Abstract
Biotic interactions can improve agricultural productivity without costly and environmentally challenging inputs. Micromonospora strains have recently been reported as natural endophytes of legume nodules but their significance for plant development and productivity has not yet been established. The aim of this study was to determine the diversity and function of Micromonospora isolated from Medicago sativa root nodules. Micromonospora-like strains from field alfalfa nodules were characterized by BOX-PCR fingerprinting and 16S rRNA gene sequencing. The ecological role of the interaction of the 15 selected representative Micromonospora strains was tested in M. sativa. Nodulation, plant growth and nutrition parameters were analyzed. Alfalfa nodules naturally contain abundant and highly diverse populations of Micromonospora, both at the intra- and at interspecific level. Selected Micromonospora isolates significantly increase the nodulation of alfalfa by Ensifer meliloti 1021 and also the efficiency of the plant for nitrogen nutrition. Moreover, they promote aerial growth, the shoot-to-root ratio, and raise the level of essential nutrients. Our results indicate that Micromonospora acts as a Rhizobia Helper Bacteria (RHB) agent and has probiotic effects, promoting plant growth and increasing nutrition efficiency. Its ecological role, biotechnological potential and advantages as a plant probiotic bacterium (PPB) are also discussed.
Nodules are new organs generated mainly in roots of leguminous plants, in cooperation with alpha and beta proteobacteria developed for biological nitrogen fixation. It was initially thought that only symbiotic nitrogen-fixing bacteria could exist inside healthy N2 fixing nodules. Recent studies have shown that they are frequently populated by a broad and heterogeneous range of both gram-positive and gram-negative bacteria1,2,3,4. Recently, the first intranodular actinobacteria have been described5,6, but from the first description in this environment, the number of actinomycetes found has increased and in fact even new species have been described. Examples of these new findings inside nodules are Curtobacterium in Trifolium and Ornithopus7,8; Microbacterium in Acacia, Glycyrrhiza, Medicago and Ornithopus7,8,9,10,11; Micromonospora in several legumes12,13,14,15,16; Streptomyces in Sphaerophisa17 and others. Notably Micromonospora, which has been isolated from more than 20 different widely distributed plant species seem to have good potential as a plant-probiotic bacteria (PPB), although this remains to be studied in depth.
At our laboratory, strains of Micromonospora have been isolated from healthy plant nodules in a variety of genera of leguminous plants including M. sativa (alfalfa)12,13,15,16. Alfalfa is one of the most widely adapted agronomic crops and a cheap source of protein-rich forage with high digestibility, which is a valuable trait in economical animal husbandry. Alfalfa should be considered a key component of sustainable agricultural systems for the future because of its high yield, nutritional quality, pest resistance, and its value in soil conservation and improvement18.
One of the major challenges for the twenty-first century will be sustainable crop production. Agricultural practices derived from the green revolution, defined by the use of pesticides, fertilizers and herbicides of chemical origin, together with the genetic improvement of plant germoplasm, produced an increase in agricultural productivity. Decades ago, the cost and risks derived of this kind of agriculture were elucidated and as a consequence19,20 a new agricultural revolution is now starting to develop in which probiotic microorganisms have become an alternative to chemicals21. The possibilities for influencing plant growth-promoting potential applying microorganisms as Plant Probiotic Bacteria (PPB) agents have been largely explored22,23,24,25. The interest of these microorganisms is clear, and today inoculants can be found on the market in several countries. Based on recent surveys, interest in the use of inoculants is also rising, suggesting that the market potential of bioinoculants will increase further in coming years26.
However, it is necessary to study their ecological role and make an adequate analysis, evaluation and selection of the microbial strains used in order to obtain the desired effect and, unfortunately, the beneficial plant-microbe interaction has often been ignored in breeding strategies, even after their importance in soil ecosystems was confirmed (reviewed by Smith and Goodman27).
In light of the foregoing, the main goal of this study was to determine the diversity and ecological function of Micromonospora and analyze its plant probiotic capabilities since there is little information about it even though its biotechnological potential and also its impact in this new agricultural revolution are relevant.
Results
Bacterial isolation and morphological characterization
Micromonospora-like colonies were isolated from surface sterilized root nodules of naturally occurring alfalfa plants on yeast mannitol agar, along with rhizobia-like bacteria after 3-week incubation at 28°C. Micromonospora strains were recovered in almost all of the nodules sampled. In all, 66 strains were isolated from the sampling sites: Aldearrubia (AL) 21 strains, Babilafuente (ALFb) 11 strains, Palaciosrubios (ALFpr) 19 strians, San José (ALFr) 4 strains and Tormes riverbank (ALF) 11 strains. All 66 actinobacterial strains had the morphology described for the genus Micromonospora; they were Gram+, filamentous, lacked aerial mycelium, and presented orange or brown colonies that darkened after around 3 weeks due to sporulation.
Genetic diversity of the Micromonospora-like isolates
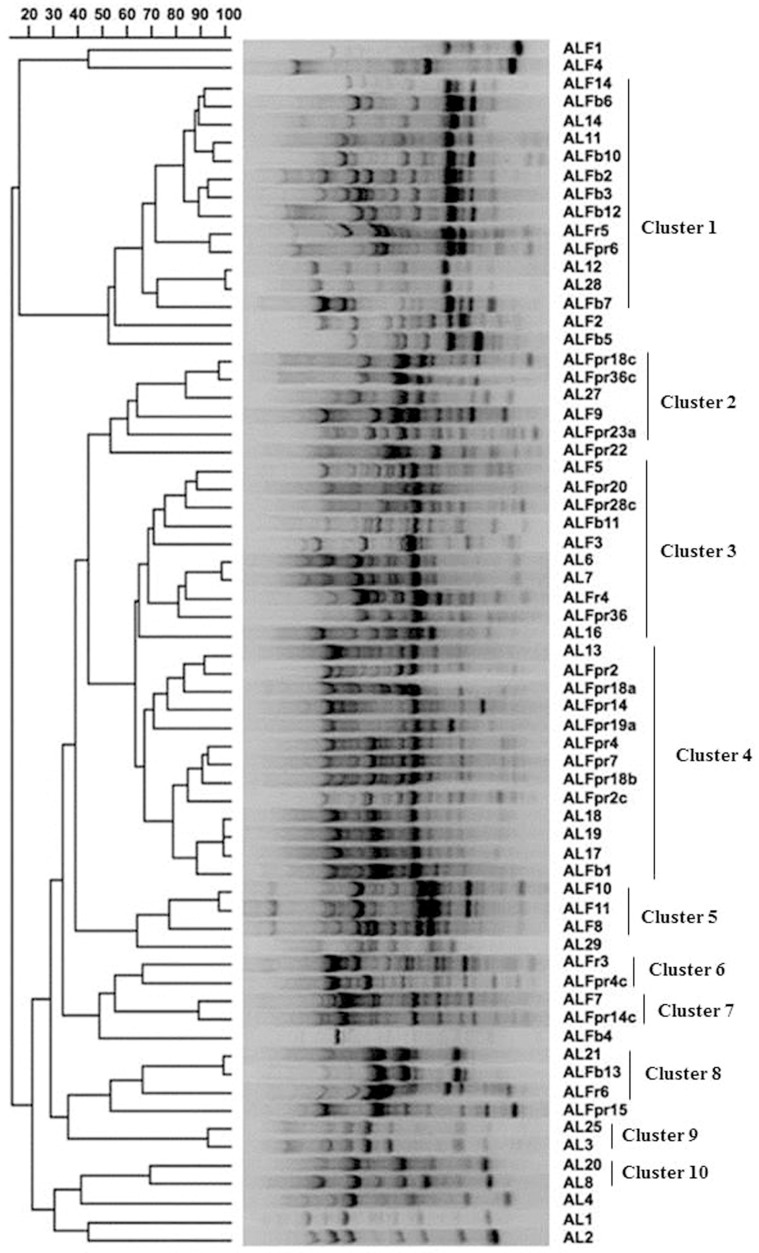
High-resolution BOX-PCR fingerprints were obtained for the 66 actinomycetes isolated from the nitrogen-fixing root nodules of M. sativa (Figure 1). The amplified fragments ranged from 0.1 to 2.2 kb. Clusters based on the similarity matrix generated with Pearson's coefficient and the UPGMA algorithm were defined at the 60% similarity level, affording 10 groups and revealing the high genetic diversity of the isolates. Figure 1 shows the diversity of the genetic profiles of the strains studied. Fifty-five strains were distributed in 10 clusters containing 2–13 strains; the remaining 11 isolates had a unique profile. No clones were found even in the strains from the same nodule. With respect to the isolation site, the strains isolated from Aldearubia (21 strains) and the 19 strains recovered from Palaciosrubios were distributed along the entire dendrogram, they have representatives in almost every cluster; the 11 strains from Babilafuente were detected in 6 groups; the 11 strains from Tormes River bank in 8 groups and the 4 strains from San José in 4 groups. Two groups contained strains from the five different sampling sites (cluster 1 and 3). Clusters 9 and 10 (2 strains each) only contain strains from Aldearrubia, the rest of the clusters contained strains from 2 or more of the locations sampled.
