Abstract
P. aeruginosa and Acinetobacter spp. are important pathogens associated with late nosocomial pneumonia in hospitalized and institutionalized individuals. The oral cavity may be a major source of these respiratory pathogens, particularly in the presence of poor oral hygiene and periodontal infection. This study investigated the prevalence of P. aeruginosa and Acinetobacter spp. in subgingival biofilm and saliva of subjects with periodontal disease or health. Samples were obtained from 55 periodontally healthy (PH) and 169 chronic periodontitis (CP) patients. DNA was obtained from the samples and detection of P. aeruginosa and Acinetobacter spp. was carried out by multiplex and nested PCR. P. aeruginosa and Acinetobacter spp. were detected in 40% and 45% of all samples, respectively. No significant differences in the distribution of these microorganisms between men and women, subgingival biofilm and saliva samples, patients ≤ 35 and > 35 years of age, and smokers and non-smokers were observed regardless periodontal status (p > 0.05). In contrast, the frequencies of P. aeruginosa and Acinetobacter spp. in saliva and biofilm samples were significantly greater in CP than PH patients (p < 0.01). Smokers presenting P. aeruginosa and high frequencies of supragingival plaque were more likely to present CP than PH. P. aeruginosa and Acinetobacter spp. are frequently detected in the oral microbiota of CP. Poor oral hygiene, smoking and the presence of P. aeruginosa are strongly associated with periodontitis.
Keywords: Pseudomonas aeruginosa, Acinetobacter spp., subgingival biofilm, saliva, periodontitis, PCR
Introduction
Periodontal diseases are bacterial infections associated with a complex microbiota of the dental biofilm that induces a local and systemic inflammatory response, leading to periodontal tissue destruction (Page and Kornam, 1997; Paster et al., 2006; Socransky et al., 1998). The microbial diversity of the human oral cavity has been recognized for decades, and over 700 species have been identified in this habitat (Paster et al., 2006). In addition to the resident oral species, studies have shown that the oral cavity harbours high proportions of various medically important pathogens, particularly in individuals with poor oral hygiene, periodontal diseases and/or immunosuppression (Ali et al., 1994, 1996; Botero et al., 2007; Colombo et al., 2002, 2009; Da Silva-Boghossian et al., 2011; Fritschi et al., 2008; Gonçalves et al., 2009; Persson et al., 2008; Slots et al., 1988, 1990; Souto and Colombo 2008; Souto et al., 2006). Conceivably, these species may disseminate to distant body sites increasing the risk for systemic infectious conditions such as bacterial pneumonia (Paju and Scannapieco, 2007; Scannapieco, 1998; Scannapieco et al., 1998).
Pseudomonas aeruginosa and Acinetobacter spp. are major respiratory pathogens associated with late-onset nosocomial pneumonia in hospitalized and institutionalized individuals (Chastre and Fagon, 2002). P. aeruginosa can be identified in a range of infections, especially those with a tendency to become chronic such as lung infections in cystic fibrosis patients (Wagner and Iglewski, 2008). This species presents many virulence properties including the ability to produce and secrete extracellular enzymes and toxins (Pihl et al., 2010; Smith and Iglewski, 2003, Woods et al., 1982) to adhere to and form biofilms on tissues and abiotic surfaces (Pihl et al., 2010; Smith and Iglewski, 2003), as well as to present resistance to many antibiotics (Slots et al., 1988, 1990). Acinetobacter spp. are a major concern in nosocomial infections due to their rapid development of multi-drug resistance, surviving desiccation and persistence in the environment for long periods of time (Fourrier et al., 1998; Karlowsky et al., 2003; Luna et al., 2007). These organisms are associated with bacteremia, pulmonary infections, meningitis, diarrhea and notorious nosocomial infections with mortality rates of 20 to 60% (Luna et al., 2007). Transmission is via person-to-person contact, contaminated water, food and hospital equipment (Agodi et al., 2007; Luna et al., 2007). The prevalence of Pseudomonas and Acinetobacter spp. in subjects with periodontal diseases may vary widely among different populations (Ali et al., 1994, 1996; Colombo et al., 2002; Persson et al., 2008; Slots et al., 1988, 1990; Slots et al., 1991; Souto et al., 2006). These pathogens have also been associated with treatment failure in patients with refractory periodontitis (Colombo et al., 1998, 2009). The role of these microorganisms in the etiology and pathogenesis of periodontal diseases is unclear. Regardless, we hypothesized that the existence of disease may increase colonization of the oral microbiota by these species. The aim of this study was to determine the carriage rate of P. aeruginosa and Acinetobacter spp. in the subgingival biofilm and saliva of subjects with periodontal health or chronic periodontitis.
