.0) Arch, ArchT and Mac all showed intracellular accumulations reminiscent of the endoplasmic reticulum (ER) aggregations observed with NpHR1.0 (ref. 39). We observed the same accumulations in the GFP versions of the constructs (Supplementary Fig. 15a); the GFP and eYFP 1.0 versions (that is, those without added trafficking motifs) had similar photocurrents (data not shown). ER aggregation was confirmed by co-staining with the ER marker KDEL (Fig. 5b). Given that the trafficking modifications applied to eNpHR3.0 generalize to the proton pumps bacteriorhodopsin and G. theta rhodopsin-3 (GtR3)33, we added the same trafficking motifs to Arch, ArchT and Mac. These new trafficking-enhanced versions, which we term (by analogy with NpHR version progression) eArch3.0, eArchT3.0 and eMac3.0, all had markedly reduced intracellular labeling and improved membrane localization with labeling of cellular processes (Fig. 5b and Supplementary Fig. 15b). Intermediate 2.0 versions (that is, with an ER export motif but not the additional neurite-trafficking signal (TS) motif that defines 3.0 versions) were potent but not as successful as the 3.0 versions (Supplementary Fig. 15c).
Because only those proteins expressed on the membrane can contribute to the measured photocurrent, we anticipated that this improved opsin trafficking should increase photocurrent size. Indeed, all three trafficking-enhanced proton pumps had dramatically increased photocurrents (P < 0.001; Fig. 5c). Whereas the 1.0 versions of the proton pumps had significantly smaller photocurrents than eNpHR3.0, eArch3.0 and eArchT3.0 photo-currents were significantly larger (P < 0.01 for each comparison; Fig. 5c and Supplementary Fig. 15d). eNpHR3.0-expressing cells had the dimmest fluorescence, but the greatest photocurrent per fluorescence, of these tools (Supplementary Fig. 15e).
Although maximal eMac3.0 photocurrents were the smallest among the enhanced opsins (and significantly smaller than eNpHR3.0; P < 0.05), Mac has been reported to have an activation spectrum sufficiently blue-shifted to allow dual-inhibition in combination with eNpHR3.0 (ref. 40). After verifying that membrane trafficking did not change the spectra (Supplementary Fig. 16a), we compared the spectra of the enhanced pumps, plotted with ChR2, for reference (Fig. 5d). eNpHR3.0 was red-shifted (peaking at 560–590 nm) relative to the three proton pumps (peaking at 520–560 nm), exhibiting the least overlap with ChR2; we saw no functionally relevant differences among the proton pumps.
We next investigated the temporal precision of hyperpolarizing photocurrents by quantifying on kinetics (τon) and τoff at the beginning and end of a 1-s light pulse. All pumps activated rapidly, with proton pumps activating significantly faster than eNpHR3.0 (all in the range of 1.5–3 ms; Fig. 5e). Both Mac variants had much slower off kinetics compared with the other pumps (P < 0.001; Fig. 5e).
We also considered the light sensitivity of the hyperpolarizing pumps by measuring photocurrents across a range of light power densities from ∼0.05 mW mm−2 to ∼20 mW mm−2 (Fig. 5f and Supplementary Fig. 16b–d; owing to small photocurrents, we eliminated Mac1.0 from this and subsequent analyses). As expected, the 3.0 pumps had much larger operational light sensitivity (that is, by absolute current magnitude) than the 1.0 counterparts, although trafficking enhancement did not affect the population sensitivity (normalized current magnitudes or EPD50). eMac3.0 was the most sensitive (EPD50 = 1.9 ± 0.4 mW mm−2 versus 5.4 ± 0.2 mW mm−2 for eNpHR3.0; P < 0.001). We note that off kinetics and population light sensitivity were therefore inversely correlated for the hyperpolarizing tools (Supplementary Fig. 16d), reminiscent of the pattern observed for depolarizing tools (Fig. 4).
Given that many behavioral neuroscience experiments may require prolonged inhibition on the order of minutes, we investigated the stability of the hyperpolarizing photocurrents. Whereas all pump photocurrents decayed across 60 s of continuous light, eNpHR3.0 currents were the most persistent and the large 3.0 proton pump currents (eArch3.0 and eArchT3.0) had the largest dropoff in vitro (Supplementary Fig. 17a). All pumps recovered photocurrents with similar efficacy under these cultured-neuron conditions (Supplementary Fig. 17b).
Inhibiting spikes in pyramidal cells in acute slice
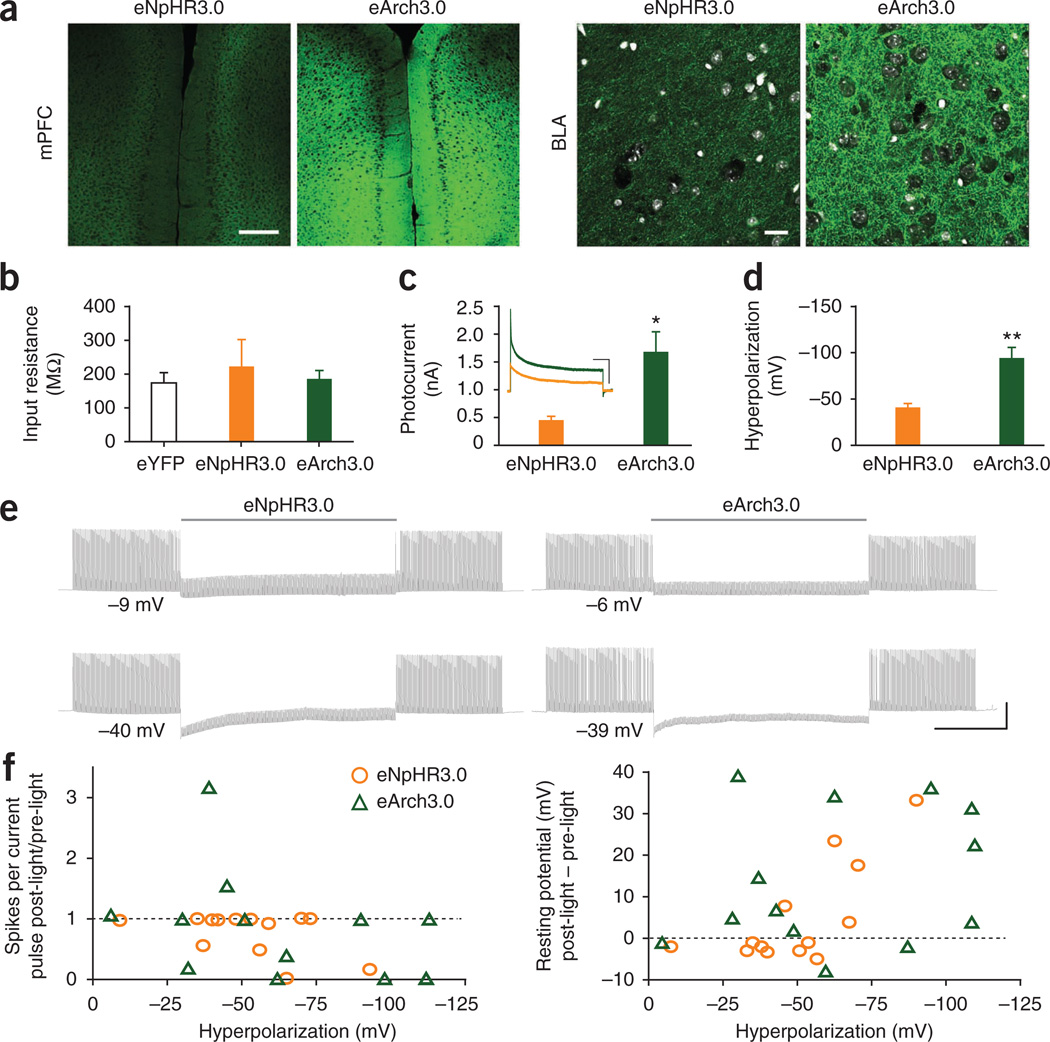
