Abstract
Polyhydroxyalkanoates (PHAs) can be produced by microorganisms and are a biodegradable alternative to fossil-fuel based plastics. Currently, the focus is on reducing production costs by exploring alternative substrates for PHAs production, and on producing copolymers which are less brittle than monomers. Accordingly, this study used a substrate consisting of wastewater from waste-glycerol fermentation, supplemented with different amounts of acetic and propionic acids. These substrates were used to feed mixed microbial communities enriched from activated sludge in a sequencing batch reactor. A reactor supplemented with 2 mL of acetic acid produced 227.8 mg/L of a homopolymer of hydroxybutyrate (3HB); 4 mL of acetic acid produced 279.8 mg/L 3HB; whereas 4 mL of propionic acid produced 673.0 mg/L of a copolymer of 3HB and 3HV (hydroxyvalerate). Ribosomal Intergenic Spacer Analysis (RISA) was used to show the differences between the communities created in the reactors. Thauera species predominated in biomass that produced 3HB; Paracoccus denitrificans in the biomass that produced 3HB-co-3HV. Because P. denitrificans produced the more desirable copolymer, it may be advantageous to promote its growth in PHAs-producing reactors by adding propionate.
Key words: biopolymers, microbial community, polyhydroxyalkanoates, RISA, Thauera sp
Introduction
The group of polyesters known as polyhydroxyalkanoates (PHAs) are an interesting alternative to fossil-fuel based plastics, because they are biodegradable and can be produced from renewable resources. PHAs have been industrially produced by pure bacterial cultures of either wild forms or recombinant strains. Recently, there has been a great interest in examining potential alternative processes for PHAs production with the aim of decreasing polymer production costs. Therefore, mixed microbial communities (MMC) and by-products from agroindustry have been intensively tested for production of these biopolymers.
To lower the final price of polymers, complex waste could be used, for example, fermented sugar cane molasses, palm oil mill effluents, industrial wastewaters, and brewery wastewater (Serafim et al., 2008). Besides these substrates, crude glycerol generated as a by-product during biodiesel production has acquired importance during the last years, as reviewed by da Silva et al. (2009). This substrate will probably be widely available in the future because the production of biodiesel in Europe is still increasing. Glycerol can be converted into PHAs more effectively if it is be supplied in the form of volatile fatty acids (VFAs). The type of VFAs used as the substrate changes the monomer composition of PHAs. The most commonly used substrate for PHAs production is acetic acid, which bacteria use to synthesize polyhydroxybutyrate (PHB). However, because the mechanical properties of PHB are not ideal, other VFAs have been tested for the production of more applicable copolymers. Among them, propionate, butyrate, and valerate were successfully used to produce co-polymers possessing different monomers (Lemos et al., 2006). Unfortunately, the function of particular microorganisms in the synthesis of specific monomers and co-polymers is still undiscovered.
In addition, the composition of the mixed microbial communities tested for production of these biopolymers is also still unknown. These communities are able to perform specific intra- and extracellular reactions, and are selected by the operational conditions imposed on the biological systems (Dias et al., 2006). Knowledge of these microorganisms that are involved in this process is important for a better understanding of the mechanisms of PHAs production. Recently, molecular techniques, which are based mainly on the analysis of the 16S gene, have been found to be useful in the study of microorganisms creating mixed communities in both natural and semi-natural environments (Otawa et al., 2006). The most known and widely applied fingerprinting techniques are denaturing gradient gel electrophoresis (DGGE) (Muyzer et al., 1993), terminal restriction fragment length polymorphism (TRFLP) (Osborn et al., 2000), and ribosomal intergenic spacer analysis (RISA) (11). DGGE relies on melting-point variation in a variable portion of a target gene, usually coding for 16S rRNA. In TRFLP, 16S rRNA gene is amplified by PCR with a 5′ fluorescent primer, and amplicons are digested using restriction enzymes, resulting in terminal restriction fragments of distinctive lengths. However, in spite of the usefulness of these two techniques, the sequence of the 16S rRNA gene is often not divergent enough to distinguish species of the same genus. On the other hand, the fragment located between genes coding for 16S rRNA and 23S rRNA, known as a ribosomal intergenic spacer (RIS), has a highly variable length (Gürtler and Stanisich, 1996). The RIS can be amplified by PCR using general primers complementary to the flanking genes coding for 16S and 23S rRNA. In contrast to rRNA coding sequences, the RIS is under minimal selective pressure during evolution and therefore this genome fragment is less conserved. According to Leblond-Bourget et al. (1996), the evolutionary rate of the RIS is assumed to be 10 times greater than the evolutionary rate of the 16S rRNA gene. RISs have been shown to vary significantly in both the sequence and size of genera, species, strains, and even between different ribosomal operons within a single bacterium (Gürtler and Stanisich, 1996). Ribosomal intergenic spacer analysis (RISA) has been successfully used to analyze a complex bacterial community involved in polyhydroxyalkanoates production (Ciesielski et al., 2009).