Figure 1. Dendrogram showing genetic relatedness of 66 Micromonospora strains isolated from M. sativa determined by analysis of BOX–PCR fingerprints using the Pearson's coefficient and UPGMA cluster methods.
According to the genetic diversity (BOX-PCR fingerprinting) and geographical origin of the isolates, we selected fifteen strains for in planta interaction studies.
Phylogenetic analysis and functional characterization of selected Micromonospora strains
Nearly complete 16S rRNA gene sequences (≥1434 nt) were obtained for the fifteen selected strains. NCBI and Eztaxon nucleotide blast searches revealed that 100% of the sequenced microorganisms were identified as belonging to the genus Micromonospora as suggested by their morphological characteristics.
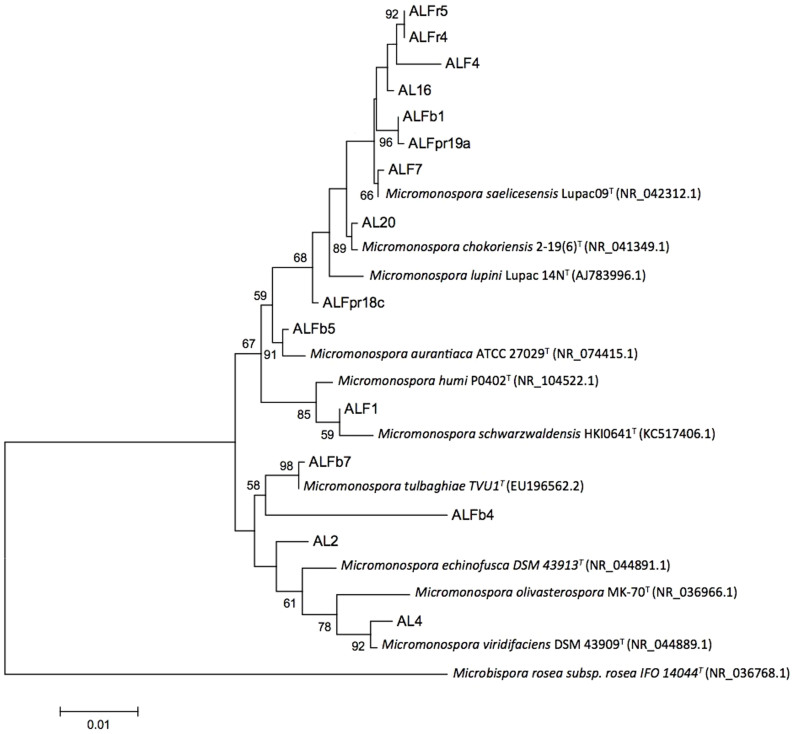
Sequence similarities between the new isolates and currently described Micromonospora species ranged from 97.78 to 100%. A significant number of the isolates sequenced (approx. 87%) showed > 99% sequence similarity with already described Micromonospora species (Table 1). The inferred phylogenetic tree based on 16S rRNA gene sequences using maximum likelihood (Figure 2) and neighbour-joining methods (Figure S1) showed that six of the isolates clustered with already described Micromonospora species. The tree topology generated by both maximum likelihood and neighbour joining methods was strongly supported by bootstrap values which were similar for both methods. However, nine of the strains did not group closely with any of the currently recognized species (AL2, ALFb4, ALFpr18c, ALFpr19a, ALFb1, AL16, ALF4, ALFr4 and ALFr5; Figure 2). Further taxonomic work will be required to elucidate the status of these last strains.
Table 1. Geographical origin and 16S rRNA gene sequence analysis of strains selected for in planta trials.
| Strain | Origin | # Accession | Most similar Micromonospora type strain. (Accession number) | Similarity (%) | Source |
|---|---|---|---|---|---|
| AL2 | Aldearrubia | KF876220 | M. chaiyaphumensis MC5-1 (AB196710) | 99.72 | This work |
| AL4 | Aldearrubia | KF876221 | M. viridifaciens DSM 43909T (X92623) | 99.52 | This work |
| AL16 | Aldearrubia | KF876222 | M. saelicesensis Lupac 09 (AJ783993) | 99.65 | This work |
| AL20 | Aldearrubia | KF876223 | M. chokoriensis 2-19/6 (AB241454) | 99.79 | This work |
| ALF1 | Tormes riverbank | KF876224 | M. humi P0402 (GU459068) | 99.51 | This work |
| ALF4 | Tormes riverbank | KF876225 | M. coxensis 2-30-b/28 (AB241455) | 99.31 | This work |
| ALF7 | Tormes riverbank | KF876233 | M. saelicesensis Lupac 09 (AJ783993) | 99.86 | This work |
| ALFb5 | Babilafuente | KF876226 | M. aurantiaca ATCC 27029 (CP002162) | 99.72 | [37] |
| ALFb7 | Babilafuente | KF876227 | M. tulbaghiae TVU1 (EU196562) | 99.93 | This work |
| ALFb1 | Babilafuente | KF876228 | M. saelicesensis Lupac 09 (AJ783993) | 99.58 | This work |
| ALFb4 | Babilafuente | KF876229 | M. echinospora ATCC 15837 (U58532) | 97.78 | This work |
| ALFpr18c | Palaciosrubios | KF876230 | M. lupini Lupac 14N (AJ783996) | 99.31 | [37] |
| ALFpr19a | Palaciosrubios | KF876231 | M. saelicesensis Lupac 09 (AJ783993) | 99.51 | This work |
| ALFr5 | San José | KF876232 | M. cremea CR30 (FN658654) | 98.62 | This work |
| ALFr4 | San José | KF876234 | M. saelicesensis Lupac 09 (AJ783993) | 99.51 | This work |
Figure 2. Maximum likelihood phylogenetic tree based on 16S rRNA gene sequences showing the relationship between the Micromonospora isolates and the closest recognized Micromonospora species.
Bar, 0.01 substitutions per nucleotide position. Bootstrap percentages (1000 replicates) above 50% are shown at nodes.