To investigate the characteristics of prolonged photocurrents under conditions more relevant to in vivo experiments and to test the functional ability of hyperpolarization to stably inhibit spiking, we turned to an acute slice preparation. We compared one of each broad class of hyperpolarizing tool (the chloride pump eNpHR3.0 against one of the proton pumps). We chose the enhanced counterpart of the best-established proton pump (Arch1.0) to date, eArch3.0. To express eNpHR3.0 and eArch3.0 in vivo, we stereotactically injected an adeno-associated viral vector (AAV serotype 2/5), with the opsin-eYFP fusion gene under control of the CaMKIIα promoter. Under matched conditions, eArch3.0 expressed much more strongly based on fluorescence, both at the injection site and in axons at downstream targets such as the basolateral amygdala (BLA; Fig. 6a). Compared with eYFP-transduced controls, cells expressing both opsins had similar baseline input resistances (Fig. 6b) and resting potentials but slightly higher membrane capacitance (Supplementary Fig. 17c), as has previously been observed for opsin-expressing HEK cells42. Also as expected from the in vitro work (Fig. 5), at matched light power densities (5 mW mm-2) eArch3.0 had significantly larger photocurrents (P = 0.01), averaging 1,680 ± 360 pA versus 450 ± 70 pA for eNpHR3.0 (Fig. 6c). Under current clamp, eArch3.0-mediated hyperpolarization was also significantly larger (−94 ± 12 mV versus −41 ± 4 mV, P = 0.005; Fig. 6d); smaller differences in hyperpolarization compared with photocurrent could be due to voltage-dependent slowing of photocycle turnover in proton pumps.
Figure 6.
Performance of hyperpolarizing tools. (a) Confocal images of eNpHR3.0 and eArch3.0 expression at the injection site in medial prefrontal cortex (mPFC) and the downstream basolateral amygdala (BLA). Scale bars, 250 µm and 25 µm. DAPI staining (white) delineates cell bodies. (b) Mean input resistances for opsin-expressing cells and eYFP controls (n = 10–22). (c) Representative traces and mean onset photocurrents for eNpHR3.0 and eArch3.0 in response to 60 s 5 mW mm−2 light pulses (n = 8–10). Vertical and horizontal scale bars represent 400 pA and 10 s, respectively. (d) Mean peak hyperpolarization generated by eNpHR3.0 and eArch3.0 with 60 s 5 mW mm−2 light pulses (n = 6–10). (e) Suppression of current injection-evoked spiking in reliably firing cells by 60 s of continuous light in cells expressing eNpHR3.0 or eArch3.0. Cells were illuminated with light power densities set to achieve approximately matched hyperpolarization. Vertical and horizontal scale bars represent 40 mV and 20 s, respectively. (f) Relationship between hyperpolarization magnitude and cell stability. Post-light recovery of evoked spiking (relative to pre-light performance) and change in resting potential plotted against light-evoked hyperpolarization. Population data are plotted as mean ± s.e.m. *P < 0.05 and **P < 0.01. eNpHR3.0 was activated with 590-nm light, and eArch3.0 was activated with 560-nm light.
Because photocurrent stability and cell responses to hyperpolarization may depend on photocurrent magnitudes, we carried out a final set of experiments using nonmatched light power densities (5–10 mW mm-2 for eNpHR3.0; 0.25–5 mW mm−2 for eArch3.0) to obtain a similar range of photocurrents for the two tools. We again illuminated cells for 60 s under voltage clamp and measured the start and end photocurrent for each cell (Supplementary Fig. 17d). These data were well fit by linear regression (eNpHR3.0 R2 = 0.68, eArch3.0 R2 = 0.88) with eArch3.0 having significantly higher slope (F1,36 = 22.2, P < 0.001), reflecting the fact that, for cells with similar onset photocurrents, eArch3.0-expressing cells had more photocurrent remaining at the end of the light pulse under these slice conditions, as seen in the illustrative traces and in contrast with the pattern of stability observed in vitro. The relationship between photocurrent (measured in voltage clamp) and hyperpolarization (measured in current clamp) is shown in Supplementary Figure 17e.
Finally, we compared the ability of eArch3.0 and eNpHR3.0 to inhibit spiking in current clamp. We elicited spiking with modestly suprathreshold current injections at 5 Hz, with 30 s baseline (pre-light), 60 s light and 30 s after light offset. Both pumps blocked spikes throughout the prolonged light stimulation (Fig. 6e). We observed that from both groups some cells became unstable after prolonged hyperpolarization especially by >50 mV, failing to spike to current injections or rebounding to a more depolarized resting potential after light offset. We quantified these factors for each cell and plotted each against the degree of hyperpolarization (Fig. 6f). Under more moderate (<50 mV) hyperpolarizations, we observed no consistent or lasting effects on excitability or membrane resistance.
DISCUSSION
Depending on experimental requirements, different properties may be critical in guiding optogenetic tool selection. For single-target optogenetic experiments (involving introduction of only one opsin gene and no additional light-activated elements), a relatively broad or nonselective spectrum is not problematic, and from a practical perspective broader wavelengths could enable more efficient activation with off-peak-wavelength light, thus relaxing the requirement for precisely tuned optical filters. All things being equal, optogenetic tools with greater response to redder light (such as the C1V1 family and eNpHR3.0) will enable control of deeper tissue (longer wavelengths will scatter less and more effectively penetrate tissue, and also these lower-energy photons may cause less phototoxicity)4. Future development of further red-shifted depolarizing and hyperpolarizing tools will be welcome.
