Over the past decade “RNA interference” has emerged as a natural mechanism for silencing gene expression. This ancient cellular antiviral response can be harnessed to allow specific inhibition of the function of any chosen target genes, including those involved in causing diseases such as cancer, AIDS, and hepatitis. RNA interference is already proving to be an invaluable research tool, allowing much more rapid characterisation of the function of known genes. More importantly, the technology considerably bolsters functional genomics to aid in the identification of novel genes involved in disease processes. But can RNA interference be used as an effective therapeutic strategy? Many people in the biotechnology industry are betting that it can, but first considerable problems relating to delivery to target cells will have to be solved. This problem has proved the undoing of previous wonder technologies such as gene therapy and antisense. This review discusses the promises and pitfalls of RNA interference in research and treatment.
This article is based on a review of the literature on RNA interference and post-transcriptional gene silencing appearing in the PubMed database, along with personal experience of working in this field for the past four years. It also draws on consensus views expressed at several international conferences on RNA interference in 2003.
Current understanding of RNA interference
The first hints of the existence of the gene silencing mechanism that is now called RNA interference emerged from work on the genetic modification of plants in the late 1980s. Attempts to deepen the violet hue of petunias by expressing higher levels of an enzyme involved in the synthesis of the pigment unexpectedly resulted in the appearance of many white flowers. The introduction of extra copies of the gene had somehow caused a decrease in its expression rather than the anticipated increase.1,2
For some time this remained an unexplained oddity. It was soon joined by similar observations in the filamentous fungus Neurospora crassa and then the nematode worm Caenorhabditis elegans. Once the large community of developmental biologists working on the worm became involved, the pace quickened. In 1998 the key observation was made that led to the coining of the term “RNA interference.”3 Fire and Mello showed that double stranded RNA was able to direct the degradation of messenger RNA (mRNA) with sequence complementary to one or other strand.
Summary points
RNA interference is an ancient natural antiviral mechanism that directs silencing of gene expression in a sequence specific manner
RNA interference can be exploited artificially to inhibit the expression of any gene of interest
The principal systems for achieving RNA interference are short synthetic double stranded RNA molecules and gene expression vectors that direct their production in the cell
Libraries of RNA interference molecules have been constructed that allow the analysis of gene function on a genome-wide scale
RNA interference systems could be used clinically to suppress gene expression as a therapeutic strategy in many diseases characterised by elevated gene function
An ancient antiviral mechanism
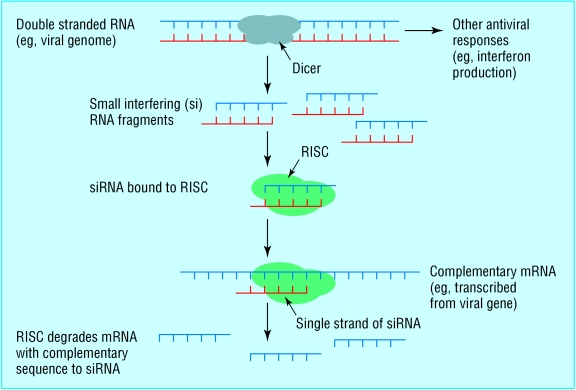
Over the next few years the mechanism underlying RNA interference was established from work on diverse organisms, especially the worm and the fruit fly.4 RNA interference was considered to be an evolutionarily ancient mechanism for protecting organisms from viruses. Many viruses have RNA, rather than DNA, as their genetic material and go through at least one stage in their life cycle in which they make double stranded RNA. All multicellular organisms possess a conserved protein machinery that recognises double stranded RNA. An enzyme called dicer degrades this into small segments around 20 nucleotide pairs in length (fig 1).
Fig 1.
