Abstract
We previously developed reporter-peptide nucleic acid (PNA)-peptides for sequence-specific radioimaging and fluorescence imaging of particular mRNAs in cells and tumors. However, a direct test for PNA-peptide hybridization with RNA in the cytoplasm would be desirable. Thiazole orange (TO) dye at the 5′ end of a hybridization agent shows a strong increase in fluorescence quantum yield when stacked upon a 5′ terminal base pair, in solution and in cells. We hypothesized that hybridization agents with an internal TO could distinguish a single base mutation in RNA. Thus, we designed KRAS2 PNA-IGF1 tetrapeptide agents with an internal TO adjacent to the middle base of the 12th codon, a frequent site of cancer-initiating mutations. Our molecular dynamics calculations predicted a disordered bulge with weaker hybridization resulting from a single RNA mismatch. We observed that single-stranded PNA-IGF1 tetrapeptide agents with an internal TO showed low fluorescence, but fluorescence escalated 5–6-fold upon hybridization with KRAS2 RNA. Circular dichroism melting curves showed ∼10 °C higher Tm for fully complementary vs single base mismatch TO-PNA-peptide agent duplexes with KRAS2 RNA. Fluorescence measurements of treated human lung cancer cells similarly showed elevated cytoplasmic fluorescence intensity with fully complementary vs single base mismatch agents. Sequence-specific elevation of internal TO fluorescence is consistent with our hypothesis of detecting cytoplasmic PNA-peptide:RNA hybridization if a mutant agent encounters the corresponding mutant mRNA.
Introduction
Lung cancer kills more people than any other form of cancer. Cancer cell growth depends on high expression of cancer genes.1 As lung cancer progresses, more and more characteristic cancer genes turn on in transformed lung cells, driving malignant cancer cell growth, such as EGFR and KRAS2.2 Overexpression of epithelial growth factor receptor (EGFR) stimulates lung cancer cell growth. However, 70% of lung cancer patients fail to respond to tyrosine kinase inhibitors (TKIs) directed against EGFR.3 Similarly, 76% of lung cancer patients fail to respond to anti-EGFR antibodies.4KRAS2 mutation, particularly in the 12th codon, makes cancer cell proliferation through the Ras-Raf-MEK pathway independent of EGFR activity.2 As a result, treating lung cancer with anti-EGFR antibodies or TKIs fails when KRAS2 has been mutated.5 PCR of biopsy tissue is the current gold standard for determination of KRAS2 mutation.6 A noninvasive imaging method would reduce patient discomfort and stress, as well as reduce healthcare costs.
Single base mutations in DNA occur frequently in nature and variations in the genome can affect the development and progression of certain diseases. Mutations in specific genes can result in proteins with altered function.7 An example of this phenomenon is oncogene activation in cancers.7 Nucleic acid probes that are designed to target oncogenes are usually only capable of recognizing one distinct sequence. In the case where several different single base substitutions occur in one location on a particular gene, single mismatches can lead to severe reduction in oligonucleotide targeting efficacy.
Noninvasive radioimaging of gene expression by detection of specific mutant or wild-type mRNAs in cells to diagnose cancer and other diseases in living systems has been achieved with antisense oligonucleotides.8 Specific imaging of target gene mRNA requires multiple steps: probe permeation into tissue, endocytosis into the cells, probe hybridization with the target mRNA, probe:mRNA accumulation in the specific cells, and effluxing of unbound probes. Nonhybridized probes must efflux from cells with little or no expression of the target mRNA in order to permit a specific image in the targeted cells.
Peptide nucleic acids (PNA) are a useful nuclease-resistant DNA derivative for the purpose of mRNA imaging. PNA includes a N-(2-aminoethyl)glycine backbone with DNA bases attached to the α-N.9 With such a structure, the bases are spaced appropriately to hybridize with DNA/RNA via Watson–Crick hydrogen bonding.10 PNA is electrically neutral, resulting in rapid and stronger hybridization compared to DNA:DNA or DNA:RNA duplexes. The stability of PNAs in biological fluids and their low cytotoxicity make them attractive for use in human diagnostic and therapeutic applications.11
Previously, we pioneered reporter-peptide nucleic acid (PNA)-spacer-insulin-like growth factor 1 (IGF1) tetrapeptides to enable IGF1R-mediated cellular uptake,8 followed by single-mismatch specific PET imaging of CCND1 mRNA12 and HER2 mRNA13 in breast cancer xenografts, and mutant KRAS2 mRNA in pancreatic cancer xenografts14 and spontaneous transgenic lung cancer.15 Mismatches lowered tumor signals to background. Incubation with recombinant IGF1 also lowered tumor signals to background.12 But a direct test for PNA hybridization with RNA in the cytoplasm is desirable.
Seitz and co-workers utilize thiazole orange (TO) dye as a surrogate base for detection of DNA/RNA hybridization in cells.16−19 Free thiazole orange has a low fluorescence quantum yield, but intercalation into DNA induces a high quantum yield.20 The single-stranded TO probe shows low fluorescence because the lack of planarity around the central methine bridge minimizes delocalization and directs the excited state to nonfluorescent decay modes. Upon TO stacking with neighboring nucleobases in the formed probe-target duplex, however, the TO rings adopt a planar conformation, favoring delocalization and maximizing fluorescence quantum yield.21
TO-PNA fluorescence enhancement occurs when TO at one end of a hybridization agent is stacked upon a matched base pair. This has facilitated applications in real-time PCR genotyping and RNA detection.22 In live cells, TO-PNA probes have been used to detect H1N1 mRNA,18 miRNA-122,23 and mutant KRAS2 mRNA.24
To apply a rigorous RNA hybridization test to our fluorophore-PNA-IGF1 tetrapeptide agents that enter cells by receptor-mediated endocytosis,25,26 traffic to the cytoplasm via early endosomes,27 and accumulate in cells with single mismatch specificity,27 we designed, modeled, synthesized, and tested a variety of KRAS2 TO-PNA-IGF1 tetrapeptide agents with the TO embedded within the PNA sequence, adjacent to the site of mutation (Figure 1).
Figure 1.

Schematic of KRAS2 PNA with internal TO hybridizing with KRAS2 RNA, resulting in TO intercalation.
Results
Design of TO-PNA-Peptide Hybridization Agents
We previously found that 12 PNA residues provided sufficient hybridization strength and specificity for KRAS2 mRNA PET imaging in tumors in mice with single mismatch specificity.14 We hypothesized that PNA 12mers with an internal TO could distinguish a single base mutation in RNA. Thus, we designed KRAS2 PNA-peptides with an internal TO adjacent to the middle base of the 12th codon, a frequent site of cancer-initiating mutations, such as G12D (Table 1; Figure 2). We further hypothesized that the introduction of a separate thiazole orange PNA residue after the initial N-GCCA of the original 12mer would provide greater stability than the 12mer with TO substituting for A. Hence, 13mer PNA-IGF1 tetrapeptides were also designed (Table 1).
Table 1.