For multiple-target optogenetic experiments (involving more than one opsin, or an opsin plus a light-sensitive element such as calcium or voltage sensors), both the value and the selectivity of the activation wavelength are critical. In targeting two distinct cell types for independent depolarization, C1V1 variants can be used in combination with blue-light-selective channelrhodopsins, as recently demonstrated11; for such experiments, lasers or narrower bandpass filters than used here are recommended. GR, ChETATC and ChETAT are less spectrally separated and therefore less useful for such combinatorial experiments. Because many available calcium or voltage sensors respond preferentially to blue light, C1V1 variants also present the possibility of optical manipulation of one cell type, with concurrent optical readout from the same or different cell types, in a single experimental preparation. These combinatorial possibilities will become increasingly practical as new indicators with different spectral sensitivities are developed and optimized43.
All proton pumps in our analysis (Arch, ArchT and Mac) were slightly blue-shifted relative to eNpHR3.0, but the still-large spectral overlap would preclude reliable independent hyperpolarization of multiple cell types in intact mammalian tissues. A preferentially blue light-activated hyperpolarizing tool has yet to be described but would be a powerful addition to the optogenetic toolbox. For combined hyperpolarization and depolarization, either in the same cells or in distinct populations in the same preparation, the relatively red-shifted spectrum of NpHR variants may be suitable for combination with blue light-selective depolarizing optogenetic tools.
For all optogenetic tools, photocurrent magnitude is a critical parameter determining whether light will successfully evoke or inhibit action potentials, which in turn depends on several other factors. First, there is a limit to the intensity of light that can safely be applied in vivo while avoiding phototoxic effects resulting in heating artifacts or tissue damage11. Second, light scatters as it passes through tissue, decreasing in power density at increasing distance from the light source; opsin-expressing tissue, if activated by a single light source, is therefore subjected to a light power density gradient4,37 as a function of distance from the source. Third, light sensitivity at the cell is crucial, and here we distinguish among three different perspectives on light sensitivity, which we term operational, population and intrinsic sensitivity.
Operational light sensitivity refers to the ability of the expressed opsins at a given light power density to exert an effect on the cell (for example, to evoke or inhibit spikes), which is largely determined by the absolute photocurrent magnitude. Many factors contribute to this photocurrent magnitude, including properties at the single protein level (see below), but also critically including the overall degree of opsin expression and the efficiency of membrane targeting. Operational light sensitivity is therefore the most practically useful number for the experimentalist but is also sensitive to many factors that alter expression and targeting (such as transgenic versus viral expression, viral type and titer, promoter, expression time and membrane trafficking), which will need to be tested in each experimental situation.
Although absolute opsin expression and membrane targeting are critical in determining operational photocurrent magnitude, it is also informative to consider the contribution of ‘normalized’ characteristics of the opsin population response to light. From the perspective of the whole cell, changes in such a ‘population light sensitivity’ (EPD50; normalized to control for influences of expression) could arise in part from differences in the duration of the conducting state of the opsin after photon absorption (off kinetics; Fig. 4). Both operational and population light sensitivity will be influenced by how efficiently each individual protein absorbs an incident photon; this constitutes ‘intrinsic light sensitivity’, a third measure given by the product of the molar absorption coefficient and the quantum yield of the molecule24. Given that EPD50 values are highly correlated with off kinetics, differences in off kinetics likely dominate population sensitivity, but differences in intrinsic light sensitivity may still come into play with additional molecular engineering or genomic discovery. Although we did not analyze the bistable SFOs11,15 here, they would be the most appropriate tool for recruiting very large volumes of tissue owing to extreme light sensitivity. The SFOs are also particularly useful for investigating the impact of asynchronous elevation of the firing rate of a defined neuronal population, such as the epochs of persistent activity observed in cortical and subcortical sequences of depolarizations44,45.
For depolarizing tools, researchers face a theoretical tradeoff between activating large or distant volumes of tissue and evoking spikes with high temporal precision or at high frequencies. Because ultimately the absolute (not normalized) photocurrent magnitude matters for performance, in practice one way to circumvent the tradeoff is by increasing the expression and membrane targeting of faster opsins, with the caveat that high expression may lead to toxicity. Other possible approaches for improving trafficking and photocurrents include separating the opsin from the fluorophore (note that we fused all of our opsins in frame with eYFP) or adding trafficking-enhancing sequences (as with C1V1 (ref. 11) and the hyperpolarizing opsins).
In addition to a critical role in determining population sensitivity, off kinetics (together with on kinetics) also will define the temporal precision of the photocurrent effects. For depolarizing channels, fast on kinetics will support short spike latencies; moreover, depolarizing channels with faster off kinetics will avoid problems resulting from prolonged depolarization, including arte-factual extra spikes and plateau potentials, both of which may degrade delivery of a precise neural signal. Just as different cell types respond differently to sustained injection of depolarizing current, different cell types will also respond differently to plateau potentials, another example of the interaction between biophysical properties of the optogenetic tool and the host cell in determining the ultimate response. With slower channelrhodopsins, which give rise to larger plateau potentials, pyramidal cells exhibit severe depolarization block at higher frequencies. On the most precise end of the spectrum, we identified ChETAA and ChIEF as perhaps best suited for eliciting rapid spike trains with high precision in fast-spiking cells, owing to kinetic stability across voltages.
Effective photocurrent size in response to pulsed light will be somewhere between the peak and steady-state photocurrent values, depending upon the interplay between the stimulation parameters and the conductance properties. Relevant properties include steady-state/peak ratio, kinetics of desensitization (τdes, the transition from peak to steady state), and kinetics of recovery from desensitization (the recovery of the peak photocurrent). Instability in photocurrent magnitude could manifest as lower temporal stationarity in spiking or higher spike-latency dispersion across pulse trains, reflecting inconsistent spike success or timing across a pulse train, respectively. Even recovery from desensitization alone is itself complex, with different (much faster) kinetics operating in the presence of continued illumination23, particularly relevant for high duty-cycle illumination. Computational modeling is needed to accurately predict photocurrent dynamics over a train for different optogenetic tools and stimulation paradigms, given the interactions among all of these properties. In principle, higher steady-state/peak ratios, slower desensitization and more rapid recovery from desensitization should all promote stable photocurrents across stimulation conditions.