The main goals of this work were to analyze changes in bacterial communities in three independent processes, and to examine how adding volatile fatty acids influences PHAs synthesis.
Methods
VFAs generation and PHAs production
Batch experiments for VFAs production were conducted under anaerobic thermophilic conditions (39 °C) in three independent reactors; each reactor had a working volume of 3 L. The reactors differed in the amount of waste glycerol (Experiment A - 12.5 g/L; B - 25.0 g/L; C -50.0 g/L). Waste glycerol was purchased from Biodiesel Manufacturing Plant (Biopaliwa S.A., Poland) and contained from 80% to 85% of glycerin. The ash concentration did not exceeded 7%, matter organic non-glycerine (M.O.N.G) concentration was less than 2%, and methanol was below 0.5%. 150 mL excess sludge from a wastewater treatment plant (Olsztyn, Northern Poland) was added to each reactor to facilitate waste glycerol conversion into VFAs. After fermentation, the mixture collected from each reactor was centrifuged at RCF = 8 693 for 10 min and the supernatant was diluted 1:2 with sterile water. One liter of fermented liquids were supplemented with 2 mL (Experiment A) and 4 mL (Experiment B) of acetic acid, and with 4 mL of propionic acid (Experiment C). The values of Chemical Oxygen Demand, and the concentrations of ammonia and total nitrogen were analyzed three times (Table 1). PHAs were produced in sequencing batch reactors (SBR) with a working volume of 5 L at a temperature of 22 °C. The reactor was inoculated with activated sludge from a wastewater treatment plant (Olsztyn, Northern Poland). The pH was maintained at 7.0 by adding NaOH 1 mol/L and HCl 1 mol/L, and oxygen was supplied by an air compressor to keep the dissolved oxygen (DO) at at 90%. The SBR was operated with a cycle of 24 h, each cycle consisted of a filling (0.25 h), aerobic (23.5 h) and settling phase (0.25 h). The cycle began with the addition of 0.5 L of the production culture medium, and the same volume of effluent was removed at the end the settling period. The process of PHAs production was run for 62 d, 139 d and 71 d for Experiments A, B and C, respectively.
Table 1.
The characteristics of fermented liquid used for PHAs production.
| Characteristic | Unit | Experiment | ||
|---|---|---|---|---|
|
| ||||
| A | B | C | ||
|
|
||||
| Chemical Oxygen Demand, O2 | mg/L | 2351 (± 518) | 4421 (± 640) | 5016 (± 858) |
| Ammonia concentration, N-NH4 | mg/L | 205 (± 21) | 169 (± 27) | 125 (± 25) |
| Total Nitrogen concentration, NT | mg/L | 253 (±19) | 202 (±24) | 161.7 (±31) |
Polyhydroxyalkanoates analysis
Biomass samples (50 mL) taken from SBR were centrifuged immediately and the supernatant was decanted. Samples were then lyophilized for 24 h in a freeze drying unit (LYOVAC GT2, STERIS GmbH, Germany). PHAs polymers were extracted by shaking the freeze-dried cells in chloroform at 50 °C for 3 h. The mixture was filtered through No. 1 Whatman filter paper by simple filtration. The PHAs dissolved in 2 mL of chloroform were precipitated with 10 volumes of a 70% solution of chilled methanol and then allowed to evaporate for at least 2 d at room temperature. Then, PHAs were suspended in 2 mL of chloroform and 2 mL of acidified methanol containing 3% v/v H2SO4. Next, the tubes with the suspension were placed in a 100 °C oven for 20 h to esterify the PHAs. 3- hydroxyacyl methyl esters were detected by gas chromatography using a previously described method (Braunegg et al., 1978; Comeau et al., 1988). The GC (Varian 3800, Varian, Inc. Corporate Headquarters, U.S.A.) was operated with a Varian FactorFour VF-5 ms column (length: 30 m; internal diameter: 0.25 mm; film thickness: 0.25 μm), a split injection ratio of 1:15, and helium as the carrier gas (1.0 mL/min). The flame ionization detector (FID) temperature was 300 °C with the injection port temperature at 250 °C. Initial column temperature was kept constant for 1 min at 80 °C, then increased at 10 °C/min to 120 °C, further increased at 45 °C/min to 270 °C, which was held for 3 min. Total time amounted to 11.33 min. 3HB and 3HV were calibrated with a standard poly(3-hydroxybutyric acid-co-3-hydroxy-valeric acid) (140 g/L of 3HV) of natural origin (Sigma-Aldrich; Ref. Nr 403121), to which the same procedure had been applied as to the reactor samples.