Natural mechanism of RNA interference. The appearance of double stranded (ds) RNA within a cell—for example, as a result of viral infection—triggers an RNA interference response. The cellular enzyme dicer binds to the dsRNA and cuts it into short pieces of 20 or so nucleotide pairs in length known as small interfering RNAs or siRNAs. These bind to a cellular enzyme complex RISC (RNA induced silencing complex) that uses one strand of the siRNA to bind to single stranded RNA molecules such as mRNA of complementary sequence. RISC then degrades the mRNA, thus silencing expression of the viral gene. In mammals, other antiviral responses to dsRNA also exist
Not content with just degrading the viral double stranded RNA, the cell uses an enzyme complex called RISC (RNA induced silencing complex) to use the short pieces of RNA produced by dicer as a template to seek out and destroy single stranded RNA with the same sequence, such as mRNA copies used by the virus to direct synthesis of viral protein. Together, dicer and RISC make up the RNA interference system whereby double stranded RNA is recognised and used as a guide to prevent expression of similar sequences by destroying mRNA transcripts, a process sometimes termed post-transcriptional gene silencing.
As well as being involved in battling viruses, RNA interference is also probably important in maintaining order in the genome by suppressing the movement of mobile genetic elements such as transposons and repetitive sequences. The RNA interference machinery may also have a role in fine tuning normal cellular gene expression.5
RNA interference in mammals
Most of the work described above was done in invertebrates. Initial attempts to induce RNA interference responses in human cells were unsuccessful. Introduction of double stranded RNA into mammalian cells induces a powerful set of quite different antiviral responses characterised by production of interferons, resulting in inhibition of all gene expression and rapid cell death, limiting the ability of a virus to replicate and spread throughout the organism. It seemed that RNA interference might have been lost all together, replaced by the more recently evolved interferon system that is not found in invertebrates.
However, there were hints that RNA interference might still exist in mammals. The breakthrough came when short, double stranded RNA molecules of less than about 30 nucleotide pairs long were shown to be unable to induce the interferon response. As the global shut down of gene expression no longer occurred with these “small interfering RNAs” (siRNAs), they could be seen to be capable of directing a sequence specific degradation of homologous mRNA in a manner very similar to that in plants, worms, and flies.6 siRNAs of about 20 nucleotide pairs in length, when introduced into mammalian cells, directly engage RISC and promote silencing of the expression of genes with the same sequence (fig 1). Parsimonious nature had kept RNA interference as a back-up system even after the evolution of the interferon system.
RNA interference as a research tool
Long before RNA interference had been established as operating in mammalian cells, researchers working on worms had recognised the great power that it promised as a research tool. The sequencing of the genomes of humans and most commonly studied model organisms has led to a situation in which the identities of very large numbers of genes are known but little is understood about their function. A cheap and easy way of ablating gene function holds out massive hope for improving our ability to untangle the complex regulatory pathways that control cellular behaviour in health and disease. RNA interference allows analysis in a matter of days of the effect of loss of gene function at the cellular level that would have taken several months or even years by previous methods such as homologous recombination.
Targeting individual genes
RNA interference is now commonly used in biological and biomedical research to study the effect of blocking expression of a given gene. This has proved particularly easy in C elegans, where simply feeding the worms with bacteria expressing the double stranded RNA has been found to cause RNA interference throughout the tissues of the worm.
Researchers working on mammalian systems have had more difficulties. Most of the work has concentrated on introducing small interfering double stranded RNAs into cells in tissue culture. A popular method has been to make these synthetically in vitro. However, as mammalian cells will not readily take up naked nucleic acids, the RNAs have to be complexed with agents such as cationic lipids to allow them to enter the cells. Synthetic siRNAs can cause efficient inhibition of expression of homologous genes, although only for a few days. As the effect is rarely complete, it is generally termed a “knock down” to distinguish it from the “knock out” achieved by deletion of the gene.