For hyperpolarizing tools, it is important to note that the three hyperpolarizing opsins with largest photocurrents (eNpHR3.0, eArch3.0 and eArchT3.0) all have relatively high EPD50 values, so none may be well-suited for hyperpolarizing a large volume of tissue; to achieve sufficient inactivation of cells far from the light source may require excessive hyperpolarization that could be detrimental to health of the cells closest to the light source. Ideally, therefore, large-volume inhibition would be hypothetically achieved with a hyperpolarizing, photon-integrating version of the SFO, with moderate photocurrents and an extremely low EPD50 value, enabling more even photocurrent responses to the gradient of light intensity across tissue. As slow off kinetics are conceptually achievable with channels but not with pumps (which use energy provided by light to actively transport ions and therefore in theory should not be able to sustain photocurrents in the absence of light), a major open area for future opsin engineering efforts would be the development of a light-activated hyperpolarizing tool, such as a light-activated K+ channel.
All hyperpolarizing pumps we examined here showed suitably fast on kinetics for single-spike precision, with the proton pumps faster than eNpHR3.0, but notably Mac1.0 and eMac3.0 had markedly slower off kinetics than the other pumps. Depolarizing optogenetic tools (more so than hyperpolarizing tools) tend to be used with many different patterns of pulsed light stimulation: examples in the literature include 20-Hz stimulation of amygdala cells sustained over a 3-min behavioral trial37 and phasic 50 Hz bursts of light to stimulate dopaminergic cells in the ventral tegmental area29. But although hyperpolarizing tools may also be stimulated with brief light pulses, more typical useful paradigms for behavioral loss-of-function experiments involve sustained hyperpolarization across many seconds or even minutes; a recent effort relied upon sustained inhibition of hippocampal pyramidal cells for 30 min38.
Optogenetic photocurrents, such as physiological synaptic currents, are mediated by specific ions that can have effects distinct from membrane voltage. The depolarizing optogenetic tools are cation channels, conducting a mix of Na+, K+, H+ and Ca2+ (refs. 16,46). As Ca2+ is an important intracellular signaling molecule, Ca2+ conductance could have effects independent of membrane voltage which may either be an advantage or a confound depending on the experiment47,48; for example, it has been suggested that Ca2+-dependent K+ channels can be recruited downstream of the directly light-activated photocurrent16. It is also possible that large or prolonged optogenetic ion fluxes could alter extracellular fields and ion concentrations sufficiently to influence either the expressing cell or proximal non-expressing cells. Such effects can also happen in the course of physiological neural activity49 in the form of ephaptic interactions after high-frequency activity, or rebound excitation after prolonged inhibition owing to increased membrane excitability or chloride balance changes.
For many applications with hyperpolarizing opsins, sustained stimulation will be desired (for example, to mimic a lesion). We did not observe consistently increased excitability in the slice after eNpHR3.0 activity (Fig. 6), nor have in vivo studies observed prolonged effects outlasting light by more than ∼20 s35,38; nevertheless it is important to consider the potential for effects after light exposure when designing either physiological or optogenetic prolonged-inhibition experiments, and particularly for inhibition it is routine and advisable to focus experimental attention on the within-light, rather than post-light, epoch. Finally, although native amounts of high-concentration extracellular ions may be unlikely to be substantially depleted by intracellularly directed optogenetic pumps, we cannot exclude the possibility that large extracellularly directed fluxes of low-concentration ions could influence surrounding non-expressing cells.
All of the experiments described here involve illumination of cell bodies and do not directly address performance in axons11,36,37,50, important for applications such as projection targeting. The relationship between where depolarization or hyperpolarization is initiated and how that voltage change will propagate is complex and will depend on experiment-specific conditions (including distribution of opsins in the cell, location and intensity of light delivery, cell type, local host channels, axon caliber and myelination and other factors). Such subcellular factors could also come into play with spatially restricted two-photon illumination. Finally, many optogenetics experiments will be performed in dynamic systems with complex feedback and nonlinearities. At the network level, activation or inhibition of a population of connected cells will evoke circuit-level responses, and as many of these optogenetic tools are sensitive to membrane voltage, local activity can feed back onto and modulate properties of the expressed opsins in a time-varying fashion. Although we attempted to eliminate these considerations by patching in the presence of sodium channel or synaptic blockers, in vivo experiments will have no such constraints.
Our data can contribute to initial guiding principles in selection and characterization of opsins. As the optogenetic toolbox will expand, we suggest that future introduction of tools include side-by-side comparison of key parameters with relevant existing opsins, for maximal utility to the scientific community. With the many variables and tradeoffs among different figures of merit, it is unlikely that any given tool will prove superior by all measurable parameters. These data should therefore not be used to support the unqualified endorsement of one opsin over another but rather to facilitate informed decisions based on tradeoffs relevant to specific experimental requirements, with due regard for caveats arising from extrapolation across preparations.
ONLINE METHODS
Experiments with mice
All experiments were conducted under protocols approved by the Stanford Administrative Panel on Laboratory Animal Care. Male C57/BL6 mice (wild type or Pvalb::cre) were injected at 3–4 weeks of age and patched 4–6 weeks later. For each opsin comparison, all experiments were performed in parallel in littermates.
Molecular cloning
Lentiviral constructs contained BamHI between the promoter and the opsin gene, NotI between the opsin gene and the fluorophore gene, and EcoRI between the fluorophore gene and the WPRE. As an exception, pLenti-CaMKIIα-ChIEF-EYFP contained EcoRI between the opsin and the fluorophore. Wild-type ChR2 (from C. reinhardtii) and the mutated variants ChETAA, ChETAAR, ChETAT, ChETATR and ChIEF were also cloned in the reverse orientation into a Cre-inducible recombinant AAV vector carrying two pairs of incompatible lox sites28–30under the EF1α promoter (double-floxed inverted open reading frame (DIO)). Opsin-fluorophore fragmentswere PCR-amplified to add AscI and NheI, using gtggcgcgccctattacttgtacagctcgtccatg (for all), tatgctagccaccatggactatggcggcgc (for ChR2 mutants), and gttatg ctagcgccaccatgtcgcggaggccatggc (for ChIEF), and then ligated to an AAV-Ef1α-DIO backbone.
ChR2 was human codon—optimized (humanized). ChIEF (non-human codon–optimized (nonhumanized)) was obtained from M. Lin (Stanford University) in a Sindbis expression plasmid. GR and FR were constructed by fusing ChR1 (from C. reinhardtii) to ChR2 (neither were humanized) as described20,21. C1V1 was generated by fusing ChR1 (nonhumanized) with VChR1 (humanized) (GenBank ACD70142.1) by overlap extension PCR as previously described11. All ChR2 and C1V1 mutations were produced by site-directed mutagenesis (QuikChange, Agilent), as previously described13. Double mutants were made with combinations of the single-mutant primers.