The 3HB and 3HV contents were determined by comparison with the peak-areas of the methyl-3-hydroxybutyrate, methyl-3-hydroxyvalerate derived from the standard. The peak-areas of each sample were measured 5 times, and the extreme values were omitted by the application of the Dixon test. Based on mean values, the concentrations of 3HB and 3HV were calculated as follows: the methyl ester concentration in the standard was multiplied by the mean area of the corresponding peaks; the product was then divided by the peak-area in the standard.
DNA extraction
DNA was directly extracted as follows: 0.02 g of biomass sample were washed in a sodium phosphate buffer (0.1 mol/L; pH 8.0), pelleted by centrifugation, suspended in a proteinase K buffer (100 mmol/L Tris-HCl; 10 mmol/L EDTA; pH 8.0) and incubated at 55 °C in the presence of sodium dodecyl sulphate, proteinase K and lysozyme. In order to improve the process of cell wall lysis, the samples were sonicated for 1 min. DNA was purified using a phenol/chloroform/isoamyl alcohol (25/24/1, v/v/v) solution, and precipitated with 2 volumes of ethanol (96%) and 0.1 volume of sodium acetate (pH 5.2). The pellet was washed with 70% ethanol, dried and re-suspended in 100 μL of TRIS/EDTA (10 mmol/L Tris-HCl, 1 mmol/L EDTA, pH 7.5).
Polymerase chain reaction
The Intergenic Sequence Region was amplified using primer pair (S-D-Bact-1522-β-S-20: 5′-TGCGGCTGGATCCCCTCCTT-3′ and L-D-Bact-132-α-A-18: 5′-CCGGGTTTCCCCATTCGG-3′) published by Normand et al. (1996). PCR was performed in an Eppendorf® Mastercycler Gradient (Eppendorf, Germany). The mixtures used for PCR amplification contained 50 ng of extracted DNA, 0.5 μmol/L of each primer, 100 μmol/L of deoxynucleoside triphosphate (Promega, Winsconsin, USA), 2 units of Pfu DNA polymerase (DNA Gdansk, Poland), 5 μL reaction buffer (500 mmol/L KCl, pH 8.5; Triton X-100), 1.5 mmol/L MgCl2, to which sterile water was added for a final volume of 50 μL. The temperature program for DNA amplification was as follows: 94 °C for 5 min; 35 cycles of denaturation at 94 °C for 30 s, annealing at 43 °C for 1 min, extension at 72 °C for 1 min and a single final elongation at 72 °C for 10 min. The PCR amplicons of PHA were checked by resolving on 1.5% agarose gels stained with ethidium bromide.
Polyacrylamide electrophoresis
After successful DNA amplification, the PCR products were separated in 5% polyacrylamide gel (29:1 acrylamide:bisacrylamide). Electrophoresis was carried out at 80 V for 100 min in 1x TBE buffer (89 mmol/L Tris base, 89 mmol/L Boric acid, 2 mmol/L EDTA; pH 8.0). The size of PCR products was estimated using a molecular weight marker (100 bp Ladder, Promega, USA). The gel was stained with SybrGold (Invitrogen) and viewed with an ultraviolet transilluminator and recorded with a CCD camera (Gel Logic 200, Eastman Kodak Company, USA). Bands were detected automatically from the digital images of the gel using KODAK 1D 3.6 Image Analysis Software (Eastman Kodak Company, USA).
