Abstract
Biochemical and structural studies of dynamin have shown that the C-terminus of the GTPase effector domain (GED) folds back and docks onto a platform created by the N- and C-terminal α-helices of the GTPase domain to form a three-helix bundle. While cross-linking studies suggested that insect cell-expressed dynamin existed as a domain-swapped dimer, X-ray structures of protein expressed in Escherichia coli failed to detect evidence of this domain swap. Here, by cross-linking several cysteine pair replacements and analyzing cross-linked species by matrix-assisted laser desorption ionization Mega time of flight, we conclude that dynamin is not domain-swapped and that GED–GTPase domain interactions occur in cis.
The large atypical GTPase dynamin is perhaps best understood for its role in fission during clathrin-mediated endocytosis (CME).1,2 Functional catalysis of fission requires dynamin’s self-assembly on the narrow necks of invaginated coated pits and coordinated GTP hydrolysis leading to global conformational changes that destabilize the underlying lipid bilayer.3,4 Dynamin consists of five functional domains: an N-terminal GTPase domain (G domain), largely α-helical middle and GTPase effector domains (GEDs) that together form the “stalk” of dynamin required for higher-order assembly, a PtdIns(4,5)P2 targeting pleckstrin homology (PH) domain, and a C-terminal proline and arginine rich domain (PRD) that interacts with many SH3 domain-containing proteins. The N- and C-terminal α-helices of the G domain pack against the C-terminal α-helix of the GED forming a three-helix bundle signaling element (BSE) that is critical for structural integrity, self-assembly, and assembly-stimulated GTPase activity.4,5
In vitro purified dynamin exists predominantly as a tetramer; however, dynamin dimers are thought to be the fundamental assembly unit. Several mutations that disrupt tetramerization and higher-order self-assembly have been identified,6−9 but monomeric dynamin mutants have not been identified. A possible explanation for this observation came from cross-linking studies in which single cysteine residues were introduced into both the N-terminal helix (R15C) and the C-terminal helix of the GED (R730C) in an otherwise reactive cysteine-less (DynRCL) dynamin construct. Consistent with the known structure of the BSE,10 DynRCL(R15C/R730C) was efficiently cross-linked by the short (3.6 Å) cysteine reactive cross-linker MTS-1-MTS.4 The resulting product, when subjected to nonreducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), migrated as an ∼180 kDa species relative to several commercially available, prestained molecular mass markers [although there was some variation depending on the source of the marker proteins (Figure S1 of the Supporting Information)]. On the basis of this migration, and given that cross-linking a minimal R15C/R730C GTPase–GED construct resulted in a faster migrating species on SDS–PAGE,5 we concluded that dynamin existed as a domain-swapped dimer in which the GED from one polypeptide docked onto the G domain of a second polypeptide.
Near-coincident publications reporting the X-ray structures of ΔPRD dynamin dimers expressed in Escherichia coli failed to reveal evidence of this domain swap.8,9 One possibility for this discrepancy was that the prokaryotic expression system is likely incapable of facilitating this presumably chaperone-assisted domain swap.11 To test this, we expressed DynRCL(R15C/R730C) in E. coli and performed cross-linking analyses in comparison to the same construct expressed in insect cells. Unexpectedly, the E. coli-derived protein was efficiently cross-linked and migrated in a manner identical to that of the insect cell-derived protein (Figure 1).
Figure 1.
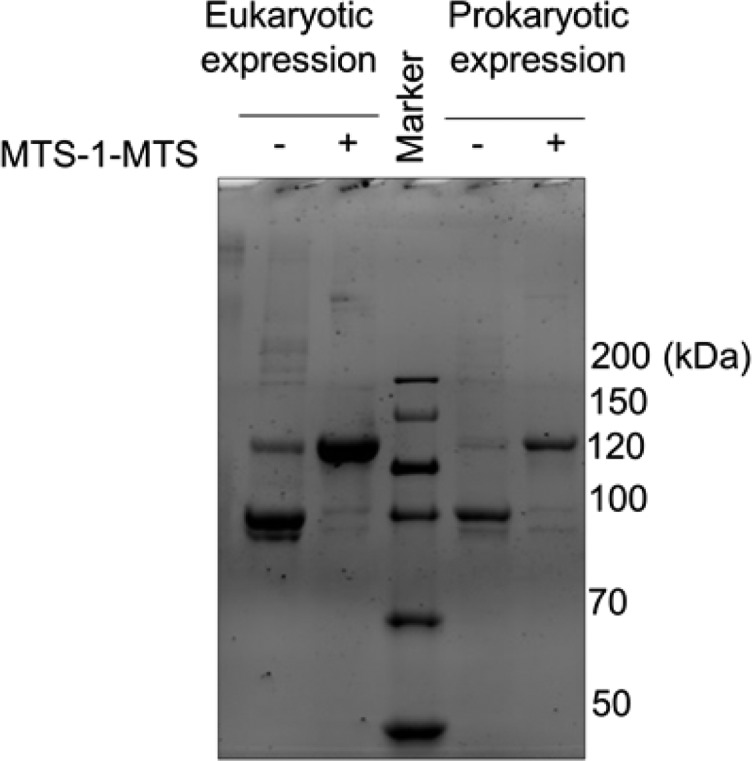
Comparison of cross-linking efficiency and SDS–PAGE migration of insect cell-derived and E. coli-expressed DynRCL(R15C/R730C). Lonza Marker: ProSieve Unstained Protein Marker II, 10–200 kDa.