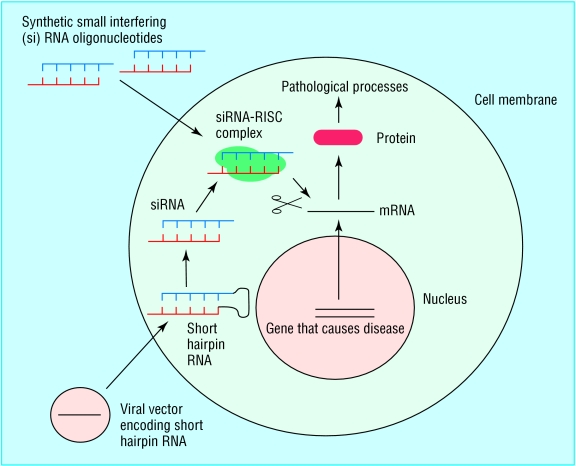
Another way of introducing siRNAs into cells is to use expression vectors such as engineered viruses to direct expression of short RNA sequences that will form hairpins owing to the presence of complementary sequences of about 20 nucleotide pairs (fig 2). These short hairpin RNAs are then processed within the cell to remove the loop and form siRNA duplexes. Viral vector mediated RNA interference can result in long term inhibition of target gene expression. Retroviruses, adenoviruses, and lentiviruses have all been used as vehicles for RNA interference constructs.
Fig 2.
Targeting disease by RNA interference. Diseases caused by aberrant gene expression include viral diseases and cancer. A gene implicated in causing the disease state can be silenced by RNA interference. Two of the most commonly used methods for artificially inducing RNA interference are shown here. Synthetic small interfering RNA molecules can be introduced into cells by using reagents such as cationic lipids to promote uptake across the cell membrane. Alternatively, engineered viral vectors can be used to deliver an expression construct to the cell, which will direct the production of a short hairpin RNA. This is then processed within the cell to form an siRNA. The siRNAs from either route then use the cellular RNA machinery to degrade mRNA with complementary sequence, in this case chosen to target the gene that causes the disease
Although a big improvement on previous methods, RNA interference has its limitations. Not every sequence works—most researchers get a success rate of about one in three. In addition, although the effects are generally thought to be highly sequence specific, some question marks remain as to whether or not some of the effects seen are “off target.” Some residual activation of the interferon system has been reported, as well as degradation of closely related, but non-identical, mRNAs.
RNA interference as a functional genomics tool
DNA microarray technology has now enabled the level of expression of every gene in the genome to be determined under any condition. This has led to a vast accumulation of information about genes whose expression is significantly altered in various disease states. For example, huge databases have been established of genes that are aberrantly regulated in cancers. In a few cases this has resulted in the identification of key genes involved in the formation of the tumour and provided important new therapeutic targets. However, most of the time the pattern of gene expression is far too complex to allow identification of the relatively small number of misexpressed genes that are involved in causing or maintaining the disease rather than the much larger number that are innocent bystanders.
The ability of RNA interference to provide relatively easy ablation of gene expression has opened up the possibility of using collections of siRNAs to analyse the significance of hundreds or thousands of different genes whose expression is known to be up-regulated in a disease, given an appropriate tissue culture model of that disease. Perhaps more important still is the possibility of using genome-wide collections of siRNAs, whether synthetic or in viral vectors, as screening tools. This has attracted much attention recently from both academic and industrial researchers. The libraries of RNA interference reagents can be used in one of two ways. One is in a high throughput manner, in which each gene in the genome is knocked down one at a time and the cells or organism scored for a desired outcome—for example, death of a cultured cancer cell but not a normal cell. Owing to the very large numbers of assays needed to look at the involvement of all 35 000 or so genes in the human genome, this approach is very labour intensive. The approach has been used successfully on a relatively small scale to investigate cell death signalling by TRAIL (tumour necrosis factor related apoptosis inducing ligand), an agent that might have therapeutic potential against various cancers.7,8 In addition, the approach was used to identify the familial cylindromatosis tumour suppressor gene (CYLD) as a de-ubiquitinating enzyme in the nuclear factor-κB pathway.9 As aspirin is known to target this pathway, this work suggested a novel therapeutic approach to this rare inherited cancer.