DNA sequencing and analysis
The dominant bands excised from the polyacrylamide gel and DNA fragments were washed and directly reamplified with the same primer. Sequencing reactions were carried out with an ABI 373 Automated DNA Sequencer (PE Applied Biosystems, Foster City, CA, USA). DNA was sequenced by the commercial company GENOMED (Warsaw, Poland). All reactions were run following the manufacturer’s protocols. The GenBank database was searched with the BLAST program (Altschul et al., 1997). Sequences were aligned using the ClustalW program (Thompson et al., 1994). The sequences were deposited in GenBank under accession numbers from JF729203 to JF729213.
Profile banding analysis
The RISA patterns were converted to a binary matrix, using presence-absence data. A pairwise similarity of the banding patterns of the different samples was calculated using Nei-Li distance (Nei and Li, 1979). UPGMA cluster analysis was conducted with DGGEstat software (Erik van Hannen; the Netherlands Institute of Ecology).
Results
Biopolymers were obtained in all three experiments. The highest PHAs amount, 673.0 mg/L, was noticed in Experiment C. In Experiments A and B, much smaller amounts of PHAs were obtained: 227.8 mg/L and 279.8 mg/L, respectively. Maximum PHA concentrations, measured in dry cell weight, were similar in all fermentations: 30.7%, 34.2%, and 30.0% in Experiments A, B, and C, respectively. In the first two experiments, only polyhydroxybutyrate was detected, whereas in Experiment C, a co-polymer was detected that contained 72% hydroxyvalerate.
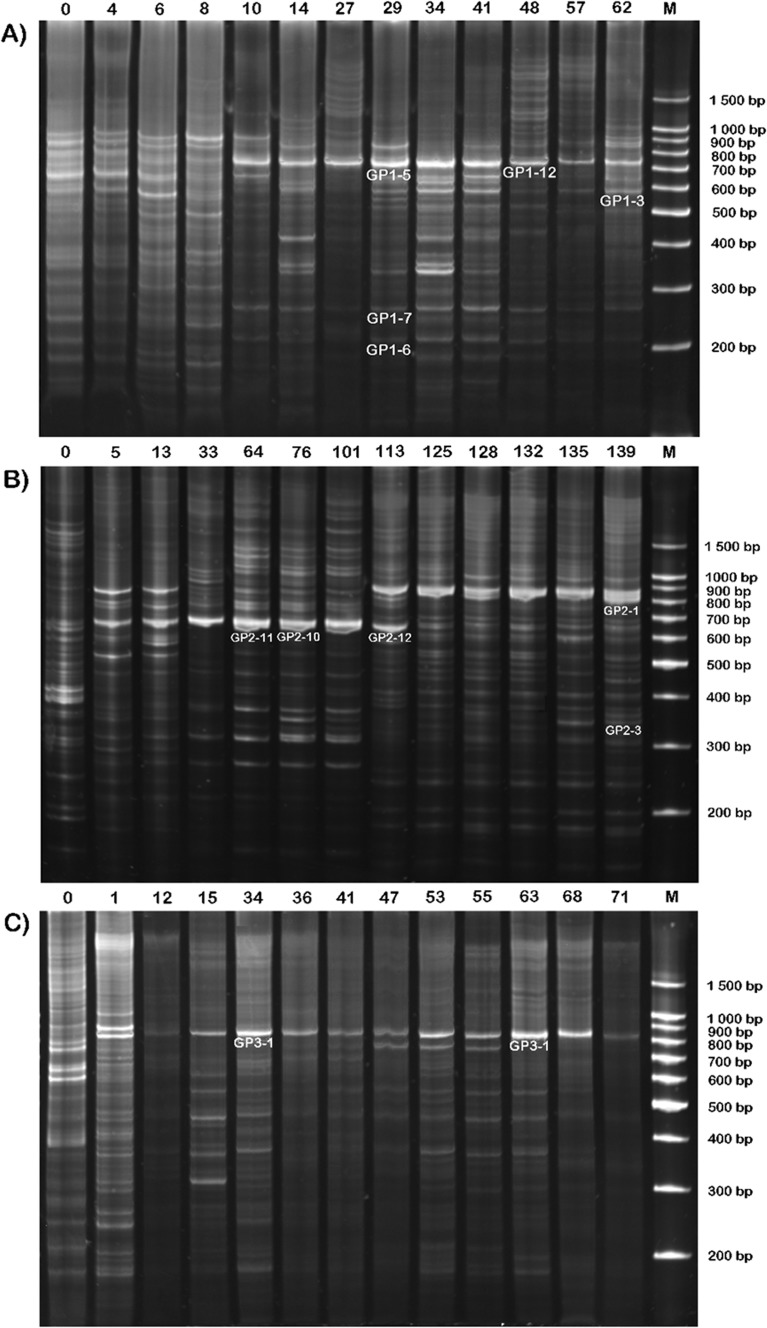
To evaluate the effect of the treatment on the diversity of uncultured microorganisms, the total DNA obtained at each sampling time was analyzed by PCR amplified ribosomal intergenic spacer analysis. Electrophoretic separation of the amplicons from each sample resulted in a distinct RISA pattern. In the electrophoregrams presented in Figure 1, bands ranging between 200 base pairs (bp) and 1500 bp were considered. For all the analyzed fermentations, the banding pattern showed some differences in band number and intensity.