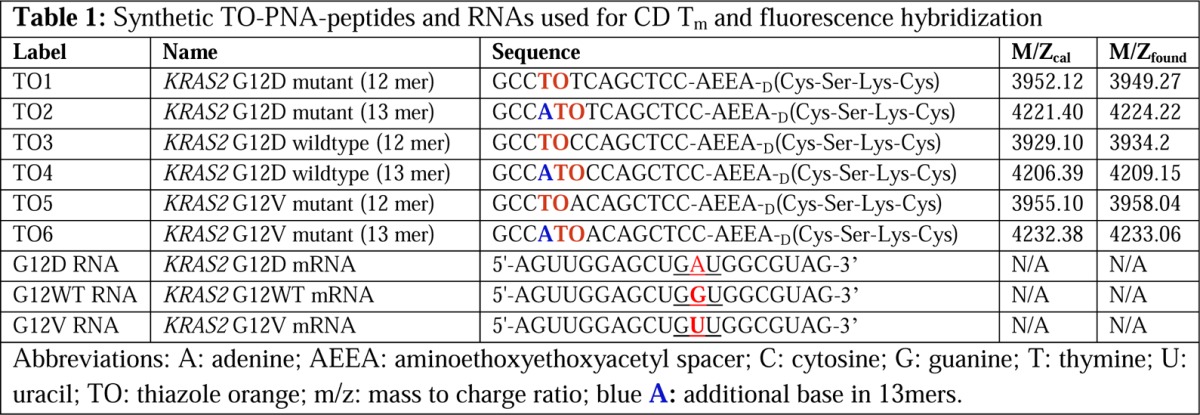
Figure 2.

Structure of TO1, KRAS2 PNA-IGF1 tetrapeptide with internal TO specific for KRAS2 G12D mRNA.
One of the primary requirements for an oligonucleotide analogue to be successful as an antigene/antisense agent is for it to be taken up by the cells in reasonable quantity so that it can reach its target in sufficient concentration. Since PNAs suffer from poor cellular uptake they have not been well developed as an antigene/antisense therapeutic agents.28,29 To improve cellular uptake, PNAs have been conjugated with cell-penetrating positively charged peptides with homology to nuclear localization sequences such as transportan, penetratin, and TAR-binding peptides,30 or the SV40 nuclear localization sequence itself.31,32
Cell penetration in these cases is not receptor-dependent and thus not cell-specific. To elevate cellular uptake of PNA by cancer cells that overexpress insulin-like growth factor receptor (IGF1R), we introduced a disulfide-cyclized d-peptide IGF1 analogue, d-(CysSerLysCys),33 at the C-terminus. The introduction of disulfide bridges into peptides allows the creation of conformational constraints that can improve the recognition between a ligand and its receptor, therefore improving biological activity.34 The flexible, hydrophilic aminoethoxyethoxyacetic acid (AEEA) spacer was introduced between the IGF1 tetrapeptide ligand and the PNA to reduce steric hindrance.8
Molecular Dynamics Predictions of PNA:RNA Duplex Stabilities
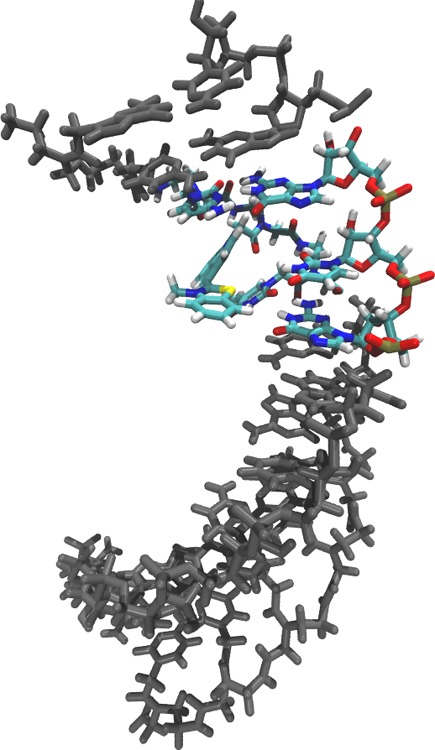
Prior to initiating chemical synthesis, the energetic effects of including a TO PNA residue in the PNA:RNA duplex were measured computationally (Figure 3). The MM-PBSA method was used to calculate the free energy of the PNA:RNA duplexes from the molecular dynamics simulations. We used normal mode approximations to calculate the entropy change of each trajectory. Typically nonlinear solvation methods can lead to overestimation of the free energy changes, complicating the calculation of absolute free energy changes.35 However, this overestimation does not impede the determination of relative free energies.36 MM-PBSA calculations were done on all the duplexes using the last 8 ns of each simulation (Supporting Information Table 2). Figure 3 illustrates the predicted destabilization induced by a mismatch just adjacent to the TO residue in a PNA:RNA duplex.
Figure 3.

MM-PBSA calculated structure of TO1, KRAS2 G12D TO-PNA hybridized to KRAS2 G12WT RNA in 0.1 M NaCl, pH 7.0. The TO residue was predicted to flip out of the helix at the site of PNA:RNA mismatch. 50 ns of simulated motion of KRAS2 G12D TO-PNA hybridized to KRAS2 G12WT RNA are shown in an mpg file (Supporting Information).
The theoretical Tm’s were calculated from the MM-PBSA results in order to compare them with the experimental Tm’s from CD temperature ramps (see below). Previously we found that using aMD to increase sampling efficiency was effective in improving the theoretical Tm’s.37 After 25 ns of cMD were carried out, an additional 50 ns of aMD for each duplex was done. Post-processing of the binding energies was done using MM-PBSA over the entire aMD run.
Synthesis and cCharacterization of TO-PNA-Peptides
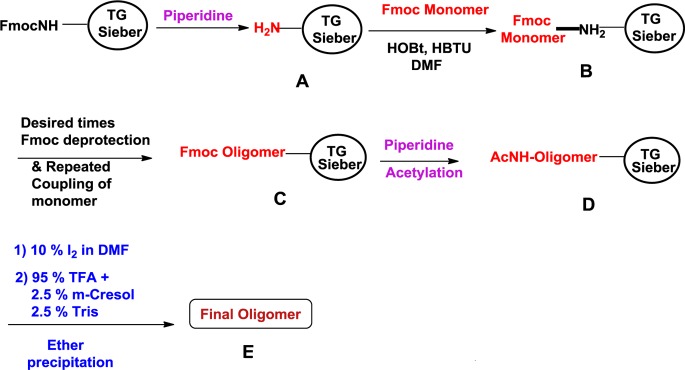
The syntheses of 12 mer/13mer TO-PNA-peptide chimeras (Table 1) were carried out (Scheme 1) by Fmoc solid phase peptide synthesis on TG Sieber resin on a Protein Technologies PS3 synthesizer at the 10 μmol scale. Cys-Cys cyclization was executed with 10 equiv of I2 in Me2NCHO for 4 h at room temperature before final cleavage.38 The PNA-peptide chimeras after cleavage from the resin were purified by preparative HPLC and characterized by MALDI-TOF (Supporting Information). The purity of TO-PNA-peptide chimeras was >95% (Supporting Information).
Scheme 1.
Thermodynamics of TO-PNA-Peptide:RNA Duplexes Measured by CD Temperature Ramps
CD spectra were obtained for TO-PNA-peptide:RNA duplexes in a JASCO J-810 spectropolarimeter with Peltier temperature control.37,39 Spectra were recorded over 320–220 nm at 25 °C. The CD spectra were recorded by using fully complementary RNA (Figure 4A) versus single base mismatch RNA (Figure 4B). CD spectra revealed that all TO-PNA-peptide:RNA duplex spectra showed a negative absorbance band around 290 nm and a strong positive CD signal around 265 nm (Figure 4) which is a distinct hallmark feature of A-like helices.40
Figure 4.