With the exception of strain AL2 lacking pectinase activity, all of the other Micromonospora strains showed the ability to degrade plant cell wall components, namely cellulose, pectin and xylan (Table 2). Even though, the cellulose activity was weak in all the strains. Other components of organic matter such as proteins (caseinase and gelatinase activities) and starch were also degraded by all the strains, being the only exception the strain ALF1, which could not degrade gelatine. Moreover, all of the tested strains showed lipase activity. They were able to degrade Tween 80. Tween 20 was strongly degraded by two strains (ALFr5 and ALFb7), weakly by nine and no hydrolytic activity was detected in four of them. Neutral and alkaline phosphatase activities were detected in all the strains but none showed acid phosphatase activity (Table 2).
Table 2. Ecological, PPB related enzimatic activities and indolacetic acid production in selected Micromonospora strains.
| Strains/Activity | AL2 | AL4 | AL16 | AL20 | ALFb1 | ALFb4 | ALFb5 | AL b7 | ALF1 | ALF4 | ALF7 | ALFpr18c | ALFpr19a | ALFr4 | ALFr5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cellulase | w | w | w | w | w | w | w | w | w | w | w | w | w | w | w |
| Xilanase | w | w | + | + | + | + | + | + | + | w | + | + | + | + | + |
| Pectinase | - | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Caseinase | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Gelatinase | + | + | + | + | + | + | + | + | - | + | + | + | + | + | + |
| Amilase | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Phosphatase (Acid) | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Phosphatase (Neutral) | + | + | + | + | + | + | + | + | + | + | + | + | + | w | + |
| Phosphatase (Alkaline) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Tween20 | w | w | w | w | - | w | w | + | w | w | - | - | w | - | + |
| Tween80 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Indolacetic Acid | 8.0 | nd | 86.4 | 74.8 | 34.4 | 28.4 | 22.9 | 16.7 | 15.0 | nd | 2.9 | 27.2 | 11.3 | 47.0 | 27.0 |
| pH 6.5 | w | + | + | w | - | w | + | + | + | - | - | + | - | - | - |
(+) positive, (-) negative and (w) weak. Indolacetic acid production is expressed in µg/ml. In the case of phosphatases, positive activity was considered when the absorbance reading was 0.2 above that of the controls.
nd, not detected.
Thirteen Micromonospora strains were able to produce IAA. AL16 and AL20 were the strains with the highest production levels (≥74.8 µg/mL) whereas the strain ALF7 showed the lowest (2.9 µg/mL), the remaining had IAA production ranging from 11.3 to 47.0 µg/L (Table 2).
The ability to grow at different pH (from 4.5 to 9) was tested. All the fifteen Micromonospora strains grew well at a pH range of 7 to 8. None of the strains grew at pH below 5.5 nor at pH 9. We found high variability when grown at pH 6.5 (Table 2).
Effect of Micromonospora on plant growth and nutrient content of alfalfa
Investigating putative plant growth-promoting effects on alfalfa of the fifteen Micromonospora strains alone and in co-inoculation with the model strain E. meliloti 1021 was addressed in this part of the study. A mesocosm experiment was conducted in a greenhouse in pots containing a sandy-clay soil (Table S1) under controlled conditions of temperature, photoperiod and humidity. At harvest, shoot and root biomass, number of nodules and shoot nutrient contents were determined (Figure S2, Tables S2 and S3). The measured plant growth and nutrient content parameters were grouped together to form a data matrix of 2,560 data points (8 parameters × 32 inoculation treatments × 10 replicates).
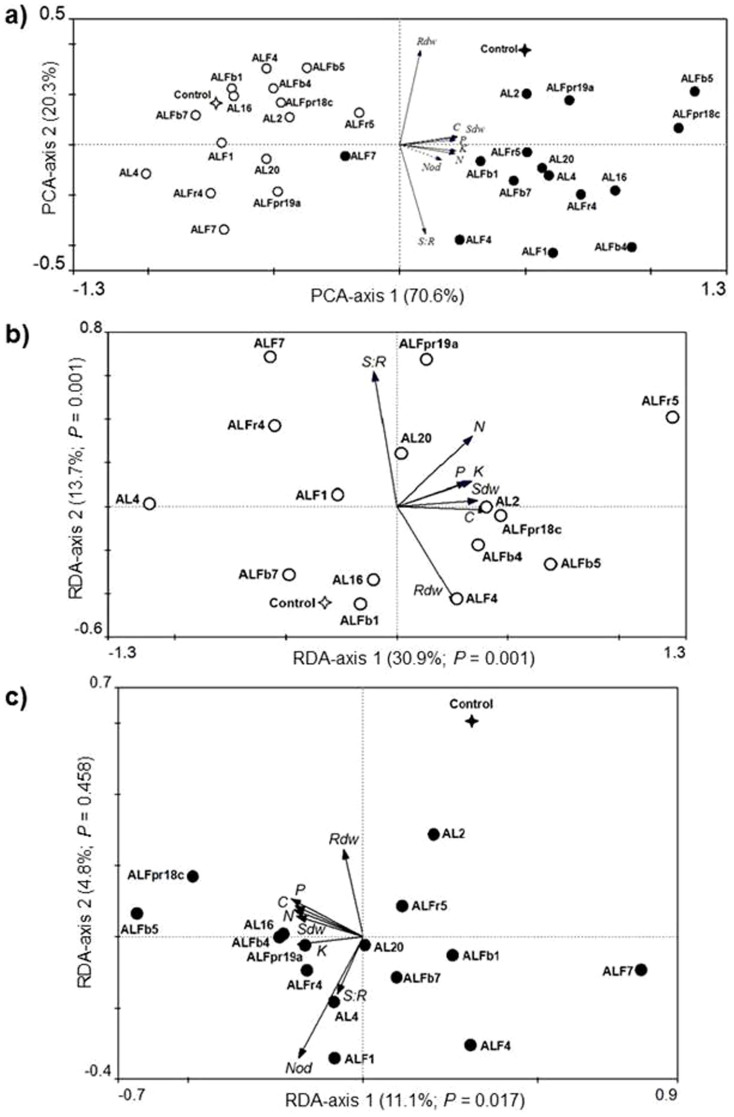
We first used an indirect analysis (PCA) of the data [excepting number of nodules (Nod) data] to summarize the variation across all the 320 alfalfa plants tested. Figure 3a shows the distance biplot resulting from the PCA analysis. The first PCA axis explained 70.6% of the variance in the data, while the second axis accounted for 20.3% only. We calculated the relative amount of the total variability that each of the two first axes should explain under the null model of random variation by using the broken-stick approach28. The values predicted for the first and second axes are 37.0% and 22.8%. Therefore, only the fraction of variability explained by the first axis surpasses the values predicted by the null model, indicating that the first axis describes non-random, interpretable variation in the data while the second does not. With the exception of Rdw (root dry weight) and S:R (shoot to root ratio), the remaining five response variables [Sdw (shoot dry weight), C, N, P and K] had high positive correlation (> 0.9) with the PC1 scores. Further examination of the PCA biplot, focusing on the disposition the centroids of the 32 dummy independent variables (inoculation treatments) projected post hoc into the ordination space, reveals that the first principal component is related to E. meliloti 1021 inoculations (with and without E. meliloti 1021). Plants inoculated with E. meliloti 1021 tended to have Sdw and shoot contents of C, N, P and K higher than the E. meliloti 1021-free plants. Similarly, within the cohorts of plants inoculated and non-inoculated with E. meliloti 1021, the inoculation with specific Micromonospora strains tended to produce higher or lower values of these parameters comparing to other Micromonospora inoculation treatments and to the controls (Figure 3a). Redundancy analyses (RDA) testing for the significance of effects of the inoculation with E. meliloti 1021, the inoculation with Micromonospora and their interaction revealed statistical significance of all the three factors (Table S4).