Mac (from L. maculans) and Arch (from H. sodomense) were obtained from Addgene as GFP fusion genes and switched to eYFP for consistency. Humanized ArchT (from Halorubrum strain TP009) was synthesized by DNA2.0. Mac, Arch and ArchT were enhanced to the 2.0 versions using the ER export element alone and to the 3.0 versions with both the ER export motif and the trafficking signal as described previously33.
All constructs were fully sequenced, and all AAV vectors were tested for in vitro expression before viral production. All cloning and mutagenesis primers are listed in Supplementary Table 2. Complete sequences are available at http://www.optogenetics.org/.
Hippocampal neuron culture and calcium phosphate trans-fections
Primary cultured hippocampal neurons were prepared from P0 Sprague-Dawley rat pups (Charles River). CA1 and CA3 were isolated, digested with 0.4 mg ml−1 papain (Worthington), and plated onto glass coverslips precoated with 1:30 Matrigel (Becton Dickinson Labware). Cultures were maintained in a 5% CO2 humid incubator with Neurobasal-A medium (Invitrogen) containing 1.25% FBS (HyClone), 4% B-27 supplement (Gibco), 2 mM Glutamax (Gibco) and 2 mg ml−1 fluorodeoxyuridine (FUDR) (Sigma), and grown on coverslips in a 24-well plate at a density of 65,000 cells per well.
For each well, a DNA-CaCl2 mix was prepared with 2 µg DNA (Qiagen endotoxin-free preparation) and 1.875 µl 2 M CaCl2 (final Ca2+ concentration 250 mM) in 15 µl H2O. To DNA-CaCl2 we added 15 µl of 2× HEPES-buffered saline (pH 7.05). After 20 min at room temperature (20–22 °C), the mix was added dropwise into each well (from which the growth medium had been removed and replaced with prewarmed minimal essential medium (MEM)) and transfection proceeded for 45–60 min at 37 °C, after which each well was washed with 3 × 1 ml warm MEM before the original growth medium was returned.
Stereotactic injections
Adeno-associated virus (AAV) sero-type 2/5 containing the various constructs was produced by the University of Carolina Chapel Hill Vector Core. Genomic titers were 1.5 × 1012 c.f.u. ml−1 for ChETAA, ChETATR and ChIEF and 4 × 1012 c.f.u. ml−1 for eYFP, eNpHR3.0 and eArch3.0. One microliter of virus was stereotactically injected bilaterally into the medial prefrontal cortex of 3–4-week-old mice at +1.7 mm anteroposterior, 0.4 mm mediolateral and 2.5 mm dorsoventral (from bregma).
Whole-cell electrophysiology recordings
Recordings in cultured neurons were performed 4–6 d after transfection in Tyrode’s solution (320 mOsm): 125 mM NaCl, 2 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 30 mM glucose and 25 mM HEPES, titrated to pH 7.3–7.4 with NaOH. Tyrode was perfused at a rate of 1–2 ml min−1 and was kept at room temperature (20–22 °C). Intracellular solution (300 mOsm) contained 130 mM K-gluconate, 10 mM KCl, 10 mM HEPES, 10 mM EGTA and 2 mM MgCl2, titrated to pH 7.3 with KOH. Characterization of excitatory tools was done with bath-applied tetrodotoxin (TTX) (1 µM; Sigma-Aldrich) and intracellular QX-314 chloride (1 mM; Tocris Bioscience). In vitro patching of hyperpolarizing tools and current clamp recordings for depolarizing tools were performed in the presence of synaptic transmission blockers 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX; 10 µM; Sigma-Aldrich) and d(-)-2-amino-5-phosphonovaleric acid (APV; 25 µM, Sigma-Aldrich) as well as gabazine for the current clamp experiments (10 µM; Sigma-Aldrich). Recordings were performed on an upright Leica DM-LFSA microscope.
Recordings of ChETAA, ChETATR and ChIEF-expressing fast-spiking cells were performed in acute slices from Pvalb::cre transgenic mice, 6 weeks (ChETAA versus ChETATR comparison) or 4 weeks (ChETAA versus ChIEF comparison) after virus injections. Artificial cerebrospinal fluid (ACSF) (300 mOsm) was composed of 123 mM NaCl, 26 mM NaHCO3, 3 mM KCl, 1.25 mM NaH2PO4·H2O, 1 mM MgCl2·6H2O, 2 mM CaCl2·2H2O and 11 mM glucose. ACSF was bubbled with 95% O2 and 5% CO2 (Praxair) to an equilibrium pH of 7.3. ACSF was perfused at a rate of 7 ml min−1 and heated to 32 °C. The intracellular solution was adjusted to 280 mOsm using water. Fast-spiking cells were identified by eYFP expression and characteristic electrophysiological properties. Recordings were performed on an upright Leica DM-LFSA microscope.
Recordings of eYFP-, eNpHR3.0- and eArch3.0-expressing pyramidal cells were performed in acute slices from wild-type C57BL/6 mice 6–7 weeks after virus injection. ACSF contained CNQX, APV and gabazine. Intracellular solution (280 mOsm) contained 135 mM K-gluconate, 5 mM KCl, 10 mM HEPES, 0.1 mM EGTA, 2 mM MgCl2, 2 mM Mg-ATP and 0.2 mM Na2-GTP, titrated to pH 7.4 with KOH. Pyramidal cells were identified by morphology and characteristic electrophysiological properties. Recordings were performed on an upright Olympus BX51 microscope. For all patching experiments, borosilicate glass (Sutter Instruments) pipette resistances were 3–6 MΩ. For cell-attached electrophysiology recordings, upon obtaining GΩ seals, holding potential was set so that no net current flowed across the membrane; the same stimulation protocols were used as for whole-cell spiking experiments. After the cell-attached recording was performed, we applied suction to the pipette to break into the cell and repeated the same experiments in whole-cell configuration to provide a direct within-cell comparison. No exogenous retinal cofactor was added to neurons in any preparation.
Data collection across opsins was randomized and distributed to minimize across-group differences in expression time, room temperature and so on.
Light delivery
All experiments were performed using single-photon activation. For cultured neurons, light was emitted from a 300 W DG-4 lamp (Sutter Instruments) and was delivered through a 40×, 0.8 numerical aperture (NA) water-immersion objective. Pulsed input signals were delivered to the DG-4 from pClamp (Axon Instruments) via a Bayonet Neill-Concelman (BNC) connection. The delay from the DG-4 trigger signal to full light output was measured using an amplified photodetector (Thorlabs) as ∼1 ms, with a 200 µs rise time. All measurements of time to peak and latency were corrected for this delay.