The other approach is to use large pools of RNA interference viral vectors and apply a selective pressure that only cells with the desired change in behaviour can survive. The identity of the genes knocked down in the surviving cells can then be identified by sequencing the RNA interference vectors that they carry. This method is being used to investigate genes involved in neurodegenerative diseases, diabetes, and cancer. It has recently been used successfully to identify several novel components of the p53 tumour suppressor gene signalling pathway.10 Both approaches show considerable promise in identifying novel genes that may make important therapeutic targets for inhibition either by conventional drug discovery methods or, more controversially, by RNA interference itself.
RNA interference as a novel therapeutic agent
RNA interference clearly has much promise in the laboratory, but how about in the clinic? In principle, RNA interference might be used to treat any disease that is linked to elevated expression of an identified gene. This might make it suitable for combating viral diseases, cancers, and inflammatory diseases, to name but three areas. Indeed, in tissue culture models, impressive results have been achieved against various cancer cells by using RNA interference to target oncogenes and against HIV, influenza, and polio viruses by targeting viral genes.11-14 However, a huge gap exists between achieving such results in vitro and in a whole animal or patient.
Delivery problems
The major challenge in turning RNA interference into an effective therapeutic strategy is the delivery of the RNA interference agents, whether they are synthetic short double stranded RNAs or viral vectors directing production of double stranded RNA, to the target cells within the body. Lessons can be learnt from two earlier technologies that held out much initial therapeutic promise but have ultimately failed to deliver effective treatments.
One of these is antisense. This uses short pieces of single stranded DNA complementary to the mRNA that was to be targeted. The resulting RNA-DNA hybrids forming in the cell can block translation of the mRNA by the protein synthesis machinery and also promote its degradation. Despite nearly two decades of work, antisense has failed to prove its efficacy in the clinic, although several clinical trials have been done. In part this reflects the fact that antisense in general provides a much less robust inhibition of gene expression than RNA interference, but also major difficulties arose in getting the antisense oligonucleotides to their target cells without them being degraded elsewhere in the body.
Additional educational resources
Journal articles
Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol 2003;4:457-67
Paddison PJ, Hannon GJ. RNA interference: the new somatic cell genetics? Cancer Cell 2002;2:17-23
Lau NC, Bartel DP. Censors of the genome. Sci Am 2003;Aug:34-41
Schmidt CW. Therapeutic interference. Modern Drug Discovery 2003;Jul:37-42
Websites
Nature Publishing Group (www.nature.com/focus/rnai/library/news_views.html)—A compendium of reviews and original articles on RNA interference
Ambion (www.ambion.com/techlib/resources/RNAi/)—A set of review and news articles on RNA interference, plus information about research tools, maintained by an RNA specialist company
Qiagen (www1.qiagen.com/siRNA/references.aspx)—A compendium of literature and citations on RNA interference, plus information about research tools, maintained by an RNA specialist company
Thomas Tuschl's laboratory (www.rockefeller.edu/labheads/tuschl/sirna.html)—An siRNA users guide: technical information on getting RNA interference to work
Another technology we can learn from is gene therapy. Gene therapy aims to replace defective genes in target tissues by delivering correct versions of them in expression vectors. Like antisense, gene therapy has failed to make significant progress in the clinic, despite enormous early hype. The problems have again centred around how to deliver the new versions of the defective gene safely and efficiently. Recent high profile safety problems with two of the most commonly used viral delivery systems, adenoviruses and retroviruses, have been a major setback for this approach.