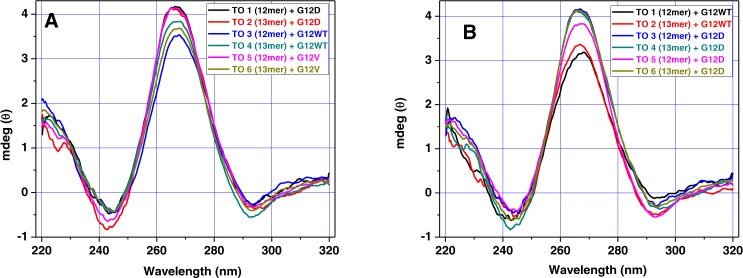
CD spectra of TO-PNA-peptide:RNA duplexes. 320–220 nm scans of annealed 1 μM TO-PNA-peptide:1 μM RNA duplexes in 100 mM Na2HPO4, 1.0 M NaCl, 5.0 mM EDTA, pH 7.2, were measured at 25 °C. (A) TO-PNA-peptide with fully complementary RNA. (B) TO-PNA-peptide with single mismatch RNA.
In the case of activated KRAS2 G12D mRNA overexpressed in multiple cancers, the Tm of the complementary KRAS2 G12D TO-PNA-peptide 12mer with a KRAS2 G12D RNA 20mer was 80 ± 2 °C, independent of the peptide ligand sequence,14 following the typical behavior of PNA:RNA duplexes.38 The CD melting studies for TO-PNA-peptides were carried out with fully complementary as well as single mismatch RNA (Figure 5 and Table 2).
Figure 5.
CD thermal denaturation plots of normalized ellipticity at 265 nm versus temperature at pH 7.2 for TO-PNA-peptide:RNA duplexes (fully complementary versus single mismatch) (A, C, E). Sigmoid decreases correlate with a duplex to single strand transition upon melting. First derivative plots determined Tm, the midpoint of the duplex to single strand transition (B, D, F).
Table 2. Melting Temperatures of TO-PNA-Peptide:RNA Duplexesa.
| complementary TO-PNA-peptide:RNA duplex |
single mismatch TO-PNA-peptide:RNA duplex |
||
|---|---|---|---|
| duplex | Tm (°C) | duplex | Tm (°C) |
| TO1 (12mer): KRAS2 G12D | 72.27 ± 0.25 | TO1 (12mer): KRAS2 G12WT | 64.30 ± 0.06 |
| TO2 (13mer): KRAS2 G12D | 70.14 ± 0.12 | TO2 (13mer): KRAS2 G12WT | 63.4 ± 0.47 |
| TO3 (12mer): KRAS2 G12WT | 73.23 ± 0.25 | TO3 (12mer): KRAS2 G12D | 60.10 ± 0.41 |
| TO4 (13mer): KRAS2 G12WT | 72.33 ± 0.12 | TO4 (13mer): KRAS2 G12D | 63.12 ± 0.36 |
| TO5 (12mer): KRAS2 G12V | 72.3 ± 0.25 | TO5 (12mer): KRAS2 G12D | 61.1 ± 0.56 |
| TO6 (13mer): KRAS2 G12V | 71.7 ± 0.21 | TO6 (13mer): KRAS2 G12D | 62.2 ± 0.36 |
CD thermal ramps from 25 to 95 °C were monitored at the peak wavelength, 265 nm. Tm of each duplex was determined from first derivative peaks ± standard deviation from n = 3 independent measurements.
The Tm for TO1 (12mer) KRAS2 G12D duplex was 72.27 ± 0.25 °C, 8 °C less than the Tm of the TO-free duplex, 80 ± 2 °C.14 The Tm for TO2 (13mer) KRAS2 G12D duplex was 70.14 ± 0.12 °C, which is 2 °C less than Tm for TO1 (12mer) KRAS2 G12D duplex.
Figure 6 shows the average CD Tm values for fully complementary versus single mismatch TO-PNA-peptide:RNA duplexes. The observed average Tm for fully complementary TO-PNA-peptide:RNA duplexes was 72.0 ± 1.0 °C, while for single mismatch TO-PNA-peptide:RNA duplexes, Tm was 62.4 ± 1.6 °C. A single mismatch, corresponding to the difference between the KRAS2 G12D sequence, vs KRAS2 G12WT or KRAS2 G12 V sequences, lowered the Tm by 9.6 ± 2.3 °C. These values revealed that introduction of TO into the PNA backbone slightly destabilized TO-PNA-peptide:RNA duplexes, with a significant difference between fully complementary versus single mismatch sequences.
Figure 6.
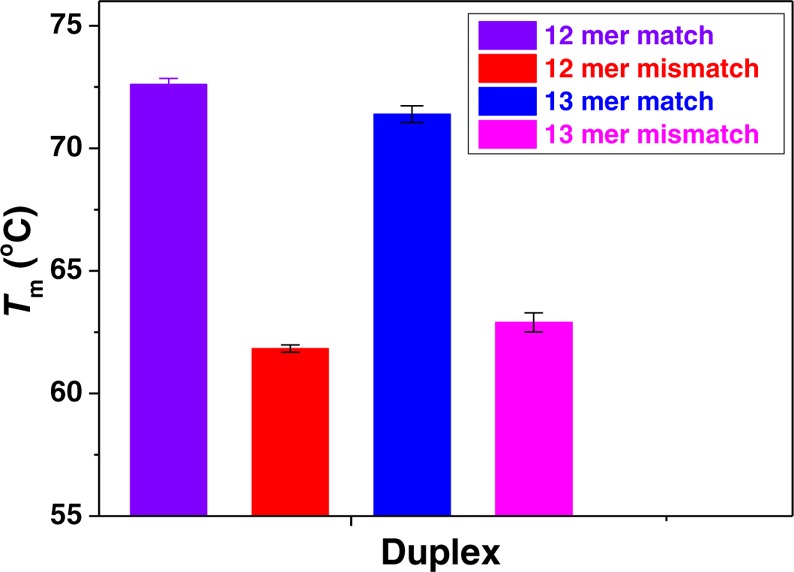
Average CD melting temperatures of TO-PNA-peptide:RNA duplexes from 25 to 95 °C were monitored at the peak wavelength 265 nm. (Violet) 12mer TO-PNA-peptide with fully complementary RNA, (Red) 12mer TO-PNA-peptide with single mismatch RNA, (Blue) 13mer TO-PNA-peptide with fully complementary RNA, (Magenta) 13mer TO-PNA-peptide with single mismatch RNA.
To test the validity of molecular dynamics predictions of TO-PNA-peptide:RNA stability, we compared n = 6 calculated Tm values vs measured CD Tm values (Figure 7). For the 12mer TO-PNA-peptides, good agreement (r2 = 0.87) between the measured CD Tm and the calculated Tm was obtained. For the 13mer TO-PNA-peptides, the correlation between the experimental and calculated Tm was poor (r2 = 0.46). This poor correlation could be derived from limitations in the MM-PBSA approximations, further sampling inefficiencies, or the accuracy of the force fields. Apparently, the strain created in the RNA backbone to accommodate the intercalated TO was not reflected in the molecular dynamics calculations.
Figure 7.

Correlation between calculated and experimental Tm. Calculated Tm from MM-PBSA for the 12mer TO-PNA-peptides (A) and 13mer TO-PNA-peptides (B) hybridized with RNA 12mers were compared with experimental Tm from CD. N = 6 values were then correlated for the 12mer (r2 = 0.87) and 13mer (r2 = 0.46).
Fluorescence Measurements of Hybridization of TO-PNA-Peptides with Synthetic RNA
We tested the ability of 12mer and 13mer TO-PNA-peptides with internal TO residues to hybridize in solution and fluoresce in the presence of complementary KRAS2 G12D RNA versus single mismatch KRAS2 G12WT RNA 20mers (Table 1). Synthetic RNAs were hybridized with TO-PNA-peptides at 37 °C and the resulting fluorescence intensity was compared between match and mismatch duplexes for 12mer and 13mer TO-PNA-peptides (Figure 8). TO-PNA-peptides alone in buffer only showed background fluorescence intensity.