Methods
Subject population and clinical diagnosis
Two hundred and twenty four adult subjects who sought dental treatment at the Dental School of the Federal University of Rio de Janeiro, Brazil were recruited for the study. Informed consent was obtained from all enrolled individuals. The study protocol was approved by the Review Committee for Human Subjects of the Clementino Fraga Filho University Hospital. Exclusion criteria included pregnancy, use of local or systemic antimicrobial agents within 6 months prior to the entry into the study, diabetes and other systemic conditions that could affect the periodontal status. All subjects had at least 10 natural teeth and were over 18 years of age. During the first visit, subjects were submitted to an anamnesis questionnaire, and information regarding age, gender and smoking status was obtained. Smoking was recorded as never-having-smoked and smoker (current or former smokers). Periodontal clinical measurements were performed at six sites per tooth and included probing depth (PD), clinical attachment level (CAL), presence or absence of supragingival biofilm (SB), bleeding on probing (BOP) and suppuration (SUP). After initial clinical evaluation, subjects were categorized as periodontally healthy (PH, n = 55) or chronic periodontitis (CP, n = 169). The PH controls had no sites with PD and/or CAL > 3 mm and no more than 10% of sites with BOP. CP patients presented at least 10% of teeth with PD and/or CAL 3 5 mm, or at least 15% of teeth with PD and/or CAL 3 4 mm, and > 10% of sites with BOP. After clinical examination and sampling, subjects with evidence of destructive periodontal disease were treated for periodontitis by full mouth scaling and root planning under local anesthetic and instructions in proper home care procedures in the Department of Dental Clinic at the Federal University of Rio de Janeiro.
Sampling
After removal of supragingival biofilm with sterile gauze, subgingival biofilm samples were taken of up to 6 sites with PD ≥ 4 mm and BOP from CP patients, and from 3 randomly selected sites with PD < 4 mm and no BOP from PH individuals using sterile periodontal curettes (Hu-Friedy, Chicago, IL). For each patient, samples were pooled and placed in Eppendorf tubes containing 500 μL of TE (Tris-HCl 10 mM, EDTA 1 mM, pH 7.6). For saliva samples, patients were not allowed to clean their teeth or to eat 30 min before sampling. They rinsed out their mouths with 10 mL of 0.9% sterile saline for 60 s, and the mouthwashes were collected in sterile plastic recipients. All samples were stored in a freezer at −20 °C.
Isolation of bacterial DNA
Bacterial DNA was extracted from saliva and subgingival biofilm samples according to the methodology described by Laine et al. (2000). Briefly, samples were centrifuged at 300 × g for 10 min. The pellet was washed twice in 0.9% saline, re-suspended in 100 μL of 50 mM NaOH and boiled for 10 min. Samples were then neutralized with 14 μL of 1 M Tris (pH 7.5) and centrifuged at 14,000 × g for 3 min. Supernatants were collected and stored at −20 °C until PCR amplification was performed.
Detection of P. aeruginosa and Acinetobacter spp. by multiplex and nested PCR
For PCR detection of P. aeruginosa, two sets of primers were used: one set (PS1, 5′-ATGAACAACGTTCTGAAATTCTCTGCT-3′ and PS2, 5′-CTTGCGGCTGGCTTTTTCCAG-3′) consisted of primers corresponding to the beginning and the end of the open reading frame of the OprI gene (De Vos et al., 1997), while the other set of primers (PAL1, 5′-ATGGAAATGCTGAAATTCGGC-3′ and PAL2, 5′-CTTCTTCAGCTCGACGCGGACG-3′) corresponded to the open reading frame of the OprL gene. A negative control with no DNA template and a positive control consisting of 20 μL of DNA obtained from the P. aeruginosa strain ATCC 27853 were used in all reactions. The PCR reaction was performed in a 100 μL reaction mixture containing 30 pmol of each primer, 10 μL of 10 X PCR buffer, 1.5 mM of MgCl2, 0.2 mM of desoxynucleotide triphosphate mixture (final concentration 0.2 mM each dATP, dCTP, dTTP and dGTP), 2 U of Platinum® Taq DNA polymerase (Invitrogen, Grand Island, NY) and 20 μL of DNA template. The reaction was performed in a thermal cycler Primus 25/96, MWG (Biotech, Ebersberg, Germany). The initial denaturation step occurred at 95 °C for 2 min, and was followed by 30 cycles of denaturation at 94 °C for 40 s, annealing at 57 °C for 1 min, extension at 72 °C for 1 min, and a final extension at 72 °C for 2 min. The products obtained were analyzed on a 1.5% agarose gel electrophoresis performed at 4 V/cm in Tris-acetate-EDTA