Figure 3. Biplot representations of the results of PCA and RDA analyses.
(a) PCA performed on the matrix with plant growth parameters and shoot nutrients content. RDA performed on the matrix of plant growth parameters and shoot nutrients content constrained by the matrix of bacterial inoculation treatments, either in (b) the cohort of plants singly inoculated with different strains of Micromonospora spp. and in (c) the cohort of plants co-inoculated with Micromonospora and E. meliloti 1021. Open circles represent the centroids of the treatments inoculated singly with each of the fifteen Micromonospora strains while full circles represent those of the treatments also inoculated with E. meliloti 1021. Star symbols represent the centroids of the control treatments without any microbial inoculation (open star) and inoculated with E. meliloti 1021 only (full star). Arrows represent variables measured on individual alfalfa plants: shoot dry weight (Sdw), root dry weight (Rdw), shoot to root ratio (S:R), number of nodules (Nod); and shoot contents of carbon (C), nitrogen (N), phosphorus (P), and potassium (K). The variable Nod is passively projected into the PCA diagram (a) but it was not included in the calculation (dotted arrow). Values on the axes indicate percentages of total variation explained by each axis and P-values of significance for the RDA canonical axes (b and c) obtained by Monte Carlo permutation tests (999 permutations).
Given the significance of the interaction effect of E. meliloti 1021 and Micromonospora inoculations (F-ratio = 2.090, P-value = 0.002; Table S4), we performed separate RDA analyses for the cohorts of plants inoculated and non-inoculated with E. meliloti 1021. In the cohort of E. meliloti 1021-free plants, redundancy analysis (RDA) revealed that the explanatory effect of the Micromonospora inoculations was highly significant according to the Monte Carlo test for significance of all canonical axis (F-ratio = 9.920, P = 0.0010). Figure 3b shows the distance biplot resulting from this RDA analysis. The proportion of variability explained by all the constrained canonical axes was 50.8%, and 30.9% and 13.7% by, respectively, the first and second canonical axes, both being significant (Figure 3b). We undertook pair-wise RDA comparisons in order to determine which of the Micromonospora-inoculated treatments produced statistically significant differences when compared with the uninoculated control treatment. Table S5 summarizes the results of this set of multivariate tests. Results indicated that only three out of the 15 Micromonospora strains (namely AL16, ALFb1 and ALFb7) produced non-significant differences with respect to the uninoculated control treatment (Table S5). Univariate (ANOVA) analyses on the response variables were performed to compare differences within the control treatment and those Micromonospora-inoculated treatments showing significant differences in the pairwise RDA comparisons (Table 3). It was found that shoot biomass production (Sdw) of alfalfa plants inoculated with Micromonospora ALFb5, ALFr5 or AL4 was significantly different (P ≤ 0.01) to that of the uninoculated control plants. The mean shoot dry weight (Sdw) of plants that were inoculated with Micromonospora ALFb5 or ALFr5 was, respectively, 19% and 35% greater than that of control plants, while in plants inoculated with the strain AL4 was 20% lower. However, root dry weights (Rdw) in four of the inoculation treatments were significantly lower (P ≤ 0.01) than in the control treatment, and only the inoculation with Micromonospora ALFb5 produced an increase marginally significant (P ≤ 0.1). Therefore, S:R ratios in treatments inoculated with Micromonospora were similar to, or higher (P ≤ 0.01) than that in the control treatment (Table 3). Regarding to the shoot nutrient contents, significant decreases respect to the control were only observed for carbon in plants inoculated with Micromonospora AL4 and ALFr4. Plants inoculated with Micromonospora AL20, ALFb5, ALFpr19a or ALFr5 had higher (P ≤ 0.1) shoot contents of N, P and K than the control plants. Besides, K shoot contents were also higher in the treatments inoculated with Micromonospora AL2, ALF4 or ALFb4 than in the control treatment (P ≤ 0.05). It is noteworthy that inoculation with any of these twelve Micromonospora strains yielded higher (P ≤ 0.1) shoot N contents than the control treatment (Table 3), with increases ranging from 22% (ALF7) to 101% (ALFr5).
Table 3. Growth parameters and shoot contents of C, N, P and K of alfalfa plants inoculated with those Micromonospora strains that alone or in co-inoculation with Ensifer meliloti 1021 showed significant multivariate (RDA) differences respect to the Micromonospora-free controls.
| (mg plant−1) | (no. plant−1) | |||||||
|---|---|---|---|---|---|---|---|---|
| Strain | S:Ra | Sdw | Rdw | Shoot C | Shoot N | Shoot P | Shoot K | Nodules |
| Single inoculation with Micromonospora spp. | ||||||||
| Controlb | 1.03 | 956 | 948 | 408 | 9.9 | 2.68 | 18.6 | N/A |
| AL2 | 1.18 | 1067 | 943 | 556 | 13.6*** | 3.22 | 25.4*** | N/A |
| AL4 | 1.22 | 763*** | 635*** | 278*** | 12.2* | 2.20 | 15.3 | N/A |
| AL20 | 1.35*** | 1015 | 763 | 399 | 12.8*** | 3.42** | 22.9* | N/A |
| ALF1 | 1.20 | 939 | 802 | 373 | 14.3*** | 2.75 | 19.2 | N/A |
| ALF4 | 0.92 | 1032 | 1129 | 405 | 13.1*** | 3.17 | 23.6** | N/A |
| ALF7 | 1.81*** | 992 | 559*** | 359 | 12.1* | 2.68 | 19.6 | N/A |
| ALFb4 | 1.01 | 1016 | 1049 | 423 | 15.5*** | 3.15 | 23.6** | N/A |
| ALFb5 | 1.00 | 1140*** | 1161* | 457 | 14.6*** | 3.47** | 24.9*** | N/A |
| ALFpr18c | 1.07 | 1030 | 978 | 475** | 16.4*** | 3.12 | 22.4 | N/A |
| ALFpr19a | 1.51*** | 972 | 662*** | 384 | 16.6*** | 3.37* | 25.5*** | N/A |
| ALFr4 | 1.45*** | 880 | 619*** | 347*** | 13.1*** | 2.63 | 20.7 | N/A |
| ALFr5 | 1.31** | 1288*** | 1027 | 515*** | 19.9*** | 3.69*** | 26.4*** | N/A |
| Co-inoculation with Ensifer meliloti 1021 | ||||||||
| Control c | 1.20 | 1572 | 1381 | 668 | 41.7 | 4.62 | 32.0 | 42 |
| ALFb5 | 1.60* | 1980*** | 1312 | 826** | 52.1*** | 5.57** | 43.2*** | 87*** |
| ALFpr18c | 1.85*** | 1948** | 1207 | 805** | 50.5** | 5.59** | 40.9*** | 71*** |
aS:R, shoot to root ratio; Sdw, shoot dry weight; Rdw, root dry weight.
bControl treatment corresponds to uninoculated alfalfa plants.
cControl treatment corresponds to alfalfa plants inoculated only with E. meliloti 1021.
Means (N = 10) are shown. Within columns, treatment means in bold type or underlined were, respectively, higher or lower than their respective control treatment (in italics) according to Dunnett's one-tailed tests at P ≤ 0.1 (*), P ≤ 0.05 (**) and P ≤ 0.01 (***).