Figure 8.
In vitro hybridization of 50 nM 12mer or 13mer TO-PNA-peptides with 50 nM synthetic fully complementary RNA (blue) or single mismatch RNA (red) or with no RNA (green) at 37 °C. (A) G12D TO-PNA-peptide. (B) G12WT TO-PNA-peptide. Mean values are shown for n = 3 independent measurements ± SD. ** p < 0.01, *** p < 0.005.
In the presence of fully complementary KRAS2 G12D RNA, the G12D TO-PNA-peptide 12mer showed a significant 7-fold increase in fluorescence intensity compared to the TO-PNA-peptide alone (Figure 8A). With the single mismatch KRAS2 G12WT RNA, the G12D TO-PNA-peptide 12mer showed only a 4-fold increase in fluorescence intensity (Figure 8A). Similar results were observed for G12WT TO-PNA-peptide 12mer. In the presence of fully complementary KRAS2 G12WT RNA, the G12WT TO-PNA-peptide 12mer showed a significant 5-fold increase in fluorescence intensity compared to the TO-PNA-peptide alone (Figure 8B). With the single mismatch KRAS2 G12D RNA, the G12WT TO-PNA-peptide 12mer showed only a 3-fold increase in fluorescence intensity (Figure 8B).
However, G12D TO-PNA-peptide 13mers and G12WT TO-PNA-peptide 13mers showed no significant discrimination when hybridized with match versus single mismatch KRAS2 RNA (Figure 8). The above results indicate that as candidates for fluorescence hybridization probes, TO-PNA-peptide 12mers are more promising than TO-PNA-peptide 13mers, since 12mers can detect RNAs with single mismatch sensitivity while 13mers fail. Comparable results for 5′-terminal TO-DNA/TO-PNA:DNA were reported earlier.16
Confocal Fluorescence Microscopy of Hybridization of TO-PNA-Peptides with KRAS2 mRNA in Human Lung Cancer Cells
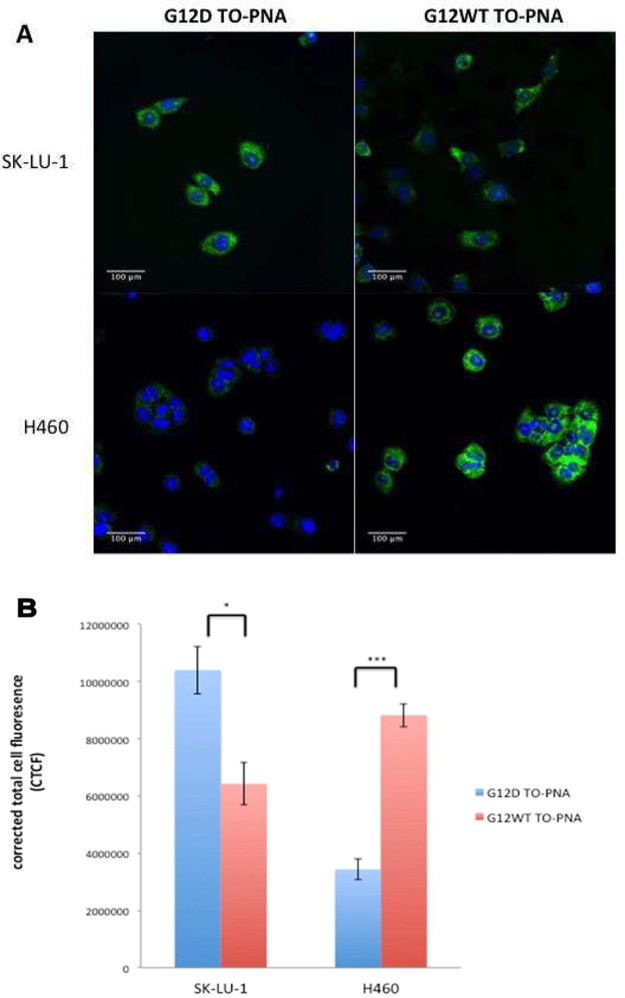
200 nM G12D and WT TO-PNA-peptide 12mers were introduced into the culture medium of SK-LU-1 KRAS2 G12D mutant lung cancer cells and H460 KRAS G12 wild type lung cancer cells. The cells were incubated 4 h at 37 °C. SK-LU-141 and H46042 cells both overexpress IGF1R. We previously observed that receptor-mediated endocytosis under these conditions results in the internalization of millions of PNA-peptides per cell, relative to the thousands of copies of overexpressed oncogene mRNAs in cancer cells.26 Excess unbound PNA-peptides efflux from cells, resulting in a dynamic equilibrium of hybridized PNA-peptides accumulated in treated cells after suitable incubation time.27
Confocal fluorescence imaging of fixed cells showed that SK-LU-1 cells treated with G12D TO-PNA-peptide showed higher cytoplasmic fluorescence intensity vs G12WT TO-PNA-peptide, while in H460 cells, G12D TO-PNA-peptide showed negligible cytoplasmic fluorescence signal compared to G12WT TO-PNA-peptide (Figure 9A). We have previously observed comparable cytoplasmic accumulation in both fixed25−27 and live27 cells, over the concentration range 4 nM – 1 μM. Therefore, TO-PNA-peptide 12mers detected complementary RNA targets in cells with single mismatch discrimination. Quantification of corrected total cell fluorescence (CTCF) of n = 9 confocal cell images agreed with the above observation that in two different cell lines, TO-PNA-peptides with matched sequences displayed higher CTCF compared to TO-PNA-peptides with one mismatched base (Figure 9B), in agreement with the lower stability of mismatched duplexes (Figure 6).
Figure 9.
Confocal fluorescent microscopy of KRAS2 G12D SK-LU-1 and KRAS2 G12WT H460 human lung cancer cells incubated with 200 nM TO-PNA-peptide 12mers after 4 h at 37 °C. (A) Fluorescent images recorded after TO excitation at 488 nm. Emission was observed at 535/40 nm. (B) Quantification of corrected total cell fluorescence (CTCF) intensity in SK-LU-1 and H460 cells treated with 200 nM G12D TO-PNA-peptide or G12WT TO-PNA-peptide 12mers. N = 9 cells were selected, three from each of three separate images. Fluorescence intensities were quantified using ImageJ. Fluorescence values were expressed as integrated density after background subtraction ± SE. * p < 0.05, *** p < 0.00001 (Student’s t test).
13mers, however, displayed no significant discrimination. The indistinguishable cytoplasmic signals agreed with the indistinguishable solution fluorescence results.
Discussion
We designed and modeled a variety of internally intercalating TO-PNA-IGF1 tetrapeptide agents that hybridized with KRAS2 RNA. Molecular dynamics calculations predicted slightly reduced thermal stability compared with normal base PNAs. In agreement with our molecular dynamics calculations, large CD Tm differences were observed between fully complementary TO-PNA:RNA duplexes versus single mismatch duplexes. We observed that the single-stranded PNA-peptide 12mer agent with an internal TO residue in place of adenine 4 showed low fluorescence, but when it hybridized with KRAS2 RNA, fluorescence escalated 5–6-fold at 37 °C. We ascribe the reduced fluorescence in the presence of a mismatched KRAS2 RNA to partial dissociation of the mismatched duplex at 37 °C.