Figure 1.
The banding profiles of ribosomal intergenic spacers from Experiments A, B, and C. The number above each lane shows the sampling day, 0 -inoculum, M - molecular weight marker (100 bp Ladder Marker, Promega, Wisconsin, USA).
Experiment A
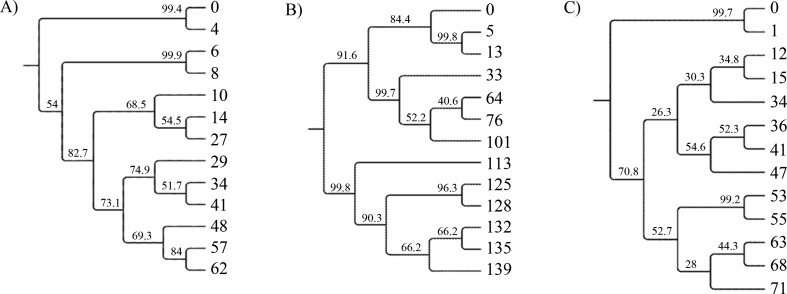
The patterns from Experiment A were very rich in the beginning of the process and were composed of more than 25 bands (Figure 1A). This started to change after 8 d, at which time the biodiversity decreased and one major band appeared (GP1-5). From the 29th day until the 41st day, some bands became larger and more intense (e.g. GP1-6 and GP1-7). After that the biodiversity decreased again and the microbial structure was stable until the end of fermentation. In order to determine relationships between the microbial communities over time, similarities were calculated on the basis of the absence or presence of bands. Cluster analysis showed a shift in the composition of the bacterial community in Experiment A (Figure 2A). The five strongest bands were excised from the gel and sequenced. The length of analyzed sequences ranged from 279 bp to 416 bp. Nucleotide sequences were compared to the GenBank database using BLASTN (Table 1). Three of them (GP1-3, GP1-6, and GP1-7) showed a high similarity to previously analyzed DNA sequences. The bands designed as GP1-5 and GP1-12 occupied the same position on the gel, although they differ in nucleotide composition. Both were identified as Thauera sp. MZ1T with low coverage/identity values (Table 2).
Figure 2.
Neighbor-joining trees representing genetic similarity of the microbial community profiles characterized in Experiments A, B, C. The numbers indicate the sampling day, 0 - inoculum. Bootstrap values are given at nodes.
Table 2.
Characteristics of the DNA sequences obtained from excised RISA bands.