For light-sensitivity measurements, light was passed through a 470/40 nm filter (for blue-light sensitive excitatory tools) or a 562/40 nm filter (for C1V1s and all inhibitory tools), and then through neutral density (ND) filters to achieve power densities ranging from ∼0.1 to 20 mW mm−2. Other properties were studied at ∼5 mW mm−2. For these experiments, the light was passed through a Lambda 10-3 filter wheel (Sutter Instruments) with a 10-position wheel for filters of different wavelengths, ND-normalized to generate closely matched power densities. Filters were 406/15 nm; 427/20 nm; 445/20 nm; 470/20 nm; 494/20 nm; 520/15 nm; 542/20 nm; 560/25 nm; and 590/20 nm. Inhibitory spectra also used a 607/45 filter. Functional performance of depolarizing tools in culture used a 470/40 nm filter (for blue light–sensitive excitatory tools) or a 562/40 nm filter (for C1V1s) and then ND filters to achieve power densities of 2 mW mm−2, 6 mW mm−2 and 20 mW mm−2.
For experiments investigating fast depolarizing tools in slice, light was emitted from the same 300 W DG-4 lamp (Sutter Instruments) and delivered through a 40×, 0.8 NA water-immersion objective. Light was passed through a 470/40 nm filter and adjusted to achieve a light power density of 5.1 mW mm−2. For experiments investigating hyperpolarizing tools in slice, a 40×, 0.8 NA LUMPlanFL/IR Objective (Olympus), XCite halogen light source (EXPO) was used. Light was passed through a 589/15 nm filter (eNpHR3.0) or a 560/14 filter (eArch3.0). For experiments comparing the photocurrent and hyperpolariza-tion magnitudes under matched conditions, light power density was adjusted to ∼5 mW mm−2. For the remaining experiments light was adjusted across a range of light power densities (5–10 mW mm−2 for eNpHR3.0; 0.25–5 mW mm−2 for eArch3.0) to achieve a comparable range of photocurrents for both.
All experiments contained at least 30 s of dark between sweeps to allow recovery to baseline. All filters are given here as wavelength in nanometers per bandwidth in nanometers. All light power densities were measured coming out of the 40× objective, at approximately the sample distance.
Data analysis
Analyses of physiological results were performed using ClampFit software (Axon Instruments) or custom software written in Matlab (Mathworks).
Access resistance (Ra) and input resistance (Rin) were monitored continually and data were only included when Ra was < 30 MΩ and Rin was > 90 MΩ. Any traces containing escaped spikes were excluded from analyses of peak photocurrent or of kinetics, but steady-state photocurrents were still measured when possible. For current clamp recordings in culture, only cells that fit those criteria and had leak currents > −150 pA (holding at −65 mV) were included for analysis. For current clamp recordings in acute slice, only cells that fit those criteria and had resting potentials < −55 mV were included for analysis. Only initially reliable cells that showed 0.9–1.1 spikes per current pulse in the pre-light baseline were analyzed for Figure 6f.
To identify the peak photocurrent, traces were smoothed using the robust Loess method with a filter width of 2 ms and the peak was defined as the extremum from laser onset to 200 ms post laser onset, less the baseline current (from the average over 500 ms before laser onset). Visual inspection ensured that no escape spikes or other anomalies occurred. Time to peak was measured from laser onset to this marked peak time. The steady-state photo-current was determined by fitting a monoexponential curve to the smoothed waveform from 2 ms after the peak to the laser offset time. Steady-state current was taken from the parameters of this fit. τoff and τdes were calculated using ClampFit. We first smoothed the trace using a lowpass Gaussian filter with a −3 dB cutoff at 1,000 Hz, and then fit a monoexponential curve to the smoothed waveform. All curves were visually inspected for goodness of fit.
Photocurrent properties of the depolarizing tools ChR2, ChETAA and ChIEF were characterized in vitro using both the lentiviral and the AAV constructs. For parameters that depend on single-molecule properties (steady-state/peak ratio, action spectrum, light sensitivity and kinetics), values were pooled across experiments after confirming that datasets were not statistically different. Photocurrent properties of the hyperpolarizing tools were assessed in two separate rounds of experiments. eNpHR3.0 photocurrent magnitudes were statistically different between the two datasets, so we only combined datasets when considering normalized values or intrinsic single-molecule properties (action spectrum, light sensitivity and kinetics) after confirming that eNpHR3.0 performed similarly across datasets.
Whole-cell spikes were defined as rising above a high threshold (−20 mV for the comparison of fast depolarizing tools in slice; 0 mV for all other comparisons) and then dropping below a low threshold (−30 mV). Subsequent spikes that occurred within 2 ms of a prior spike were ignored. To detect spikes elicited by light, we defined a window of time from 1ms to 50 ms after the pulse onset. Above 20 Hz, this window was truncated to 1 ms after the current pulse onset to 1 ms after the subsequent pulse onset. The window around the last light pulse was truncated to the same duration. Cell-attached spikes were identified using the threshold function in ClampFit. Very small, broad events were not included as spikes. Where the spike data was ambiguous, the trace was inspected manually. For each whole-cell pulse train we calculated the proportion of light pulses that elicited ≥1 spike (pulse efficacy) and that elicited > 1 spike (multiple spike likelihood).
Plateau potentials were defined as the offset of the spike waveform from the baseline. For the depolarizing tools in vitro, all cells that fired ≥ 1 spike were included for analysis. For the fast-spiking cells in slice, only traces that had 100% pulse efficacy were included for analysis. Temporal stationarity, the extent to which spiking is sustained at the same reliability over time, was calculated by dividing the light pulses into quartiles and computing the pulse efficacy each quartile. Latency and latency spread across pulse trains were determined as follows: for each light pulse, we measured the time delta from the light pulse onset to the spike time. Latency is the average of these time deltas, and latency spread is the s.d. of these time deltas. Note that latency spread therefore is a measure of how variable the latencies are in each cell, whereas the error bars on latency are the standard error of mean latencies across cells. Traces in which the cell fired < 5 action potentials were excluded from analysis.
Statistical analysis
All statistical analysis was performed using GraphPad Prism version 5.04 for Windows (GraphPad Software). For two-sample comparisons of a single variable (such as kinetics of ChETAA versus ChIEF in slice), we first tested whether the data followed a Gaussian distribution (Shapiro-Wilk normality test). If the data were detectably non-Gaussian, we performed a non-parametric Mann-Whitney test. If the data well-approximated a Gaussian, we performed an independent, two-sample t-test (equal variance). In the case of unequal variance (determined by an F test), we applied Welch’s correction. All tests were two-tailed with confidence levels of 95%.