However, the PNA-peptide 13mers with TO inserted between adenine 4 and thymine 5 showed insufficient mutant discrimination upon RNA binding. Perhaps intercalation of TO upon hybridization of the 13mer to KRAS2 RNA created excess strain on the ribose-phosphodiester backbone, limiting the achievable fluorescence quantum yield upon hybridization to RNA. Apparently, less strain was experienced by the RNA upon hybridizing with the 12mer, which required the displaced uracil to flip out of the PNA:RNA helix.
Fluorescence measurements of treated human lung cancer cells similarly showed elevated cytoplasmic accumulation of fully complementary vs single base mismatch 12mer agents. We ascribe the reduced cytoplasmic fluorescence in the presence of a mismatched KRAS2 TO-PNA-peptide to partial dissociation of the mismatched duplex at 37 °C. Sequence-specific elevation of TO fluorescence is consistent with the model of cytoplasmic TO-PNA-peptide:RNA hybridization. These studies gave direct evidence that PNA conjugated with an IGF1 receptor peptide trafficks to the cytoplasm and hybridizes specifically with a target mRNA.
For reliable clinical application of PNA-peptide agents with internal TO residues, more study is needed on the mechanism of endosome trafficking, release of agents in the cytoplasm, and efflux of unbound agent from cells. Brighter TO derivatives are desirable, ideally a near-infrared analogue, with an even greater reduction in quantum yield adjacent to a mismatch in the RNA.
Conclusion
We observed that single-stranded PNA-IGF1 tetrapeptide 12mers with an internal TO showed low fluorescence, but fluorescence escalated 5–6-fold upon hybridization with KRAS2 RNA. Circular dichroism melting curves showed ∼10 °C higher Tm for fully complementary vs single base mismatch TO-PNA agent duplexes with KRAS2 RNA. Fluorescence measurements of treated human lung cancer cells similarly showed elevated cytoplasmic fluorescence intensity with fully complementary vs single base mismatch 12mer TO-PNA agents, in agreement with the lower Tm of a mismatched duplex. Sequence-specific elevation of internal TO fluorescence is consistent with our hypothesis of detecting cytoplasmic PNA:RNA hybridization if a mutant agent encounters the corresponding mutant mRNA.
Methods
Molecular Dynamics Simulations
System equilibration and production MD simulations were performed using the Amber 12 suite of programs.43 The LeaP module of Amber 12 was used to create parameter and topology files for the MD simulations using the ff99SB force field.43,44 PNA force fields were created in our earlier study of hypoxanthine-containing wobble base PNAs.37 RNA molecules were set to predicted protonation states at pH 7.0. Na+ and Cl– counterions were added to each system to achieve neutrality and a salt concentration of 0.1 M. TIP3P water molecules were added with a minimum spacing of 30 Å (3.0 nm) from the box edges to the PNA:RNA molecules. Energy minimization on each system was performed in a two-step process. First the solute atoms were restrained while the water and ion molecules were allowed to relax over 1000 steps. The entire system was then subjected to energy minimization using the steepest descent method for the first 1000 steps, followed by the full conjugate gradient method for an additional 24 000 steps. Each molecular system was then heated to 300 K for 100 ps followed by a 50 ps constant pressure simulation to adjust the density to 1 g/mL. An additional 500 ps simulation was run prior to production simulations to allow for further temperature and pressure equilibration.
Production runs were performed using a canonical ensemble (NVT) scheme. Langevin dynamics with a collision frequency of 2.0 were used for temperature regulation, (ii) the SHAKE algorithm was used for all hydrogen atoms, and (iii) the particle mesh Ewald method was employed to treat long-range electrostatics and van der Waals forces (cutoff of 8 Å) with an integration step of 2.0 fs. All Amber 12 equilibration and production runs were performed using double precision.45 All production simulations were repeated in triplicate with random seeding for initial velocities and extended to 25 ns. Structural features were determined using the Curves+ software package,46 while visualization of trajectories was performed in VMD.47
Accelerated Molecular Dynamics Simulations
Accelerated MD (aMD) is one method to increase the sampling of a molecular system to overcome the computational cost of long time scales. aMD modifies the system’s original potential energy surface (V(r)) by adding a boost potential (ΔV(r)) when V(r) falls below a certain threshold energy (E).48 The boost potential is defined as
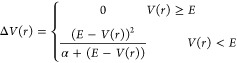 |
where α modulates the depth and roughness of the energy basins on the modified potential. In order to enhance sampling of internal and diffusive degrees of freedom, a dual boosting potential was applied to the backbone torsion angles and the overall boost potential as
where Vt(r) is the total potential of the torsional terms, Vt(r) and ΔVT(r) are the boost potentials applied to the torsional terms, and the total potential energy, respectively. The parameters were set as follows: Et = 1.2{⟨ΔVt(r)⟩}, αt = 0.20 kcal/mol, ET = 0.2 kcal/mol·number atoms, plus the ensemble averaged potential energy from the conventional MD simulations, and αT = 0.2 kcal/mol·number atoms.49,50 aMD runs simulated 50 ns.
MM-PBSA Calculations
The binding energies for each RNA:PNA duplex were calculated using the molecular mechanics-Poisson–Boltzmann surface area (MM-PBSA) method in Amber 12.50,51 The MM-PBSA method calculates the binding free energy by the free energies of solvation for the complex (ΔGduplex), PNA (ΔGPNA), and RNA (ΔGRNA).
Each term is calculated by determining the enthalpic energy of the solute using molecular mechanics (EMM), the polar solvation free energy (ΔGSOLV), the nonpolar solvation free energy (ΔGnp), and the entropic contribution (ΔS):
The enthalpic term is taken as the average over the molecular mechanics force field terms for the solute. The solute vibrational entropy is estimated using either normal-mode analysis or quasi-harmonic approximation. ΔGsolv is calculated using the Poisson–Boltzmann (PB) equation. The nonpolar term (ΔGnp) is estimated from the solvent accessible surface area (SASA).52
where γ is the surface tension, set to 0.0072 kcal/Å2. β is an offset value used to correct for the nonpolar contribution to the solvation free energy term.53 Free energy values were converted to theoretical melting temperatures using the following equation:54
where ΔH is calculated from the sum of the enthalpic terms in the MM-PBSA calculation, ΔS is the entropy value from normal-mode analysis, R is the gas constant in kcal/mol·K, and ct is the concentration (M) of each individual RNA/PNA strand in the CD experiments.
For each RNA:PNA system, MM-PBSA calculations were performed using the last 8.0 ns of the simulation with 100 ps intervals. The entropy was determined using normal-mode analysis using 50 frames over the 8.0 ns. All energy values represent 3 independent MD simulation runs for each RNA:PNA duplex. aMD MM-PBSA calculations were performed over the entire 50 ns simulation with 50 ps intervals. aMD entropy values were determined using the same method as for conventional MD (cMD) simulations. All calculations were performed with the MMPBSA.py.MPI module in Amber 11 with an ionic strength equal to 1.0 M. For MM-PBSA calculations the PB equation was solved numerically by the PBSA program included with in the AmberTools 12 module. The hydrophobic contribution was approximated by the LCPO method implemented within the Sander module.55
Synthesis of TO-PNA Monomer
The synthesis of TO-Aeg-PNA monomer (8) was achieved in seven steps (Scheme 2) by using an adaptation of previously developed methods.56−58
Scheme 2.