| Band name/accession number | Closest relative/Accession number | Coverage/identity | Length of analyzed DNA sequence | Estimated length of RIS amplicon (bp) |
|---|---|---|---|---|
| GP1-3 (JF729203) | uncultured Nitrosomonas (GQ330299) | 100/100 | 399 bp of RIS | 490 |
| GP1-5 (JF729204) | Thauera sp. MZ1T (CP001281) | 70/86 | 284 bp of RIS | 585 |
| GP1-6 (JF729205) | Uncultured Gemmatimonadaceae bacterium (HQ212581) | 100/82 | 150 bp of RIS + 129 bp of 23S rRNA gene | 150 |
| GP1-7 (JF729206) | Uncultured Actinobacterium clone BAL3SP-450-1 (HM124463) | 100/87 | 179 bp of RIS + 129 bp of 23S rRNA gene | 180 |
| GP1-12 (JF729207) | Thauera sp. MZ1T (CP001281) | 47/86 | 416 of RIS | 594 |
| GP2-1 (JF729208) | Thauera sp. MZ1T (CP001281) | 100/91 | 526 of RIS | 690 |
| GP2-3 (JF729209) | Bacillus cereus strain HY-1 (EU915686) | 99/100 | 115 bp of RIS + 131 bp of 23S rRNA gene | 200 |
| GP2-10 (JF729210) | Thauera sp. MZ1T (CP001281) | 100/91 | 526 of RIS | 670 |
| GP2-11 (JF729211) | Thauera sp. MZ1T (CP001281) | 100/87 | 535 of RIS | 526 |
| GP2-12 (JF729212) | Prochlorothrix scandica NIVA-8/90 (HQ316170) | 94/85 | 359 of RIS | 504 |
| GP3-1 (JF729213) | Paracoccus denitrificans PD1222 (CP000490) | 97/97 | 614 of RIS | 715 |
Experiment B
Sample 0, taken from the inoculum, differs significantly from the samples taken in the next days. The microbial community was stable from the 5th day until the 33rd day of fermentation. On the 33rd day, many bands lost intensity or disappeared. The strong bands, designated as GP2-10, GP2-11, and GP2-12, survived and were present in the community until the 101st day of the process. There were distinct changes in the community composition on the 113th day, and from this time the high-intensity GP2-1 band was observed (Figure 1B). Changes in bacterial composition, obtained with the genetic distance method between neighboring samples, are presented in Figure 2B. Clustering analysis showed three main groups; the first one comprised inoculum and samples taken at day 5 and day 13; the second, samples from the 33rd to the 101st day; and the third, the rest of the samples. Four bands were excised and sequenced from the gel showing microbial fingerprints from Experiment B. The two bands that were most intense from the 33rd to the 101st day (GP2-10 and GP2-11) were most similar to Thauera sp. MZ1T, although the nucleotide composition of these two bands was slightly different. The DNA sequence of band GP2-1 was also similar to Thauera sp. MZ1T, although its amplicon was longer than those of the GP2-10 and GP2-11 bands. The remaining bands, GP2-3 and GP212, were most similar to Bacillus cereus HY-1 and Prochlorothrix scandica NIVA-8/90, respectively (Table 2).
Experiment C
Similar to the previously described fermentations, sample 0, from the inoculum, had many bands of different lengths. After just one day, the microbial structure changed rapidly and at least two distinct bands appeared in the profile. Small changes were noted from the 15th to the 68th day, then the intensity of all the bands decreased on the 71st day. One distinct band, described as GP3-1, was present throughout the whole experiment (Figure 1C). Cluster analysis showed the trend of progressive community transition (Figure 2C). The dominant band (GP3-1) was excised and its nucleotide composition was determined. The taxon corresponding to this band showed high similarity to Paracoccus denitrificans PD1222 (Table 2).
Discussion
One critical factor in the development of a competitive process for PHAs production using the MMC approach is the selection of organisms with a high storage capacity. Therefore, it is necessary to track the microbial population changes during the process. In terms of microbial population dynamics, limited information is available about the organisms that accumulate PHAs and the process by which they synthesize them (Lemos et al., 2008). Initial information about microorganisms that synthesize PHAs has been provided by Dionisi et al. (Dionisi et al., 2005; Dionisi et al., 2006). The authors used denaturant gradient gel electrophoresis (DGGE) to discover that the most important organisms selected under the applied conditions belonged to the Betaproteobacteria (Thauera, Alcaligenes, Comamonas, Achromobacter), but also several Alphaproteobacteria (Xantobacter, Curtobacterium) and Gammaproteobacteria (Kluyvera, Pseudomonas, Acinetobacter) were present. Among them, the Thauera genus was the most common. In the work of Lemos et al. (2008), the application of a cell sorting RT-PCR approach allowed for identification of four genera of bacteria (Amaricoccus, Azoarcus, Thauera and Paraccoccus) that were abundant in MMC under aerobic dynamic feeding. Villano et al. (2010) described the composition of the bacterial structure in biomass enriched from activated sludge, where Betaproteobacteria were predominant. Recently Jiang et al. (2011) discovered that in an enriched mixed community the predominant bacterial species were Plasticicumulans acidivorans and Thauera selenatis.