In the cohort of E. meliloti 1021-inoculated plants, redundancy analysis (RDA) revealed that the explanatory effect of the Micromonospora inoculations was significant according to the Monte Carlo test for significance of the first axis (F-ratio = 18.046, P = 0.017) and all canonical axes (F-ratio = 2.207, P = 0.001). However, the second canonical axis was non-significant (F-ratio = 8.209, P = 0.458), indicating that it explained no more variation than random and, thus, does not need to be further considered in the interpretation of the results29. The proportion of variability explained by all the constrained canonical axes was 18.7%. Figure 3c shows the distance biplot resulting from this RDA analysis. Pair-wise RDA comparisons indicated that only Micromonospora ALFb5 and ALFpr18c produced significant differences with respect to the Micromonospora-free control treatment (Table S5). Univariate (ANOVA) analyses indicated that inoculation with Micromonospora ALFb5 or ALFpr18c produced significantly (P ≤ 0.05) more nodules and higher shoot biomass (Sdw) than the control treatment (Table 3). The mean Sdw of plants co-inoculated with E. meliloti 1021 and either Micromonospora ALFb5 or ALFpr18c were, respectively, 26% and 24% greater than that of plants only inoculated with E. meliloti 1021 (control treatment). These two co-inoculated treatments also showed higher shoot-to-root (S:R) ratios than the control treatment (P ≤ 0.1). In comparison with control plants, co-inoculated plants afforded significantly higher (P ≤ 0.05) shoot contents of C (24% and 20%), N (10% and 21%), P (21%) and K (35% and 28%).
Discussion
The number and diversity of Micromonospora strains recovered from alfalfa nodules strongly suggest that this actinobacteria is commonly associated with the symbiotic organ of legumes. Besides other microorganisms, almost all nodules selected had a population of one or more Micromonospora strains. Moreover, for each isolation experiment two sterile, non-crushed nodule was rolled over YMA agar and incubated under the same conditions as the homogenized samples in order to assess the effectivity of the sterilization procedure. No colonies appeared on any YMA plate indicating that sterilization was effective.
BOX–PCR fingerprinting has been shown to be a useful tool to discriminate highly related strains and has been applied to study the genetic diversity of different bacterial taxa, including Micromonospora30,31. A high degree of genetic variation was observed among the 66 isolates, when analysed by BOX–PCR fingerprinting, indicating that they represented different bacterial genotypes. It should also be noted that none of them were clones of one another, supporting the idea of the existence of high genetic diversity among Micromonospora strains in legume root nodules. Our results are coherent with data from Lupinus angustifolius and Pisum sativum root nodules13,15.
The BOX grouping provided a useful background for determining the taxonomic relationship of the strains isolated since these groups served to select strains for 16S rRNA gene sequencing. Even though several strains have more than 99% similarity with described species, others had 16S rRNA gene sequence similarities below 99%, indicating that they were not related to any of the already known species of Micromonospora and probably represent new ones. This case has been observed previously by Trujillo and co-workers, who described two new Micromonospora species: M. lupini and M. saelicesensis, whose 16S rRNA genes were highly similar to already described species12. Our results (BOX–PCR fingerprinting and 16S rRNA gene sequences) also suggest that the diversity of Micromonospora was independent of the location where they were isolated since in several BOX-PCR groups there are strains from more than one of the five locations sampled (Figure 1).
The high diversity and ubiquity of this actinobacteria inside legume root nodules suggest that its presence is not fortuitous, but that Micromonospora might have an important ecological role in nature. To discern ecological roles of Micromonospora in interaction with alfalfa, we evaluated in a mesocosm experiment their effects on plant growth and nutrition, both in nodulated and non-nodulated alfalfa plants. Multivariate statistics showed that E. meliloti 1021-nodulated plants tended, of course, to have higher values of aerial dry matter and nutrient content than the non-nodulated ones (Figure 3a). But we also found a significant effect of the inoculation with Micromonospora as well as a significant interaction between both E. meliloti 1021 and Micromonospora inoculations, indicating different behaviour of the Micromonospora strains according to the nodulation status of the plant (Table S4). In non-nodulated plants, twelve out of the fifteen Micromonospora strains produced significant multivariate differences with respect to the uninoculated control (Table S5; Figure 3b), while in nodulated plants only the treatments co-inoculated with the strains ALFb5 and ALFpr18c differed significantly from the Micromonospora-free control treatment (Table S5; Figure 3c). Several actinobacteria, including strains of Micromonospora sp., had been shown to promote both shoot and root growth and nodulation in alfalfa as well as in the actinorhizal plant species Ochetophila (Discaria) trinervis when co-inoculated with the corresponding nitrogen-fixing micro-symbiont, Ensifer or Frankia32,33,34. Contrary to our results, these authors found that actinobacteria alone exerted no effect on plant growth. It should be emphasized that, unlike us, Solans and co-workers grew the plants in soilless, gnotobiotic conditions32,33,34.
Three main empirical facts from our greenhouse experiment must be highlighted. The first one is that Micromonospora does not induce larger root systems. The most common effect of PPBs on plant is the formation of larger root systems, which allow exploring a greater volume of soil for water and nutrients35. Root biomasses (Rdw) in Micromonospora-inoculated treatments were similar to or lower than in control plants (Table 3; Figure 3a,b). The second fact is that most Micromonospora strains increased the N shoot content in non-nodulated plants (Table 3; Figure 3b). It has been reported in recent years the existence of putative N2-fixing Micromonospora13,36. Therefore, in a previous work we conducted an exhaustive experimental study centred on two of the representative strains here assayed, namely ALFb5 and ALFpr18c, in order to discern if they could fix N2 either as free-living diazotrophs or in symbiosis with alfalfa plants37. Neither of the strains grew in nitrogen-free media or reduced acetylene under micro-aerobic conditions. Incorporation of 15N into the microbial biomass or alfalfa tissues was not detected. Also, attempts to amplify putative nifH genes in these strains were unsuccessful. Besides, we tracked the presence of structural genes for N2-fixation in two other Micromonospora strains that have their genome sequenced38,39, finding no evidence for them37. These results seem to rule out N2 fixation by Micromonospora as source of nitrogen for plants, focusing the explanation for higher N shoot contents on enhanced nutrient uptake efficiency and/or more plant-available nitrogen in soil.
Although there are few published studies on the impact of PPB on nutrient uptake systems, concomitant improvement of mineral nutrition (including N, P and K) and increase of root surface area has been described in several plant species35. With regards to N nutrition, it has been hypothesized that PPB could directly stimulate nitrate transport systems in plants40, but recent genetic studies on Arabidopsis thaliana indicate that while there are two NO3− transporter genes (NRT2.5 and NRT2.6) that are strongly upregulated in response to inoculation with the PPB Phyllobacterium brassicacearum strain STM196, plant growth promotion is not linked to changes in NO3− uptake rate or NO3− distribution between roots and shoots41. However, most actinobacteria are saprophytes able to produce a wide range of extracellular hydrolytic enzymes2,42,43,44. All the strains we studied synthesize hydrolytic enzymes able to cleave complex nitrogen-containing polymeric substrates, such as caseinase and gelatinase (Table 2), strongly suggesting that Micromonospora can favour plant nutrition by enhancing nitrogen mineralization in soils. Nonetheless, further research is needed to fully explain the rationale for improved nitrogen nutrition in plants inoculated with Micromonospora. Moreover, all the fifteen Micromonospora showed neutral and alkaline phosphatase activities (Table 2), which can enhance the mineralization of organic phosphate in neutral or alkaline soils45 like the one used in our greenhouse experiment (pH 7.47; Table S1), thus making soil P more available to plants as suggested by higher shoot P content in some Micromonospora-inoculated treatments than in the controls (Table 3; Figure 3b, c).