The synthesis began with commercially available 2-(methylthio)benzothiazole (1) which was treated with iodomethane to yield the N-alkyl compound 3. The quinoline derivative 4 was obtained by reaction of 4-methylquinoline (2) with bromoacetic acid. The condensation of N-alkyl compound 3 and the quinoline derivative 4 in the presence of Et3N yielded TO carboxylic acid 5. The TO carboxylic acid 5 was coupled with commercially available Fmoc-Aeg-OtBu (6) in the presence of PyBOP59 and HOBt gave tBu ester compound 7 with moderate yield. When compound 7 was treated with 20% CF3CO2H in CH2Cl2 to deprotect the tBu-ester group, a complex reaction mixture was observed that was difficult to purify. Hence, a new synthetic route (Scheme 3) was adopted.
Scheme 3.
The tBu group of compound 6 was deprotected by CF3CO2H followed by treatment with allyl alcohol in the presence of thionyl chloride to yield the allyl ester 9. This was coupled with TO carboxylic acid 5 in the presence of PyBOP and HOBt to give the TO allyl ester 10. Finally, the allyl protecting group in compound 10 was removed by Pd0-catalyzed allyl transfer to N-methyl aniline60 to yield TO-Aeg-PNA monomer 8 which was used in solid phase peptide synthesis.
Circular Dichroism (CD) Spectra and Thermal Melting of PNA:RNA Duplexes
CD spectra were obtained for TO-PNA-peptide:RNA duplexes using a JASCO J-810 spectropolarimeter with Peltier temperature control.37,39 Spectra were recorded over 320–220 nm at 25 °C. Gel-purified RNA icosamers were purchased from Thermo Scientific (Dharmacon). Prior to measurement, samples were heated to 95 °C for 10 min and then cooled for 10 min. All spectra were subjected to baseline correction. Final spectra were calculated as the average of 3 independent measurements. Stock RNA samples were suspended in H2O and TO-PNA-peptide molecules in 20 mM sodium phosphate buffer (pH 7.2), TO-PNA-peptide:RNA solutions were diluted in 20 mM Na2HPO4, 1.0 M NaCl, 5 mM EDTA, pH 7.2, to reach a final concentration of 1 μM:1 μM. Thermal denaturation experiments were carried out by increasing the temperature from 25 to 95 °C at a rate of 0.5 °C/min while monitoring the CD signal at 265 nm. Tm values were determined based on the assumption of a two-state model. Melting curves were normalized by subtracting baseline slopes.54 The first derivative of the melting curves was computed with respect to the temperature (dθ/dT), and the maximum was used to determine the Tm. We have previously reported no perturbation of Tm due to peptide moieties on the PNAs.38
Fluorescence Intensity Measurements of TO-PNA-Peptide:RNA Duplexes
Hybridization was carried out of TO-PNA-peptides with synthetic RNAs. 50 nM G12D TO-PNA-peptide and G12WT TO-PNA-peptide were incubated with 50 nM RNAs (Thermo Scientific Dharmacon) in 3 mM Na2HPO4 buffer (Molecular Research Center) for 1 h at 37 °C. Single stranded TO-PNA-peptides were incubated with buffer only. At the end of 1 h incubation, fluorescent intensities were measured with a plate reader (infiniteM200PRO, TECAN). Background intensity values consisting of only buffer were subtracted from every fluorescence measurement. Each well was excited at 495 nm and the fluorescence intensities were collected at 531 nm.
Confocal Fluorescence Microscopy of TO-PNA-Peptide:RNA Duplexes in Cells
Fluorescent imaging was carried out of TO-PNA-peptides in KRAS wild type and mutant cells. SK-LU-1 cells (ATCC) were cultured in EMEM medium with 10% fetal bovine serum (FBS). H460 cells (ATCC) were cultured in RPMI 1640 medium with 10% FBS. 20 000 cells were seeded in an 8-chamber well slide (Millipore) in 10% growth medium and incubated overnight. 200 nM TO-PNA-peptides were added to each well in serum-free medium and were incubated for 4 h at 37 °C to allow equilibration of bound TO-PNA-peptide. At the end of incubation, cells were washed 3× with PBS containing Ca2+ and Mg2+ (Fisher) followed by fixation with 4% paraformaldehyde in PBS. Cells were then washed one more time with PBS containing Ca2+ and Mg2+. Chambers were removed and the slides were mounted with Prolong Gold Antifade reagent with DAPI (Life Technologies). All images were taken on a Nikon C1 Plus two point-scanning laser confocal microscope using 40× oil objective. Excitation was at 488 nm, and emission was observed at 535/40 nm.
Quantification of Confocal Images Using ImageJ
Nine cells were selected individually using drawing tools in ImageJ, followed by measurement of the area and the integrated density of each cell. Three different background areas were selected for each image and were measured for mean pixel values. The corrected total cell fluorescence for each cell = integrated density of the cell – area of the cell × mean pixel values of three background areas. Standard errors were calculated, and significance was assessed by Student’s t test.
Acknowledgments
We thank Dr. Yolanda Covarrubias for assistance with confocal microscopy in the Kimmel Cancer Center Molecular Imaging Facility, supported in part by NCI P30 CA56036. This work was supported by NIH 1 R01 CA148565 to E.W. IP owned by EW/MLT, licensed to MTTI.
Supporting Information Available
Methods and analytical data for synthesis of thiazole orange-PNA monomer and solid phase assembly of PNA-AEEA-IGF1 tetrapeptide sequences containing thiazole orange are shown. 50 ns of simulated motion of KRAS2 G12D TO-PNA hybridized to KRAS2 G12WT RNA are shown in an mpg file, which illustrates the TO residue flipping out of the helix at the site of PNA:RNA mismatch. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
Mahesh V. Sonar, Department of Chemistry, State University of New York, Binghamton NY 13902
Author Present Address
Matthew E. Wampole, Discovery Science, Thomson-Reuters, Boston MA 02210
The authors declare the following competing financial interest(s): We wish to inform the Editor of potential conflicts of interest. Prof. Eric Wickstrom and Prof. Mathew L. Thakur hold shares in GeneSeen LLC, which might ultimately benefit from the results of this investigation, but did not support the work.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Bishop J. M. (1991) Molecular themes in oncogenesis. Cell 64, 235–48. [DOI] [PubMed] [Google Scholar]
- Riely G. J.; Kris M. G.; Rosenbaum D.; Marks J.; Li A.; Chitale D. A.; Nafa K.; Riedel E. R.; Hsu M.; Pao W.; Miller V. A.; Ladanyi M. (2008) Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin. Cancer Res. 14, 5731–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwish S.; Ludovini V.; Pistola L.; Bianconi F.; Betti M.; Chiari R.; Sidoni A.; Giuffrida D.; Tofanetti F. R.; Flacco A.; Crino L. (2008) EGFR, KRAS, PIK3CA mutations and response to tyrosine kinase inhibitors (TKIs) in advanced NSCLC patients. J. Clin. Oncol. (Meeting Abstracts) 26, 22003. [Google Scholar]