The substrate composition may have a direct influence on the microbial community structure of the biomass, which subsequently affects the PHA producing capacity. To produce PHAs from waste materials composed of sugars, the sugars must first be transformed into volatile fatty acids (VFAs) by anaerobic fermentation. Because this step needs additional energy, most studies are performed using organic acids. Acetate is a commonly used feedstock, which is converted into a homopolymer in the form of polyhydroxybutyrate. To incorporate different monomer units, other organic acids such as propionate have been tested (Lemos et al., 2006). A mixture of acetate and propionate could allow the synthesis of a co-polymer of polyhydroxybutyrate (3HB) and polyhydroxyvalerate (3HV) (Dionisi et al., 2001). According to the results presented here, in a mixed microbial community fed with acetate, the dominant bacteria belonged to the Thauera sp. These bacteria were noticed in both fermentations where the amount of acetic acid was higher (Experiments A and B). Paracoccus denitrificans was detected instead of Thauera sp. in Experiment C, where the level of acetic acid was much lower and propionic acid was much higher. In this experiment, On the basis of these findings, we can conclude that acetate could be easily utilized by bacteria belonging to Thauera species, whereas P. denitrificans is responsible for PHAs synthesis in the presence of propionate. This conclusion is supported by the observations of other authors. Similar results were described in a report by Lemos et al. (2008), who showed that in microbial communities fed with acetate, the predominant bacteria belonged to the Thauera sp., independent of the Solids Retention Time (SRT) and Organic Loading Rate (OLR). When propionate was used as a carbon source, this microorganism was present in a lesser amount. Thauera sp. was also the dominant bacterium when MMC were fed with lactate and acetate (Jiang et al., 2011).
The use of RISA made it possible to track the development of bacterial populations from the very beginning. This allowed us to determine the succession and stability of bacterial communities throughout the process. The fingerprints we obtained, when submitted to a detailed single-band analysis, allowed us to determine when PHAs producers appeared in the biomass. In Experiment A, Thauera sp. increased their cell numbers on the 10th day of operation, whereas in Experiment B, it was on the 33rd day. In Experiment C, P. denitrificans was detected on the 15th day. The most interesting changes in the bacterial community were observed in Experiment B, which was monitored for 139 days. After 100 days, the most intense bands, GP-10 and GP2-11, were replaced by another taxon, also from the Thauera genus (GP2-1), which then predominated. This phenomena is difficult to explain, because none of the fermentation conditions changed at that time. This could have resulted from the fact that numerous different strains from this genus were present in the activated sludge and the enriched MMC. There were at least five different taxons of Thauera in Experiment A. The richness of the bacterial community structure that was detected with RISA suggests that it is more sensitive than DGGE and could be an excellent tool for detailed monitoring of bacterial community changes.
To sum up, a mixed microbial community fed on a substrate of organic acids supplemented with acetate synthesized a homopolymer of hydroxybutyrate; a community fed on organic acids with propionate synthesized a copolymer of 3HB and 3HV. Ribosomal Intergenic Spacer Analysis allowed us to describe the bacterial succession process. Detailed analysis of the dominant DNA bands showed the predominant bacterial taxons responsible for PHAs synthesis and accumulation: Thauera sp. in the reactor supplemented with acetate, and Paracoccus denitrificans in the reactor with propionate. As the 3HB-co-3HV is more flexible, the promotion of P. denitrificans growth should be further investigated.
Acknowledgments
The study was financed under project no. N N523 420237 of the Polish Ministry of Science and Higher Education.