For multiway comparisons of a single variable (such as kinetics) we first tested whether the data followed a Gaussian distribution (Shapiro-Wilk normality test). In cases in which distributions were detectably non-Gaussian, we used a square root transformation to stabilize the variance and make the data approximately normal and then compared all data against one specified ‘control’, correcting for family-wise error using Dunnett’s test. If the transformed data were still non-Gaussian, we used the nonparametric Dunn’s test. In all cases, we maintained overall significance levels of alpha = 0.05 (95% confidence interval). Comparisons between larger numbers of tools will therefore have a more conservative alpha (more stringent requirement for significance). This may also result in different significance values assigned to the same comparison, depending on how many comparisons are being performed in parallel. In particular, as some of the same ChR2 and ChETAA data were included in two comparisons, discrepancies in reported significance values can be attributed to the context of the comparison.
For comparisons across multiple variables (such as spiking performance across frequencies), we performed two-way ANOVAs followed by post-tests between pairs or against a specified ‘control’. We used a conservative Bonferroni’s correction to control the false positive rate. To test the relationship between two properties (such as τoff versus EPD50), we performed a nonparametric, two-tailed Spearman correlation with a confidence level of 95%. To estimate the slope, we performed a least-squares regression (either linear or linear on log-log transformed data), minimizing relative distance squared (1/Y2).
To test the dependency of a property on an experimental condition (for example, photocurrent versus light power density), we performed regressions as follows. First, for analysis of time to peak versus light power density, we performed linear regression on log-log transformed data and we compare whether, for each, the best-fit slope differs significantly from 0. Second, for analysis of recovery from desensitization, we used a nonlinear regression to fit the mean photocurrent recovery data with a two-phase association curve, constraining Y0 = 0 and plateau = 1. This fit was used to generate the curves and the R-squared values. In a separate analysis, we fit the data for each individual cell, to calculate the time required for 50% recovery. Third, for analysis of light sensitivities, we fit the raw population means with a one-site specific binding curve: Y = Bmax × X/(Kd + X). In a separate analysis, we normalized the photocurrents for each cell, and plotted the population means and standard errors for each tool. This population data was fit the same way to generate the curves and the R2 values. For each individual cell, we obtained an equilibrium binding constant (Kd), which we refer to as EPD50 (50% effective light power density).
Population significance thresholds were always set at P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) for the entire family of comparisons. All data in graphs are shown as mean ± s.e.m.
Immunohistochemistry
Six or four weeks after injection, mice were perfused transcardially with PBS followed by 4% paraformaldehyde (PFA). After an overnight post-fix in PFA, brains were equilibrated in 30% sucrose in PBS for at least 24 h. We obtained 40-µm sections using a frozen microtome, DAPI-stained them (1:50,000), and ‘cover-slipped’ them with Polyvinyl Alcohol mounting medium containing the antifading agent 1,4-diazabicyclo[2.2.2]octane (PVA-DABCO) (Sigma-Aldrich). Transfected primary hippocampal cultures were fixed for 15 min with 4% PFA. For staining with KDEL, cultures were then permeabilized for 30 min with 0.4% saponin in 2% normal donkey serum (NDS). Primary antibody incubations were performed overnight at 4 °C using a monoclonal antibody marking endogenous ER-resident proteins containing the KDEL retention signal (KDEL 1:200, Abcam). Secondary antibodies (Jackson ImmunoResearch) were applied in 2% NDS for 1 h at room temperature.
Equipment and settings
All images were obtained on a Leica confocal microscope (DM600B) as 1,024 × 1,024 pixel resolution (pixel dimensions were 3.03 µm2). Images were acquired using the following objectives: 10×, 0.40 NA (air), 40×, 1.25 NA (oil) and 63×, 1.40 NA (oil). Excitation and emission wavelengths were as follows: eYFP in Figure 5b, 514 nm/512–600 nm (excitation/ emission); eYFP for all other figures, 488 nm/500–545 nm; GFP, 488 nm/500–600 nm; Cy5, 633 nm/650–750 nm. For the following figures we used line averaging: Figure 3e,h (across two lines), Figure 5b (across four lines), Figure 6a (across three lines), Supplementary Figure 15a (across four lines). Consistent settings were used for all images in each given figure panel. The brightness and contrast of all eYFP images for Figure 5b were uniformly and identically modified in Photoshop (Adobe). All other images were unprocessed after acquisition.
Quantification of fluorescence in transfected cells
Fluorescence images were acquired from the same cells that were patched to enable quantification of expression levels and photocurrent/ fluorescence relationships. Images were acquired with Metamorph, maintaining constant settings, and processed offline using ImageJ. Hand-drawn regions of interest encompassed the soma and proximal dendrites.
Supplementary Material
ACknowledgments
We thank A. Andalman, T. Davidson, I. Diester, S. Evans, I. Goshen, D. Mattis, I. Witten, L. Grosenick, S.-Y. Kim and C. Perry for helpful discussions, M. Lin (Stanford University) for ChIEF clones, and all memebers of the Deisseroth laboratory for their support. All viruses were packaged at University of North Carolina Vector Core. Supported by Bio-X and the Stanford Medical Scientist Training Program (J.M.), the US National Institute of Mental Health (1F32MH088010-01, K.M.T.), and the International Fulbright Science and Technology Award and a Stanford Graduate Fellowship (E.A.F.). K.D. is supported by National Institute of Mental Health, National Institute on Drug Abuse, National Institute of Neurological Disorders and Stroke, Howard Hughes Medical Institute, the Defense Advanced Research Projects Agency Reorganization and Plasticity to Accelerate Injury Recovery Program, the Keck Foundation, the McKnight Foundation and the Gatsby Charitable Foundation.
Footnotes
Note: Supplementary information is available on the Nature Methods website.