- Lynch T.; Lilenbaum R.; Bonomi P.; Hanna N.; Ansari R.; Govindan R.; Janne P. A. (2004) Phase II trial of cetuximab as therapy for recurrent non-small cell lung cancer (NSCLC). J. Clin. Oncol. 22, 7084. [DOI] [PubMed] [Google Scholar]
- Pao W.; Wang T. Y.; Riely G. J.; Miller V. A.; Pan Q.; Ladanyi M.; Zakowski M. F.; Heelan R. T.; Kris M. G.; Varmus H. E. (2005) KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J. Y.; Kim J. H.; Lee Y. H.; Chung K. Y.; Kim S. K.; Gong S. J.; You N. C.; Chung H. C.; Roh J. K.; Kim B. S. (1997) Correlation between K-ras gene mutation and prognosis of patients with nonsmall cell lung carcinoma. Cancer 79, 462–7. [DOI] [PubMed] [Google Scholar]
- Bertram J. S. (2000) The molecular biology of cancer. Mol. Aspects Med. 21, 167–223. [DOI] [PubMed] [Google Scholar]
- Tian X.; Chakrabarti A.; Amirkhanov N. V.; Aruva M. R.; Zhang K.; Mathew B.; Cardi C.; Qin W.; Sauter E. R.; Thakur M. L.; Wickstrom E. (2005) External imaging of CCND1, MYC, and KRAS oncogene mRNAs with tumor-targeted radionuclide-PNA-peptide chimeras. Ann. N.Y. Acad. Sci. 1059, 106–44. [DOI] [PubMed] [Google Scholar]
- Nielsen P. E.; Egholm M.; Berg R. H.; Buchardt O. (1991) Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 254, 1497–500. [DOI] [PubMed] [Google Scholar]
- Egholm M.; Buchardt O.; Christensen L.; Behrens C.; Freier S. M.; Driver D. A.; Berg R. H.; Kim S. K.; Norden B.; Nielsen P. E. (1993) PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature 365, 566–568. [DOI] [PubMed] [Google Scholar]
- Demidov V. V.; Potaman V. N.; Frank-Kamenetskii M. D.; Egholm M.; Buchard O.; Sonnichsen S. H.; Nielsen P. E. (1994) Stability of peptide nucleic acids in human serum and cellular extracts. Biochem. Pharmacol. 48, 1310–3. [DOI] [PubMed] [Google Scholar]
- Tian X.; Aruva M. R.; Zhang K.; Cardi C. A.; Thakur M. L.; Wickstrom E. (2007) PET imaging of CCND1 mRNA in human MCF7 estrogen receptor-positive breast cancer xenografts with an oncogene-specific [64Cu]DO3A-PNA-peptide radiohybridization probe. J. Nucl. Med. 48, 1699–1707. [DOI] [PubMed] [Google Scholar]
- Paudyal B.; Zhang K.; Chen C.-P.; Mehta N.; Wampole M. E.; Mitchell E. P.; Gray B. D.; Mattis J. A.; Pak K. Y.; Thakur M. L.; Wickstrom E. (2013) Determining efficacy of breast cancer therapy by PET imaging of HER2 mRNA. Nucl. Med. Biol. 40, 994–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A.; Zhang K.; Aruva M. R.; Cardi C. A.; Opitz A. W.; Wagner N. J.; Thakur M. L.; Wickstrom E. (2007) Radiohybridization PET imaging of KRAS G12D mRNA expression in human pancreas cancer xenografts with [64Cu]DO3A-peptide nucleic acid-peptide nanoparticles. Cancer Biol. Ther. 6, 948–956. [DOI] [PubMed] [Google Scholar]
- Chen C.-P., Sethi D., Wampole M. E., Sanders J. M., Jin Y.-Y., Thakur M. L., and Wickstrom E. (2012) in XX International Roundtable on Nucleosides, Nucleotides, and Nucleic Acids, Montreal, Canada.
- Kohler O.; Seitz O. (2003) Thiazole orange as fluorescent universal base in peptide nucleic acids. Chem. Commun. (Camb) 2938–9. [DOI] [PubMed] [Google Scholar]
- Hovelmann F.; Bethge L.; Seitz O. (2012) Single labeled DNA FIT probes for avoiding false-positive signaling in the detection of DNA/RNA in qPCR or cell media. ChemBioChem 13, 2072–81. [DOI] [PubMed] [Google Scholar]
- Kummer S.; Knoll A.; Socher E.; Bethge L.; Herrmann A.; Seitz O. (2011) Fluorescence imaging of influenza H1N1 mRNA in living infected cells using single-chromophore FIT-PNA. Angew. Chem., Int. Ed. 50, 1931–4. [DOI] [PubMed] [Google Scholar]
- Socher E.; Bethge L.; Knoll A.; Jungnick N.; Herrmann A.; Seitz O. (2008) Low-noise stemless PNA beacons for sensitive DNA and RNA detection. Angew. Chem., Int. Ed. 47, 9555–9. [DOI] [PubMed] [Google Scholar]
- Glazer A. N.; Rye H. S. (1992) Stable dye-DNA intercalation complexes as reagents for high-sensitivity fluorescence detection. Nature 359, 859–61. [DOI] [PubMed] [Google Scholar]
- Karunakaran V.; Perez Lustres J. L.; Zhao L.; Ernsting N. P.; Seitz O. (2006) Large dynamic Stokes shift of DNA intercalation dye Thiazole Orange has contribution from a high-frequency mode. J. Am. Chem. Soc. 128, 2954–62. [DOI] [PubMed] [Google Scholar]
- Tonelli A.; Tedeschi T.; Germini A.; Sforza S.; Corradini R.; Medici M. C.; Chezzi C.; Marchelli R. (2011) Real time RNA transcription monitoring by Thiazole Orange (TO)-conjugated Peptide Nucleic Acid (PNA) probes: norovirus detection. Mol. Biosyst. 7, 1684–92. [DOI] [PubMed] [Google Scholar]
- Torres A. G.; Fabani M. M.; Vigorito E.; Williams D.; Al-Obaidi N.; Wojciechowski F.; Hudson R. H.; Seitz O.; Gait M. J. (2012) Chemical structure requirements and cellular targeting of microRNA-122 by peptide nucleic acids anti-miRs. Nucleic Acids Res. 40, 2152–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam Y.; Rubinstein A.; Nissan A.; Halle D.; Yavin E. (2012) Detection of endogenous K-ras mRNA in living cells at a single base resolution by a PNA molecular beacon. Mol. Pharm. 9, 685–93. [DOI] [PubMed] [Google Scholar]
- Basu S.; Wickstrom E. (1997) Synthesis and characterization of a peptide nucleic acid conjugated to a D-peptide analog of insulin-like growth factor 1 for increased cellular uptake. Bioconjugate Chem. 8, 481–488. [DOI] [PubMed] [Google Scholar]
- Tian X.; Aruva M. R.; Qin W.; Zhu W.; Duffy K. T.; Sauter E. R.; Thakur M. L.; Wickstrom E. (2004) External imaging of CCND1 cancer gene activity in experimental human breast cancer xenografts with 99mTc-peptide-peptide nucleic acid-peptide chimeras. J. Nucl. Med. 45, 2070–2082. [PubMed] [Google Scholar]
- Sethi D.; Chen C.-P.; Jing R.-Y.; Thakur M. L.; Wickstrom E. (2012) Fluorescent peptide-PNA chimeras for imaging monoamine oxidase A mRNA in neuronal cells. Bioconjugate Chem. 23, 158–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonham M. A.; Brown S.; Boyd A. L.; Brown P. H.; Bruckenstein D. A.; Hanvey J. C.; Thomson S. A.; Pipe A.; Hassman F.; Bisi J. E.; et al. (1995) An assessment of the antisense properties of RNase H-competent and steric-blocking oligomers. Nucleic Acids Res. 23, 1197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray G. D.; Basu S.; Wickstrom E. (1997) Transformed and immortalized cellular uptake of oligodeoxynucleoside phosphorothioates, 3′-alkylamino oligodeoxynucleotides, 2′-O-methyl oligoribonucleotides, oligodeoxynucleoside methylphosphonates, and peptide nucleic acids. Biochem. Pharmacol. 53, 1465–1476. [DOI] [PubMed] [Google Scholar]