In the cohort of plants nodulated by E. meliloti 1021 only two strains of Micromonospora (ALFb5 and ALFpr18c) produced statistically significant multivariate differences with respect to the Micromonospora-free control group (Table S5; Figure 3c). The success of the interaction between a PPB strain and the plant relies on a set of adaptation mechanisms by both partners, among which the phytochemical profile of the root exudates plays a fundamental role in the bacterial colonization of the root as well as in the regulation of PPB plant beneficial properties46. The composition of root exudates has been shown to differ in legumes depending on their nodulation status47,48,49, so that the biochemical environment in the rhizosphere of E. meliloti 1021-nodulated alfalfa plants might be less advantageous for Micromonospora compared with that of non-nodulated plants. Considering the soil pH, legumes are known to acidify the rhizosphere because of the release of protons following excess uptake of cations over anions during N2 fixation50,51,52. Only six out of the 15 Micromonospora strains tested in this study grew vigorously in vitro at pH 6.5 (Table 2) and none at lower pH values (4.5 or 5.5). Indeed, strains ALFb5 and ALFpr18c are among those able to grow at acidic pH (6.5) while the strain ALFr5, a strain that only excelled in the cohort of non-nodulated plants, does not (Table 2; Figure 3b,c). Therefore, a more acidic rhizosphere in the N2-fixing plants may partially explain differential effects of certain Micromonospora strains on growth of nodulated and non-nodulated plants. Nonetheless, given the great influence that the nodulation has on N acquisition capacity and growth of legumes, it is plausible that the effect of most of the Micromonospora is not marked enough to be statistically significant in symbiotically N2-fixing alfalfa plants.
And third, although in plants nodulated by E. meliloti 1021 only Micromonospora ALFb5 and ALFpr18c were found to have significant, globally positive effects on plant growth and nutrition (Table 3), it was observed a trend towards the improvement in nodulation intensity (number of nodules) with the presence of Micromonospora (Figure 3c; Figure S2c). Furthermore, all the fifteen Micromonospora strains could be re-isolated from nodules of random plants of each co-inoculated treatment, suggesting that none of the Micromonospora strains here assayed had incompatibility with E. meliloti 1021.
Plant growth-promoting bacteria can increase nodulation in legumes through different mechanisms, including the production or degradation of phytohormones involved in nodule initiation and organogenesis53, or by affecting the interaction between plant and rhizobia54,55,56. Auxins are involved in the initiation and normal development of both determinate57 and indeterminate nodules, like Medicago root nodules58. IAA production has been associated with the induction of increased nodule numbers in Medicago truncatula plants inoculated with an E. meliloti strain that overproduces IAA59 and also with nodule-like structures even in non-leguminous plants60,61,62. Moreover, rhizobial cellulases have been shown to be crucial for legume nodulation63. The ability of Micromonospora strains to produce cellulases could thus explain the increase in the number of nodules observed in co-inoculated plants compared to the control plants only inoculated with E. meliloti 1021. However, IAA production by Micromonospora may not be directly related in our study to an increase in nodulation despite of the literature.
Conclusion
In this study 66 Micromonospora strains were isolated, characterized using BOX-PCR and sequencing of 16S rRNA genes and selected some of them for studying their interaction with alfalfa. Our results, together with those from other authors, indicate that Micromonospora are ubiquitous in legume root nodules, presenting a very high genetic diversity. Most of them exhibit in vitro a great ability to degrade organic polymers as well as presenting a direct mechanism for plant growth promotion (IAA production). We have shown that Micromonospora could play an important ecological role in interaction with the host plant by enhancing aerial growth and nutrient contents, being an increase of N uptake by the plant a general phenomenon in the Micromonospora-alfalfa interaction. It remains to be elucidated whether these positive effects also occur in other plant species. Micromonospora engaged in tripartite interactions with E. meliloti 1021 and alfalfa increase nodulation, and some of their strains can also significantly promote the growth and nutrition of N2-fixing plants. Contrary to most of plant growth-promoting bacteria, beneficial effects of Micromonospora do not rely on induction of plant root growth. All the above data suggests that, in general, Micromonospora can be considered as excellent PPB, although a correct selection of strain is of capital importance because of the detrimental effect that some Micromonospora may have for plant growth (i.e. strain AL4 in non-nodulated plants; Table 3). Aditionally, Micromonospora is a sporulating bacterium so that it can endure in soil and harsh environments. Thus, some of their strains seem to be excellent candidates for the production of bioinoculants, which would make the use of environmentally unfriendly chemical fertilizers less intensive in a broad range of agroecosystems.
Methods
Isolation and ecological characterization of Micromonospora strains
Isolations of Micromonospora were done from surface sterilized root nodules of naturally occurring alfalfa plants from five different regions of Castilla y León (Spain). Functional characterization of the isolated strains included: hydrolytic activities toward casein, starch, gelatin, xylan, Tween 80 and 20, cellulose and pectin; presence of acid, neutral and alkaline phosphatase activities; production of indole acetic acid (IAA); and growth under different environmental conditions. For further details on isolation and functional characterization of the isolates, see Materials and Methods in Supplementary Information.
Genetic and phylogenetic characterization of Micromonospora strains
BOX–PCR fingerprinting profiles from bacterial genomic DNA were obtained according to Trujillo et al.13. Similarity matrices of electrophoretic band profiles were calculated using the Pearson Correlation Coefficient followed by dendrogram construction using the UPGMA algorithm. Strain clusters were defined at the 60% level of similarity. Three different strains were used as probes, processing them in every PCR and electrophoresis run in our experiments. When gel patterns were analyzed with the software BioNumerics (Applied Maths, Sint-Martens-Latem, Belgium), probe strains band patterns always were observed to be identical.
One representative strain of each BOX cluster was selected for 16S rDNA sequencing and phylogenetic analysis. The sequences were aligned and compared with those deposited in public databases, and then neighbour-joining and maximum-likelihood phylogenetic trees were constructed. Sequence data has been submitted to the GenBank database under accession numbers from KF876220 to KF876234. See Materials and Methods in Supplementary Information for full details.
Micromonospora-Ensifer meliloti 1021-alfafa interaction assay
The fifteen representative Micromonospora strains (Table 1) were tested in interaction with alfalfa plants, either alone or in co-inoculation with E. meliloti 1021. Alfalfa plants were individually grown in pots (1L volume) containing tyndallized soil in a greenhouse under controlled environmental conditions. The experimental design included 32 treatments with 10 replicates per treatment. Treatments were defined by a factorial combination of two E. meliloti 1021 inoculation treatments (non-inoculated or inoculated) and sixteen Micromonospora inoculation treatments (non-inoculated or inoculated with each of the fifteen representative strains). Plants were harvested 14 weeks after inoculations and the following parameters were determined: Shoot (Sdw) and root (Rdw) dry weight; shoot content in carbon (C), nitrogen (N), phosphorus (P) and potassium (K); and number of root nodules (Nod). See Materials and Methods in Supplementary Information for full details.
Statistical analysis
Plant growth and nutrient content data were analysed using multivariate (PCA and RDA) and univariate (ANOVA) analyses with the CANOCO 4.5 (Microcomputer Power, Ithaca, NY) and SPSS for Windows v21.0 (IBM Corp., Armonk, NY) programs. The inoculation treatments were coded as dummy variables and used as independent variable in the multivariate analyses. Significance in RDA analyses was tested by Monte Carlo permutation tests (999 unrestricted permutations) for the first canonical axis as well as for the sum of all canonical axes. In univariate comparisons, post-hoc Dunnett's one-tailed t-tests were used to identify inoculation treatments with means significantly different from the control at P ≤ 0.1, P ≤ 0.05 an d P ≤ 0.01. For further details on the statistical analyses, see Materials and Methods in Supplementary Information.