Author ContriButions
J.M., K.M.T., E.A.F., C.R., R.P., O.Y. and K.D. contributed to study design and data interpretation. J.M. coordinated all experiments and data analysis. J.M., K.M.T., E.A.F., D.J.O., R.P. and L.E.F. contributed to acquisition of electrophysiological data. C.R. cloned all constructs, cultured primary neurons, performed transfections and managed viral packaging processes. D.J.O. wrote custom analysis scripts and analyzed all electrophysiological data. M.H. contributed to data analysis. J.M., K.M.T., C.R., L.A.G. and V.G. contributed to the histological processing and fluorescence imaging. K.D. supervised all aspects of the work. J.M., K.M.T., E.A.F. and K.D. wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Deisseroth K. Optogenetics. Nat. Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deisseroth K. Controlling the brain with light. Sci. Am. 2010;303:48–55. doi: 10.1038/scientificamerican1110-48. [DOI] [PubMed] [Google Scholar]
- 3.Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu. Rev. Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Wang C, Kane MA, Napoli JL. Multiple retinol and retinal dehydrogenases catalyze all-trans-retinoic acid biosynthesis in astrocytes. J. Biol. Chem. 2011;286:6542–6553. doi: 10.1074/jbc.M110.198382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 7.Li X, et al. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. USA. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bi A, et al. Ectopic expression of a microbial-type rhodopsin restores visual responses in mice with photoreceptor degeneration. Neuron. 2006;50:23–33. doi: 10.1016/j.neuron.2006.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishizuka T, Kakuda M, Araki R, Yawo H. Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci. Res. 2006;54:85–94. doi: 10.1016/j.neures.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Nagel G, et al. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr. Biol. 2005;15:2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 11.Yizhar O, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gradinaru V, et al. Targeting and readout strategies for fast optical neural control in vitro and in vivo . J. Neurosci. 2007;27:14231–14238. doi: 10.1523/JNEUROSCI.3578-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunaydin LA, et al. Ultrafast optogenetic control. Nat Neurosci. 2010;13:387–392. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- 14.Berndt A, et al. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc. Natl. Acad. Sci. USA. 2011;108:7595–7600. doi: 10.1073/pnas.1017210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat. Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 16.Kleinlogel S, et al. Ultra light-sensitive and fast neuronal activation with the Ca(2+)-permeable channelrhodopsin CatCh. Nat Neurosci. 2011;14:513–518. doi: 10.1038/nn.2776. [DOI] [PubMed] [Google Scholar]
- 17.Zhang F, et al. Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri . Nat. Neurosci. 2008;11:631–633. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Govorunova EG, Spudich EN, Lane CE, Sineshchekov OA, Spudich JL. New channelrhodopsin with a red-shifted spectrum and rapid kinetics from Mesostigma viride . MBio. 2011;2:e00115–11. doi: 10.1128/mBio.00115-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin JY, Lin MZ, Steinbach P, Tsien RY. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys. J. 2009;96:1803–1814. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, et al. Molecular determinants differentiating photocurrent properties of two channelrhodopsins from chlamydomonas. J. Biol. Chem. 2009;284:5685–5696. doi: 10.1074/jbc.M807632200. [DOI] [PubMed] [Google Scholar]
- 21.Wen L, et al. Opto-current-clamp actuation of cortical neurons using a strategically designed channelrhodopsin. PLoS ONE. 2010;5:e12893. doi: 10.1371/journal.pone.0012893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stehfest K, Hegemann P. Evolution of the channelrhodopsin photocycle model. ChemPhysChem. 2010;11:1120–1126. doi: 10.1002/cphc.200900980. [DOI] [PubMed] [Google Scholar]
- 23.Bamann C, Kirsch T, Nagel G, Bamberg E. Spectral characteristics of the photocycle of channelrhodopsin-2 and its implication for channel function. J. Mol. Biol. 2008;375:686–694. doi: 10.1016/j.jmb.2007.10.072. [DOI] [PubMed] [Google Scholar]
- 24.Sugiyama Y, et al. Photocurrent attenuation by a single polar-to-nonpolar point mutation of channelrhodopsin-2. Photochem. Photobiol. Sci. 2009;8:328–336. doi: 10.1039/b815762f. [DOI] [PubMed] [Google Scholar]
- 25.Hedrick T, Waters TH. Spiking patterns of neocortical L5 pyramidal neurons in vitro change with temperature. Front Cell Neurosci. 2011;5:1. doi: 10.3389/fncel.2011.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin JY. A user’s guide to channelrhodopsin variants: features, limitations and future developments. Exp. Physiol. 2010;96:19–25. doi: 10.1113/expphysiol.2009.051961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cardin JA, et al. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 2009;459:663–667. doi: 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai HC, et al. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science. 2009;324:1080–1084. doi: 10.1126/science.1168878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atasoy D, Aponte Y, Su HH, Sternson SMA. FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J. Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chater TE, Henley JM, Brown JT, Randall AD. Voltage- and temperature-dependent gating of heterologously expressed channelrhodopsin-2. J. Neurosci. Methods. 2010;193:7–13. doi: 10.1016/j.jneumeth.2010.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 33.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PLoS ONE. 2007;2:e299. doi: 10.1371/journal.pone.0000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Witten IB, et al. Cholinergic interneurons control local circuit activity and cocaine conditioning. Science. 2010;330:1677–1681. doi: 10.1126/science.1193771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stuber GD, et al. Excitatory transmission from the amygdala to nucleus accumbens facilitates reward seeking. Nature. 2011;475:377–380. doi: 10.1038/nature10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tye KM, et al. Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature. 2011;471:358–362. doi: 10.1038/nature09820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goshen I, et al. Dynamics of retrieval strategies for remote memories. Cell. 2011;147:678–689. doi: 10.1016/j.cell.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 39.Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 2008;36:129–139. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chow BY, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han X, et al. A high-light sensitivity optical neural silencer: development and application to optogenetic control of non-human primate cortex. Front. Syst. Neurosci. 2011;5:18. doi: 10.3389/fnsys.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmermann D, et al. Effects on capacitance by overexpression of membrane proteins. Biochem. Biophys. Res. Commun. 2008;369:1022–1026. doi: 10.1016/j.bbrc.2008.02.153. [DOI] [PubMed] [Google Scholar]
- 43.Zhao Y, et al. An expanded palette of genetically encoded Ca(2+) indicators. Science. 2011;333:1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goto Y, O’Donnell P. Network synchrony in the nucleus accumbens in vivo . J. Neurosci. 2001;21:4498–4504. doi: 10.1523/JNEUROSCI.21-12-04498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez-Vives MV, McCormick DA. Cellular and network mechanisms of rhythmic recurrent activity in neocortex. Nat Neurosci. 2000;3:1027–1034. doi: 10.1038/79848. [DOI] [PubMed] [Google Scholar]
- 46.Nagel G, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goold CP, Nicoll RA. Single-cell optogenetic excitation drives homeostatic synaptic depression. Neuron. 2010;68:512–528. doi: 10.1016/j.neuron.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lindsay TH, Thiele TR. Lockery, S.R. Optogenetic analysis of synaptic transmission in the central nervous system of the nematode Caenorhabditis elegans . Nat Commun. 2011;2:306. doi: 10.1038/ncomms1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor CP, Dudek FE. Synchronous neural afterdischarges in rat hippocampal slices without active chemical synapses. Science. 1982;218:810–812. doi: 10.1126/science.7134978. [DOI] [PubMed] [Google Scholar]
- 50.Ren J, et al. Habenula “cholinergic” neurons co-release glutamate and acetylcholine and activate postsynaptic neurons via distinct transmission modes. Neuron. 2011;59:445–452. doi: 10.1016/j.neuron.2010.12.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.