- Soomets U.; Hallbrink M.; Langel U. (1999) Antisense properties of peptide nucleic acids. Front. Biosci. 4, D782–6. [DOI] [PubMed] [Google Scholar]
- Branden L. J.; Mohamed A. J.; Smith C. I. (1999) A peptide nucleic acid-nuclear localization signal fusion that mediates nuclear transport of DNA. Nat. Biotechnol. 17, 784–7. [DOI] [PubMed] [Google Scholar]
- Cutrona G.; Carpaneto E. M.; Ulivi M.; Roncella S.; Landt O.; Ferrarini M.; Boffa L. C. (2000) Effects in live cells of a c-myc anti-gene PNA linked to a nuclear localization signal. Nat. Biotechnol. 18, 300–3. [DOI] [PubMed] [Google Scholar]
- Pietrzkowski Z.; Wernicke D.; Porcu P.; Jameson B.; Baserga R. (1992) Inhibition of cellular proliferation by peptide analogues of insulin-like growth factor 1. Cancer Res. 52, 6447–6451. [PubMed] [Google Scholar]
- Bolin D. R.; Cottrell J.; Garippa R.; O’Neill N.; Simko B.; O’Donnell M. (1993) Structure-activity studies of vasoactive intestinal peptide (VIP): cyclic disulfide analogs. Int. J. Pept. Protein Res. 41, 124–32. [DOI] [PubMed] [Google Scholar]
- Hou T.; Wang J.; Li Y.; Wang W. (2011) Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Modeling 51, 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brice A. R.; Dominy B. N. (2011) Analyzing the robustness of the MM/PBSA free energy calculation method: application to DNA conformational transitions. J. Comput. Chem. 32, 1431–40. [DOI] [PubMed] [Google Scholar]
- Sanders J. M.; Wampole M. E.; Chen C.-P.; Sethi D.; Singh A.; Dupradeau F. Y.; Wang F.; Gray B. D.; Thakur M. L.; Wickstrom E. (2013) Effects of hypoxanthine substitution in peptide nucleic acids targeting KRAS2 oncogenic mRNA molecules: theory and experiment. J. Phys. Chem. B 117, 11584–11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X.; Wickstrom E. (2002) Continuous solid-phase synthesis and disulfide cyclization of peptide-PNA-peptide chimeras. Org. Lett. 4, 4013–4016. [DOI] [PubMed] [Google Scholar]
- Chakrabarti A.; Desai P.; Wickstrom E. (2004) Transposon Tn7 protein TnsD binding to Escherichia coli attTn7 DNA and its eukaryotic orthologs. Biochemistry 43, 2941–2946. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr. (2001) Biophysical analysis of nucleic acids. In Current Protocols in Nucleic Acid Chemistry Unit 7.1, Wiley Online Library. [DOI] [PubMed] [Google Scholar]
- Loboda A., Nebozhyn M., Dai H. (2014) Methods of predicting cancer cell response to therapeutic agents, U.S. Patent No 2014 0030255 A1.
- Sun Y.; Zheng S.; Torossian A.; Speirs C. K.; Schleicher S.; Giacalone N. J.; Carbone D. P.; Zhao Z.; Lu B. (2012) Role of insulin-like growth factor-1 signaling pathway in cisplatin-resistant lung cancer cells. Int. J. Radiat. Oncol., Biol., Phys. 82, e563–e572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D. A., Darden T. A., Cheatham T. E. III, Simmerling C. L., Wang J., Duke R. E., Luo R., Walker R. C., Zhang W., Merz K. M., Roberts B., Hayik S., Roitberg A., Seabra G., Swails J., Goetz A. W., Kolossvai I., Wong K. F., Paesani F., Vanicek J., Wolf R. M., Liu J., Wu X., Brozell S. R., Steinbrecher T., Gohlke H., Cai Q., Ye X., Wang J., Hsieh M.-J., Cui G., Roe D. R., Mathews D. H., Seetin M. G., Salomon-Ferrer R., Sagui C., Babin V., Luchko T., Gusarov S., Kovalenko A., and Kollman P. A. (2012) Amber 12, University of California, San Francisco.
- Duan Y.; Wu C.; Chowdhury S.; Lee M. C.; Xiong G.; Zhang W.; Yang R.; Cieplak P.; Luo R.; Lee T.; Caldwell J.; Wang J.; Kollman P. (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24, 1999–2012. [DOI] [PubMed] [Google Scholar]
- Gotz A. W.; Williamson M. J.; Xu D.; Poole D.; Le Grand S.; Walker R. C. (2012) Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 8, 1542–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery R.; Moakher M.; Maddocks J. H.; Petkeviciute D.; Zakrzewska K. (2009) Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Res. 37, 5917–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graphics 14(33–8), 27–8. [DOI] [PubMed] [Google Scholar]
- Hamelberg D.; Mongan J.; McCammon J. A. (2004) Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J. Chem. Phys. 120, 11919–29. [DOI] [PubMed] [Google Scholar]
- Hamelberg D.; de Oliveira C. A.; McCammon J. A. (2007) Sampling of slow diffusive conformational transitions with accelerated molecular dynamics. J. Chem. Phys. 127, 155102. [DOI] [PubMed] [Google Scholar]
- Kollman P. A.; Massova I.; Reyes C.; Kuhn B.; Huo S.; Chong L.; Lee M.; Lee T.; Duan Y.; Wang W.; Donini O.; Cieplak P.; Srinivasan J.; Case D. A.; Cheatham T. E. III. (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 33, 889–97. [DOI] [PubMed] [Google Scholar]
- Gohlke H.; Case D. A. (2004) Converging free energy estimates: MM-PB(GB)SA studies on the protein-protein complex Ras-Raf. J. Comput. Chem. 25, 238–50. [DOI] [PubMed] [Google Scholar]
- Wereszczynski J.; McCammon J. A. (2012) Statistical mechanics and molecular dynamics in evaluating thermodynamic properties of biomolecular recognition. Q. Rev. Biophys. 45, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitkoff D.; Sharp K. A.; Honig B. (1994) Correlating solvation free energies and surface tensions of hydrocarbon solutes. Biophys Chem. 51, 397–403discussion 404–9. [DOI] [PubMed] [Google Scholar]
- Mergny J. L.; Lacroix L. (2003) Analysis of thermal melting curves. Oligonucleotides 13, 515–37. [DOI] [PubMed] [Google Scholar]
- Weiser J.; Shenkin P. S.; Still W. C. (1999) Approximate solvent-accessible surface areas from tetrahedrally directed neighbor densities. Biopolymers 50, 373–80. [DOI] [PubMed] [Google Scholar]
- Svanvik N.; Westman G.; Wang D.; Kubista M. (2000) Light-up probes: thiazole orange-conjugated peptide nucleic acid for detection of target nucleic acid in homogeneous solution. Anal. Biochem. 281, 26–35. [DOI] [PubMed] [Google Scholar]
- Privat E.; Asseline U. (2001) Synthesis and binding properties of oligo-2′-deoxyribonucleotides covalently linked to a thiazole orange derivative. Bioconjugate Chem. 12, 757–69. [DOI] [PubMed] [Google Scholar]
- Carreon J. R.; Mahon K. P. Jr.; Kelley S. O. (2004) Thiazole orange-peptide conjugates: sensitivity of DNA binding to chemical structure. Org. Lett. 6, 517–9. [DOI] [PubMed] [Google Scholar]
- Coste J.; Le-Nguyen D.; Castro B. (1990) PyBOP®: A new peptide coupling reagent devoid of toxic by-product. Tetrahedron Lett. 31, 205–208. [Google Scholar]
- Ciommer M.; Kunz H. (1991) Synthesis of glycopeptides with partial structure of human glycophorin using the fluorenylmethoxycarbonyl/ allyl ester protecting group combination. Synlett 1991, 593–595. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.