The regions connecting the stalk to either the BSE or the PH domain were disordered and thus not detected in the high-resolution X-ray structures of dynamin.8,9 In particular, the large gap and flexibility of linkage between the PH domain and the stalk made it difficult to discern which PH domain linked to which stalk in the crystal structure. Thus, a second possibility for the discrepancy between cross-linking studies and structure was that the domain swap occurred in this region, such that the GED from one polypeptide was docked along the entire middle domain of the second polypeptide.12 To test this, we introduced a series of cysteine pair point mutations along the GED and middle domains, including mutations that lie within the dimer interface (Figure 2A), and expressed these proteins in insect cells. Cross-linked species were subjected to nonreducing SDS–PAGE, and their migration was compared to that of unstained molecular mass marker proteins. As expected, each of these species was efficiently cross-linked by MTS-1-MTS (Figure 2B). Cross-linking of cysteines within the G domain (M6C/Q283C) resulted in a more rapidly migrating species on nonreducing SDS–PAGE. Cross-linking cysteine residues between the middle domain and the GED (E341C/K694C and E482C/T676C) resulted in species that migrated more slowly than non-cross-linked dynamin, but close to the size of a monomer. On the basis of their migration, we would conclude that these species correspond to intramolecular cross-links between GED and the middle, and that a domain swap did not occur between the PH domain and the stalk. Using a longer (∼16 Å) cross-linking reagent, MTS-11-MTS, we were able to generate intermolecular cross-links in constructs bearing cysteine residues near the dimer interface (E341C/K694C and E482C/T676C). These presumably dimeric species migrated at apparent molecular masses well above the 200 kDa marker.
Figure 2.
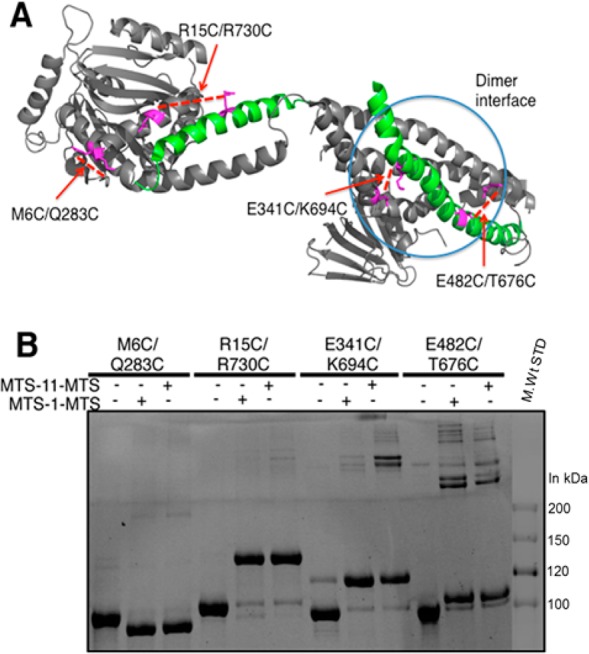
(A) Cysteine pairs within cross-linkable distances are indicated on the structure of Dyn1 (Protein Data Bank entry 3SNH). Mutants E341C/K694C and E482C/T676C also lay within the oligomerization interface, serving as positive controls for dimerization. (B) Nonreducing SDS–PAGE showing migration of cross-linked species obtained using MTS-1-MTS (3.6 Å) or MTS-11-MTS (∼16 Å).