Author Contributions
Conceived and designed the experiments: P.M.-H., E.M.-M., J.M.I. Performed the experiments: P.M.-H. Analyzed the data: P.G.-V. Wrote the paper: P.M.-H., E.M.-M., J.M.I.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by Junta de Castilla y León Grant SA306A11-2 and MICINN Grant AGL2010-17380. P.M.-H. was supported by a fellowship from CSIC JAE-PRE. We thank N. Skinner for revising the English version of the manuscript.
References
- El-Tarabily K. A. & Sivasithamparam K. Non-streptomycete actinomycetes as biocontrol agents of soil-borne fungal plant pathogens and as plant growth promoters. Soil Biol. Biochem. 38, 1505–1520 (2006). [Google Scholar]
- Hirsch A. M. & Valdés M. Micromonospora: An important microbe for biomedicine and potentially for biocontrol and biofuels. Soil Biol. Biochem. 42, 536–542 (2010). [Google Scholar]
- Velázquez E. et al. In: Beneficial Plant-Microbial Interactions 194, 214–236 (CRC Press, 2013). [Google Scholar]
- Venkateshwaran M., Volkening J. D., Sussman M. R. & Ané J.-M. Symbiosis and the social network of higher plants. Curr. Opin. Plant Biol. 16, 118–127 (2013). [DOI] [PubMed] [Google Scholar]
- Coombs J. T. J. & Franco C. M. M. C. Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl. Environ. Microbiol. 69, 5603–5608 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs J. T. & Franco C. M. M. Visualization of an endophytic Streptomyces species in wheat seed. Appl. Environ. Microbiol. 69, 4260–4262 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresu R. et al. Coexistence of predominantly nonculturable rhizobia with diverse, endophytic bacterial taxa within nodules of wild legumes. FEMS Microbiol. Ecol. 63, 383–400 (2008). [DOI] [PubMed] [Google Scholar]
- Sturz A., Christie B., Matheson B. & Nowak J. Biodiversity of endophytic bacteria which colonize red clover nodules, roots, stems and foliage and their influence on host growth. Biol. Fertil. Soils 25, 13–19 (1997). [Google Scholar]
- Hoque M. S., Broadhurst L. M. & Thrall P. H. Genetic characterization of root-nodule bacteria associated with Acacia salicina and A. stenophylla (Mimosaceae) across south-eastern Australia. Int. J. Syst. Evol. Microbiol. 61, 299–309 (2011). [DOI] [PubMed] [Google Scholar]
- Li L. et al. Biogeography of symbiotic and other endophytic bacteria isolated from medicinal Glycyrrhiza species in China. FEMS Microbiol. Ecol. 79, 46–68 (2011). [DOI] [PubMed] [Google Scholar]
- Stajković O., De Meyer S., Miličić B., Willems A. & Delić D. Isolation and characterization of endophytic non-rhizobial bacteria from root nodules of alfalfa (Medicago sativa L.). Botanica Serbica 33, 107–114 (2009). [Google Scholar]
- Trujillo M. E., Kroppenstedt R. M., Fernández-Molinero C., Schumann P. & Martínez-Molina E. Micromonospora lupini sp. nov. and Micromonospora saelicesensis sp. nov., isolated from root nodules of Lupinus angustifolius. Int. J. Syst. Evol. Microbiol. 57, 2799–2804 (2007). [DOI] [PubMed] [Google Scholar]
- Trujillo M. E. et al. The genus Micromonospora is widespread in legume root nodules: the example of Lupinus angustifolius. ISME J. 4, 1265–1281 (2010). [DOI] [PubMed] [Google Scholar]
- García L. C., Martínez-Molina E. & Trujillo M. E. Micromonospora pisi sp. nov., isolated from root nodules of Pisum sativum. Int. J. Syst. Evol. Microbiol. 60, 331–337 (2010). [DOI] [PubMed] [Google Scholar]
- Carro L., Pukall R., Spröer C., Kroppenstedt R. M. & Trujillo M. E. Micromonospora cremea sp. nov. and Micromonospora zamorensis sp. nov., isolated from the rhizosphere of Pisum sativum. Int. J. Syst. Evol. Microbiol. 62, 2971–2977 (2007). [DOI] [PubMed] [Google Scholar]
- Martínez-Hidalgo P. Endophytic actinobacteria isolated from nodules of Medicago sativa: Analysis of their biodiversity and agronomic potential as PGPR. http://hdl.handle.net/10366/121386 (2012). Date of access: 07/07/2014.
- Deng Z. S. et al. Diversity of endophytic bacteria within nodules of the Sphaerophysa salsula in different regions of Loess Plateau in China. FEMS Microbiol. Ecol. 76, 463–475 (2011). [DOI] [PubMed] [Google Scholar]
- Radović J., Sokolović D. & Marković J. Alfalfa-most important perennial forage legume in animal husbandry. Biotechnology in Animal Husbandry 25, 465–475 (2009). [Google Scholar]
- Bohlool B. B., Ladha J. K., Garrity D. P. & George T. Biological nitrogen fixation for sustainable agriculture: A perspective. Plant Soil 141, 1–11 (1992). [Google Scholar]
- Berg G. Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18 (2009). [DOI] [PubMed] [Google Scholar]
- Olivares J., Bedmar E. J. & Sanjuán J. Biological Nitrogen Fixation in the Context of Global Change. Mol. Plant Microbe In. 26, 486–494 (2013). [DOI] [PubMed] [Google Scholar]
- Emmert E. A. B. & Handelsman J. Biocontrol of plant disease: a (Gram-) positive perspective. FEMS Microbiol. Lett. 171, 1–9 (1999). [DOI] [PubMed] [Google Scholar]
- Whipps J. M. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52, 487–511 (2001). [DOI] [PubMed] [Google Scholar]
- Compant S. et al. Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl. Environ. Microbiol. 71, 1685–1693 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibach-Paldi S., Burdman S. & Okon Y. Key physiological properties contributing to rhizosphere adaptation and plant growth promotion abilities of Azospirillum brasilense. FEMS Microbiol. Lett. 326, 99–108 (2011). [DOI] [PubMed] [Google Scholar]
- McSpadden Gardener B. B. & Fravel D. R. Biological Control of Plant Pathogens: Research, Commercialization, and Application in the USA. http://www.plantmanagementnetwork.org/pub/php/review/biocontrol; 10.1094/PHP-2002-0510-01-RV (2002). Date of access: 07/07/2014. [Google Scholar]
- Smith K. P. & Goodman R. M. Host variation for interactions with beneficial plant-associated microbes. Annu. Rev. Phytopathol. 37, 473–491 (1999). [DOI] [PubMed] [Google Scholar]
- Legendre P. & Legendre L. Numerical Ecology 2nd. (Elsevier Science, 1998). [Google Scholar]
- Legendre P., Oksanen J. & Braak t. e. r., C J. F. Testing the significance of canonical axes in redundancy analysis. Methods Ecol. Evol. 2, 269–277 (2011). [Google Scholar]
- Lanoot B. et al. BOX-PCR fingerprinting as a powerful tool to reveal synonymous names in the genus Streptomyces. Emended descriptions are proposed for the species Streptomyces cinereorectus, S. fradiae, S. tricolor, S. colombiensis, S. filamentosus, S. vinaceus and S. phaeopurpureus. Syst. Appl. Microbiol. 27, 84–92 (2004). [DOI] [PubMed] [Google Scholar]
- Maldonado L. A., Fragoso-Yáñez D., Pérez-García A., Rosellón-Druker J. & Quintana E. T. Actinobacterial diversity from marine sediments collected in Mexico. Antonie Van Leeuwenhoek 95, 111–120 (2009). [DOI] [PubMed] [Google Scholar]