RNA interference in vivo
Despite the problems of delivery, RNA interference has been used effectively in the mouse to block expression of a hepatitis C virus protein in the liver.15 In addition, the same group has used specific RNA interference to block hepatitis B virus infection in mice.16 They achieved delivery by injecting large amounts of synthetic double stranded RNA or DNA encoding a short hairpin RNA into the portal vein. A similar approach was taken to target the Fas protein, an important inducer of programmed cell death, resulting in protection of mice from fulminant hepatitis caused by injection with agonistic Fas-specific antibodies.17
The problems seen with the use of viral vectors in gene therapy mean that many researchers in RNA interference are favouring the use of synthetic siRNA duplexes rather than gene expression vectors that will direct the production of such molecules within the target cell (fig 2). A large number of biotechnology companies have programmes to develop synthetic RNA interference therapies for various diseases. These include Sirna Therapeutics (Boulder, Colorado) for macular degeneration; Avocel (Sunnyvale, California) for hepatitis C; Alnylam Pharmaceuticals (Cambridge, Massachusetts) for Parkinson's disease; CytRx (Los Angeles, California) for obesity, type 2 diabetes, and ALS; Acuity Pharmaceuticals (Philadelphia, Pennsylvania) for macular degeneration and diabetic retinopathy; and Sequitur (Natick, Massachusetts) for hepatic insufficiency, respiratory syncytial virus, asthma, and cancer.
Given sufficient research into delivery methods, some of these diseases will probably eventually be treated effectively by RNA interference based therapeutics. Success is more likely in those diseases with a simple genetic basis rather than in complex multigene disorders such as cancer. Diseases involving sites where delivery of synthetic RNA is more straightforward will also be more likely to be effectively treated. The bitter experiences with antisense and gene therapy mean that the likely problems should not be underestimated, but perhaps this time the reality may—eventually—live up to the hype.
Funding: Cancer Research UK.
Competing interests: None declared.
References
- 1.Van der Krol AR, Mur LA, Beld M, Mol JN, Stuitje AR. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 1990;2: 291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 1990;2: 279-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998;391: 806-11. [DOI] [PubMed] [Google Scholar]
- 4.Hannon GJ. RNA interference. Nature 2002;418: 244-51. [DOI] [PubMed] [Google Scholar]
- 5.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science 2003;301: 336-8. [DOI] [PubMed] [Google Scholar]
- 6.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001;411: 494-8. [DOI] [PubMed] [Google Scholar]
- 7.Aza-Blanc P, Cooper CL, Wagner K, Batalov S, Deveraux QL, Cooke MP. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol Cell 2003;12: 627-37. [DOI] [PubMed] [Google Scholar]
- 8.Tewari M, Vidal M. RNAi on the apoptosis TRAIL: the mammalian cell genetic screen comes of age. Dev Cell 2003;5: 534-5. [DOI] [PubMed] [Google Scholar]
- 9.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 2003;424: 797-801. [DOI] [PubMed] [Google Scholar]
- 10.Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heinerikx M, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathways. Nature 2004;428: 431-7. [DOI] [PubMed] [Google Scholar]
- 11.Damm-Welk C, Fuchs U, Wossmann W, Borkhardt A. Targeting oncogenic fusion genes in leukemias and lymphomas by RNA interference. Semin Cancer Biol 2003;13: 283-92. [DOI] [PubMed] [Google Scholar]
- 12.Ge Q, McManus MT, Nguyen T, Shen CH, Sharp PA, Eisen HN, et al. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc Natl Acad Sci U S A 2003;100: 2718-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gitlin L, Karelsky S, Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 2002;418: 430-4. [DOI] [PubMed] [Google Scholar]
- 14.Coburn GA, Cullen BR. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J Virol 2002;76: 9225-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature 2002;418: 38-9. [DOI] [PubMed] [Google Scholar]
- 16.McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, et al. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol 2003;21: 639-44. [DOI] [PubMed] [Google Scholar]
- 17.Song E, Lee SK, Wang J, Ince N, Ouyang N, Min J, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med 2003;9: 347-51. [DOI] [PubMed] [Google Scholar]