It is well-known that protein tertiary structure packing and disulfide bond formation can alter the migration properties during SDS–PAGE due to altered SDS binding.13 For instance, gel shifts and the resulting slowly migrating species have been observed in tripartite motif (TRIM) protein upon cross-linking.14 Similarly, the E. coli outer membrane protein (ompA) shows altered migration depending on the extent of denaturation of its β-barrel packing and the amount of SDS bound.15 In the case of dynamin, the GED domain packs against the middle domain, forming a helical bundle via strong hydrophobic interactions, which would be locked in place by cross-linking. Considering these ambiguities and the fact that the R15C/R730C cross-linked species migrated significantly slower than the others, it remained possible, although unlikely given the extended, M-shaped structure of dynamin dimers,8,9 that a domain swap occurred between the stalk and BSE.
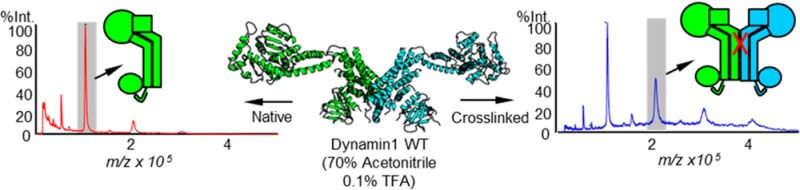
To unambiguously determine the oligomeric state of the cross-linked dynamin species, we subjected them to MALDI (matrix-assisted laser desorption ionization) Mega-time of flight (SCAAC, University of Texas at Arlington, Arlington, TX), which is compatible with large macromolecules (10–1500 kDa) and provides accurate mass determination in the range of our expected products. Proteins were prepared at high concentrations and desalted to remove buffer/salt interference and obtain a good signal-to-noise ratio. Most noncovalent interactions are disrupted under the desalting solvent conditions [70% acetonitrile and 0.1% trifluoroacetic acid (TFA) at pH ∼2.0] used for sample preparation. Ionization further disrupts noncovalent associations.16 As a positive control for the detection of dynamin dimers by mass spectrometry, we cross-linked wild-type (WT) dynamin, which contains seven cysteines, with MTS-2-MTS to generate a minor dimeric population (Figure 3A, inset). We observed a clear increase in a 2 × [M + H]+ peak, with an apparent molecular mass of 201680 (as well as apparent 300 and 400 kDa species corresponding to trimers and tetramers, respectively), in the cross-linked WT sample compared to non-cross-linked control, which exhibited the expected predominant molecular mass of 100111 Da (compare blue and red traces in Figure 3A). These data confirm that Mega-time of flight was indeed capable of detecting and distinguishing cross-linked dimeric forms of dynamin. In contrast, there was no shift in the molecular mass of the cross-linked DynRCL(R15C/R730C) protein (Figure 3B) relative to that of the non-cross-linked control, indicating that despite the reduced extent of migration on SDS–PAGE this species also corresponded to an intramolecular cross-linked monomer.
Figure 3.
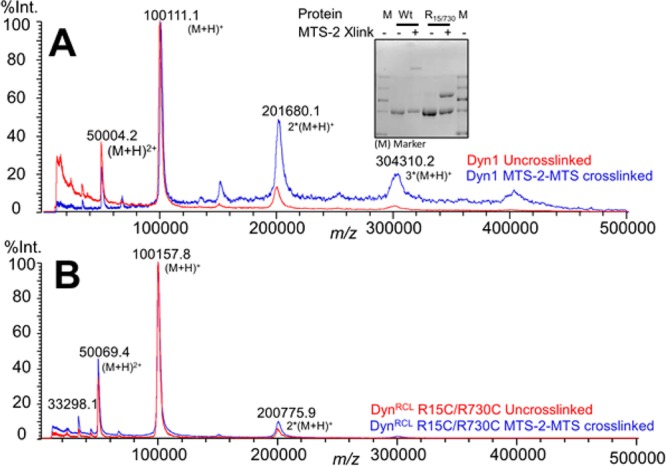
Matrix-assisted laser desorption ionization (MALDI) Mega-time of flight analysis of cross-linked and non-cross-linked dynamin samples. (A) Cross-linking wild-type Dyn1 with MTS-2-MTS generates a minor high-molecular mass band (inset) that is readily detected as a dimer peak via MALDI mass spectrometry (201680 kDa) compared to non-cross-linked control (blue vs red). (B) Cross-linking DynRCL(R15C/R730C) does not result in any change in the MALDI spectra (blue) compared to that of the un-cross-linked sample (red). The inset is a SDS–PAGE gel showing cross-linked and un-cross-linked proteins used for MALDI mass spectrometry.