References
- Altschul SF, Madden TL, Schäffer A, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bormann EJ, Leißner M, Beer B. Growth-associated production of poly(hydroxybutyric acid) by Azotobacter beijerinckii from organic nitrogen substrates. Appl Microbiol Biotechnol. 1998;49:84–88. [Google Scholar]
- Braunegg G, Sonnleitner B, Lafferty RM. A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in microbial biomass. Eur J Appl Microbiol. 1978;6:29–37. [Google Scholar]
- Ciesielski S, Klimiuk E, Mozejko J, Nowakowska E, Pokój T. Changes in microbial communities structure during adaptation towards polyhydroxyalkanoates production. Pol J Microbiol. 2009;58:131–139. [PubMed] [Google Scholar]
- Comeau Y, Hall KJ, Oldham WK. Determination of poly-β-hydroxybutyrate and poly-β-hydroxyvalerate in activated sludge by gas-liquid chromatography. Appl Environ Microbiol. 1988;54:2325–2327. doi: 10.1128/aem.54.9.2325-2327.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva GP, Mack M, Contiero J. Glycerol: A promising and abundant carbon source for industrial microbiology. Biotechnol Adv. 2009;27:30–39. doi: 10.1016/j.biotechadv.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Dias JML, Lemos PC, Serafim LS, Oliveira C, Eiroa M, Albuquerque MGE, Ramos AM, Oliveira R, Reis MAM. Recent advances in polyhydroxyalkanoate production by mixed aerobic cultures: From the substrate to the final product. Macromol Biosci. 2006;6:885–906. doi: 10.1002/mabi.200600112. [DOI] [PubMed] [Google Scholar]
- Dionisi D, Majone M, Tandoi V, Beccari M. Sequencing Batch Reactor: Influence of periodic operation on performance of activated sludges in biological wastewater treatment. Ind Eng Chem Res. 2001;40:5110–5119. [Google Scholar]
- Dionisi D, Majone M, Vallini G, Di Gregorio S, Beccari M. Effect of the applied organic load rate on biodegradable polymer production by mixed microbial cultures in a sequencing batch reactor. Biotechnol Bioeng. 2006;93:76–88. doi: 10.1002/bit.20683. [DOI] [PubMed] [Google Scholar]
- Dionisi D, Beccari M, Di Gregorio S, Majone M, Papini MP, Vallini G. Storage of biodegradable polymers by an enriched microbial community in a sequencing batch reactor operated at high organic load rate. J Chem Technol Biotechnol. 2005;80:1306–1318. [Google Scholar]
- Gürtler V, Stanisich VA. New approaches to typing and identification of bacteria using the 16S–23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Marang L, Kleerebezem R, van Loosdrecht MCM. Polyhydroxybutyrate production from lactate using a mixed microbial culture. Biotechnol Bioeng. 2011;108:2022–2035. doi: 10.1002/bit.23148. [DOI] [PubMed] [Google Scholar]
- Leblond-Bourget N, Philippe H, Mangin I, Decaris B. 16S rRNA and 16S to 23S internal transcribed spacer sequence analyses reveal inter- and intraspecific Bifidobacterium phylogeny. Int J System Bacteriol. 1996;46:102–111. doi: 10.1099/00207713-46-1-102. [DOI] [PubMed] [Google Scholar]
- Lemos PC, Serafim LS, Reis MAM. Synthesis of polyhydroxyalkanoates from different short-chain fatty acids by mixed cultures submitted to aerobic dynamic feeding. J Biotechnol. 2006;122:226–238. doi: 10.1016/j.jbiotec.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Lemos PC, Levantesi C, Serafim LS, Rossetti S, Reis MAM, Tandoi V. Microbial characterisation of polyhydroxyalkanoates storing populations selected under different operating conditions using a cell-sorting RT-PCR approach. Appl Microbiol Biotechnol. 2008;78:351–360. doi: 10.1007/s00253-007-1301-5. [DOI] [PubMed] [Google Scholar]
- Madison LL, Huisman GW. Metabolic engineering of poly(3-hydroxyalkanoates): From DNA to plastic. Microbiol Mol Biol Rev. 1999;63:21–53. doi: 10.1128/mmbr.63.1.21-53.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes encoding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand P, Ponsonnet C, Nesme X, Neyra M, Simonet P. ITS analysis of prokaryotes. In: Akkermans ADL, van Elsas JD, de Bruijn FJ, editors. Molecular Microbial Ecology Manual. Kluwer Academic Publishers; Dordrecht: 1996. pp. 1–12. [Google Scholar]
- Osborn AM, Moore RB, Timmis KN. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ Microbiol. 2000;2:39–50. doi: 10.1046/j.1462-2920.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- Otawa K, Asano R, Ohba Y, Sasaki T, Kawamura E, Koyama F, Nakamura S, Nakai Y. Molecular analysis of ammonia-oxidizing bacteria community in intermittent aeration sequencing batch reactors used for animal wastewater treatment. Environ Microbiol. 2006;8:1985–1996. doi: 10.1111/j.1462-2920.2006.01078.x. [DOI] [PubMed] [Google Scholar]
- Serafim LS, Lemos PC, Albuquerque MGE, Reis MAM. Strategies for PHA production by mixed cultures and renewable waste materials. Appl Microbiol Biotechnol. 2008;81:615–628. doi: 10.1007/s00253-008-1757-y. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions- specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villano M, Beccari M, Dionisi D, Lampis S, Miccheli A, Vallini G, Majone M. Effect of pH on the production of bacterial polyhydroxyalkanoates by mixed cultures enriched under periodic feeding. Process Biochem. 2010;45:714–723. [Google Scholar]