- Solans M. Discaria trinervis – Frankia symbiosis promotion by saprophytic actinomycetes. J. Basic Microbiol. 47, 243–250 (2007). [DOI] [PubMed] [Google Scholar]
- Solans M., Vobis G. & Wall L. G. Saprophytic actinomycetes promote nodulation in Medicago sativa-Sinorhizobium meliloti symbiosis in the presence of high N. J. Plant Growth Regul. 28, 106–114 (2009). [Google Scholar]
- Solans M., Vobis G., Cassán F., Luna V. & Wall L. G. Production of phytohormones by root-associated saprophytic actinomycetes isolated from the actinorhizal plant Ochetophila trinervis. World J. Microbiol. Biotechnol. 27, 2195–2202 (2011). [Google Scholar]
- Vacheron J. et al. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 4, 356–356; 10.3389/fpls.2013.00356 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés M. et al. Non-Frankia Actinomycetes isolated from surface-sterilized roots of Casuarina equisetifolia fix nitrogen. Appl. Environ. Microbiol. 71, 460–466 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Hidalgo P., Olivares J., Delgado A., Bedmar E. & Martínez-Molina E. Endophytic Micromonospora from Medicago sativa are apparently not able to fix atmospheric nitrogen. Soil Biol. Biochem. 74, 201–203 (2014). [Google Scholar]
- Alonso-Vega P. et al. Genome Sequence of Micromonospora lupini Lupac 08, Isolated from Root Nodules of Lupinus angustifolius. J. Bacteriol. 194, 4135–4135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch A. M. et al. Complete Genome Sequence of Micromonospora Strain L5, a Potential Plant-Growth-Regulating Actinomycete, Originally Isolated from Casuarina equisetifolia Root Nodules. Genome Announcements 1, e00759–13–e00759–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantelin S. & Touraine B. Plant growth-promoting bacteria and nitrate availability: impacts on root development and nitrate uptake. J. Exp. Bot. 55, 27–34 (2003). [DOI] [PubMed] [Google Scholar]
- Kechid M. et al. The NRT2.5 and NRT2.6 genes are involved in growth promotion of Arabidopsis by the plant growth-promoting rhizobacterium (PGPR) strain Phyllobacterium brassicacearum STM196. New Phytol. 198, 514–524 (2013). [DOI] [PubMed] [Google Scholar]
- Quecine M. C. et al. Chitinolytic activity of endophytic Streptomyces and potential for biocontrol. Lett. Appl. Microbiol. 47, 486–491 (2008). [DOI] [PubMed] [Google Scholar]
- Adams A. S. et al. Cellulose-degrading bacteria associated with the invasive woodwasp Sirex noctilio. ISME J. 5, 1323–1331 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Menezes A. B., Lockhart R. J., Cox M. J., Allison H. E. & McCarthy A. J. Cellulose degradation by micromonosporas recovered from freshwater lakes and classification of these actinomycetes by DNA Gyrase B Gene Sequencing. Appl. Environ. Microbiol. 74, 7080–7084 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson A. E., Barea J.-M., McNeill A. M. & Prigent-Combaret C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 321, 305–339 (2009). [Google Scholar]
- Drogue B., Doré H., Borland S., Wisniewski-Dyé F. & Prigent-Combaret C. Which specificity in cooperation between phytostimulating rhizobacteria and plants? Res. Microbiol. 163, 500–510 (2012). [DOI] [PubMed] [Google Scholar]
- Atemkeng M. F., Remans R., Michiels J., Tagne A. & Ngonkeu E. L. M. Inoculation with Rhizobium etli enhances organic acid exudation in common bean (Phaseolus vulgaris L.) subjected to phosphorus deficiency. African Journal of Agricultural Research 6, 2235–2242 (2011). [Google Scholar]
- Soerensen K. U., Terry R. E., Jolley V. D., Brown J. C. & Vargas M. E. The interaction of iron-stress response and root nodules in iron-efficient and inefficient soybeans. J. Plant Nutr. 11, 853–862 (1988). [Google Scholar]
- Terry R. E., Hartzook A., Jolley V. D. & Brown J. C. Interactions of iron nutrition and symbiotic nitrogen fixation in peanuts. J. Plant Nutr. 11, 811–820 (1988). [Google Scholar]
- Haynes R. J. Soil acidification induced by leguminous crops. Grass Forage Sci. 38, 1–11 (1983). [Google Scholar]
- Israel D. W. & Jackson W. A. Ion balance, uptake, and transport processes in N2-fixing and nitrate- and urea- dependent soybean plants. Plant Physiol. 69, 171–178 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.-C., Lund L. J. & Page A. L. Acidity produced by leguminous plants through symbiotic dinitrogen fixation. J. Environ. Qual. 18, 529–534 (1989). [Google Scholar]
- Fox S. L., O'Hara G. W. & Bräu L. Enhanced nodulation and symbiotic effectiveness of Medicago truncatula when co-inoculated with Pseudomonas fluorescens WSM3457 and Ensifer (Sinorhizobium) medicae WSM419. Plant Soil 348, 245–254 (2011). [Google Scholar]
- El-Sayed El-Desoky Radwan T., Mohamed Z. K. & Massena Reis V. Production of indole-3-acetic acid by different strains of Azospirillum and Herbaspirillum spp. Symbiosis 32, 39–53 (2002). [Google Scholar]
- Merzaeva O. V. & Shirokikh I. G. The production of auxins by the endophytic bacteria of winter rye. Appl. Biochem. Microbiol. 46, 44–50 (2010). [PubMed] [Google Scholar]
- Madhaiyan M., Poonguzhali S., Ryu J. & Sa T. Regulation of ethylene levels in canola (Brassica campestris) by 1-aminocyclopropane-1-carboxylate deaminase-containing Methylobacterium fujisawaense. Planta 224, 268–278 (2006). [DOI] [PubMed] [Google Scholar]
- Takanashi K., Sugiyama A. & Yazaki K. Auxin distribution and lenticel formation in determinate nodule of Lotus japonicus. Plant Signal Behav. 6, 1405–1407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D. et al. Rhizobial infection is associated with the development of peripheral vasculature in nodules of Medicago truncatula. Plant Physiol. 162, 107–115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pii Y., Crimi M., Cremonese G., Spena A. & Pandolfini T. Auxin and nitric oxide control indeterminate nodule formation. BMC Plant Biol. 7, 21; 10.1186/1471-2229-7-21 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge R. W., Ride K. M. & Rolfe B. G. Nodule-like structures induced on the roots of rice seedlings by addition of the synthetic auxin 2,4-dichlorophenoxyacetic acid. Aust. J. Plant Physiol. 20, 705–717 (1993). [Google Scholar]
- Christiansen-Weniger C. Endophytic establishment of diazotrophic bacteria in auxin-induced tumors of cereal crops. Cr. Rev. Plant Sci. 17, 55–76 (1998). [Google Scholar]
- Narula N., Deubel A., Gans W., Behl R. K. & Merbach W. Paranodules and colonization of wheat roots by phytohormone producing bacteria in soil. Plant Soil Environ. 52, 119–129 (2006). [Google Scholar]
- Robledo M. et al. Rhizobium cellulase CelC2 is essential for primary symbiotic infection of legume host roots. Proc. Natl. Acad. Sci. U S A 105, 7064–7069 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information