These mass spectrometry data, together with results obtained by varying the cross-linking sites, unambiguously establish that GED interacts in cis with the middle and G domains of the same polypeptide and that dynamin is not domain-swapped. This resolves the controversy between high-resolution structural studies8,9 and previous conclusions based on cross-linking across the G domain–GED interface.4 Importantly, these results in no way alter the main conclusions regarding the nucleotide-driven conformational changes in the BSE reported by Chappie et al.4
Acknowledgments
We thank the Shimadzu Center for Advanced Analytical Chemistry (University of Texas at Arlington) for MALDI Mega-time of flight access, Vasyl Lukiyanchuk for the design of dicysteine constructs, and Kim Reed and Aparna Mohanakrishnan for technical assistance.
Supporting Information Available
Methods and supplemental Figure S1. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
S.S. conducted all the experiments, prepared the figures, and drafted the manuscript. J.-P.M. generated the DynRCL double-cysteine constructs. S.L.S. designed the experiments, helped in interpreting the results, and wrote the manuscript.
This work was supported by National Institutes of Health Grant R01GM42455 and Welch Foundation Grant I-1823 to S.L.S. J-P.M. was supported by a postdoctoral fellowship from the Academy of Finland.
The authors declare no competing financial interests.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Schmid S. L.; Frolov V. A. (2011) Dynamin: Functional Design of a Membrane Fission Catalyst. Annu. Rev. Cell Dev. Biol. 27, 79–105. [DOI] [PubMed] [Google Scholar]
- Ferguson S. M.; De Camilli P. (2012) Dynamin, a membrane-remodelling GTPase. Nat. Rev. Mol. Cell Biol. 13(2), 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnyrova A. V.; Bashkirov P. V.; Akimov S. A.; Pucadyil T. J.; Zimmerberg J.; Schmid S. L.; Frolov V. A. (2013) Geometric Catalysis of Membrane Fission Driven by Flexible Dynamin Rings. Science 339, 1433–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie J. S.; Mears J. A.; Fang S.; Leonard M.; Schmid S. L.; Milligan R. A.; Hinshaw J. E.; Dyda F. (2011) A Pseudoatomic Model of the Dynamin Polymer Identifies a Hydrolysis-Dependent Powerstroke. Cell 147, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie J. S.; Acharya S.; Liu Y.-W.; Leonard M.; Pucadyil T. J.; Schmid S. L. (2009) An Intramolecular Signaling Element that Modulates Dynamin Function In Vitro and In Vivo. Mol. Biol. Cell 20, 3561–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B. D.; Yarar D.; Schmid S. L. (2004) An assembly-incompetent mutant establishes a requirement for dynamin self-assembly in clathrin-mediated endocytosis in vivo. Mol. Biol. Cell 15, 2243–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R.; Surka M.; Chappie J.; Fowler D. M.; Foss T. R.; Song B. D.; Schmid S. L. (2007) The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J. 24, 559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faelber K.; Posor Y.; Gao S.; Held M.; Roske Y.; Schulze D.; Haucke V.; Noe F.; Daumke O. (2011) Crystal structure of nucleotide-free dynamin. Nature 477, 556–560. [DOI] [PubMed] [Google Scholar]
- Ford M. G.; Jenni S.; Nunnari J. (2011) The crystal structure of dynamin. Nature 477, 561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie J. S.; Acharya S.; Leonard M.; Schmid S. L.; Dyda F. (2010) G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature 465, 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzer W. J.; Hartl F. U. (1997) Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature 388, 343–349. [DOI] [PubMed] [Google Scholar]
- Chappie J. S.; Dyda F. (2013) Building a fission machine: Structural insights into dynamin assembly and activation. J. Cell Sci. 126, 2773–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath A.; Glibowicka M.; Nadeau V. G.; Chen G.; Deber C. M. (2009) Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 1760–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar A. K.; Diaz-Griffero F.; Li Y.; Li X.; Sodroski J. (2008) Biochemical and Biophysical Characterization of a Chimeric TRIM21-TRIM5 α Protein. J. Virol. 82, 11669–11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi S.; Kameyama K.; Takagi T. (1998) Characterization of a heat modifiable protein, Escherichia coli outer membrane protein OmpA in binary surfactant system of sodium dodecyl sulfate and octylglucoside. Biochim. Biophys. Acta 1375, 101–109. [DOI] [PubMed] [Google Scholar]
- Heck A. J. R.; van den Heuvel R. H. H. (2004) Investigation of intact protein complexes by mass spectrometry. Mass Spectrom. Rev. 23, 368–389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