buffer (TAE). The gel was stained with 0.5 μg/mL ethidium bromide and visualized on an ultraviolet transilluminator (Bioamerica, Miami, USA) A 100 bp ladder digest (Invitrogen) was used as a standard molecular weight. The expected products after amplification were 250 bp and 510 bp in length. A pair of ubiquitous bacterial primers (5′-AGA GTT TGA TCC TGG CTC AG-3′ and 5′-ACG GCT ACC TTG TTA CGA CTT-3′) designed by Willis et al. (1999) was used to indicate the presence of bacterial DNA in the clinical samples, particularly the ones that were P. aeruginosa-negative. For the reaction using the universal 16S rDNA primers, PCR amplification included a 25 μL reaction mixture containing 0.2 mM of forward and reverse universal primers, 2.5 μL of 10× PCR buffer, 2 mM MgCl2, 1.25 U of Platinum® Taq DNA polymerase, and 200 mM of each deoxynucleosidetriphosphate (Invitrogen). The PCR program included an initial denaturation step at 97 °C for 1 min, followed by 30 cycles of a denaturation step at 97 °C for 45 s, a primer annealing step at 55 °C for 45 s, an extension step at 72 °C for 1 min and a final step of 72 °C for 4 min. The presence of bacterial DNA was determined by an amplicon of 1,505 bp in size visualized on a 1.5% agarose gel. Acinetobacter spp. detection was carried out by a nested PCR (Broderick et al., 2004; Vanbroekhoven et al., 2004). The first amplification was performed with the bacterial 16S rRNA universal primers 27f, 5′-AGA GTT TGA TCC TGG CTC AG-3′ and 1492r, 5′-TAC GGC TAC CTT GTT ACG ACT T-3′ (amplicon of 1450 bp). Approximately 100 ng of sample DNA was added into a 50 μL PCR mixture containing 0.5 pmol of each primer, 400 μM of each dNTP, 3 mM MgCl2, 10× PCR buffer (20 mM Tris-HCl [pH 8.4], 50 mM KCl), and 1.5 U Platinum® Taq DNA polymerase (Invitrogen). The amplification program included an initial step of 95 °C for 5 min followed by 35 cycles of denaturation at 94 °C for 30 s, 55 °C for 1 min, 72 °C for 1 min, and a final step of 72 °C for 5 min [34]. The second amplification was performed with the primers Ac436f, 5′-TTT AAG CGA GGA GGA GG-3′ and Ac676r, 5′-ATT CTA CCA TCC TCT CCC-3′ (amplicon of 280 bp) in a reaction volume of 50 μL containing 2 pmol of primer Ac436f, 1 pmol of primer Ac676r, 200 μM of each dNTP, 2 mM MgCl2, 10× PCR buffer, and 1.5 U Platinum® Taq DNA polymerase (Invitrogen). The amplification programme was carried out according to Vanbroekhoven et al. (2004).
Statistical analysis
All statistical tests were performed using the Statistical Package for the Social Sciences (SPSS Inc®.v.10 Chicago, IL, version 17). Full-mouth clinical measurements were computed for each subject and then average across subjects within the groups. Differences on clinical parameters between groups were sought using Mann-Whitney and χ2 tests. The frequency of detection of P. aeruginosa and Acinetobacter spp. were computed for each subject, and significant differences between groups were sought using the χ2 test. Associations between demographic and periodontal parameters, and the frequency of P. aeruginosa and Acinetobacter spp. were examined by the χ2 test. Predictor variables for periodontitis were investigated using multivariate logistic regression analysis, from which ORs with 95% CI were reported. Statistical significance was reached at the 5% level.
Results
The demographic and periodontal clinical features of the subject groups are shown in Table 1. CP patients comprised of a significantly higher proportion of smokers (p < 0.01, Chi-square test) and older individuals (p < 0.01; Mann-Whitney test) than PH subjects. No significant differences between groups were observed for gender. As expected, all clinical parameters of periodontal tissue destruction and inflammation (tooth loss, PD, CAL, BOP, SB and SUP) were significantly greater in the CP group compared to controls (p < 0.01; Mann-Whitney test).
Table 1.
Demographic and full-mouth clinical parameters (mean ± SD) of Periodontally Healthy (PH) and Chronic Periodontitis (CP) subjects of the study population.
| Clinical parameters | PH (N = 55) | CP (N = 169) |
|---|---|---|
| (%) Males | 31 | 35 |
| (%) Smokers** | 6 | 25 |
| Age (years)* | 31.1 ± 11 | 40.2 ± 14 |
| Number of missing teeth* | 0.95 ± 1.6 | 5.6 ± 5.7 |
| Probing depth (mm)* | 1.9 ± 0.5 | 2.9 ± 1.1 |
| Clinical attachment level (mm)* | 2.0 ± 0.6 | 3.7 ± 1.5 |
| % sites with: | ||
| Bleeding on probing* | 2.5 ± 4.7 | 43 ± 28 |
| Supragingival biofilm* | 11.5 ± 11 | 45 ± 30 |
| Suppuration* | 0 | 2 ± 7 |
Refers to p < 0.01, Mann-Whitney test;
refers to p < 0.01, Chi-square test.
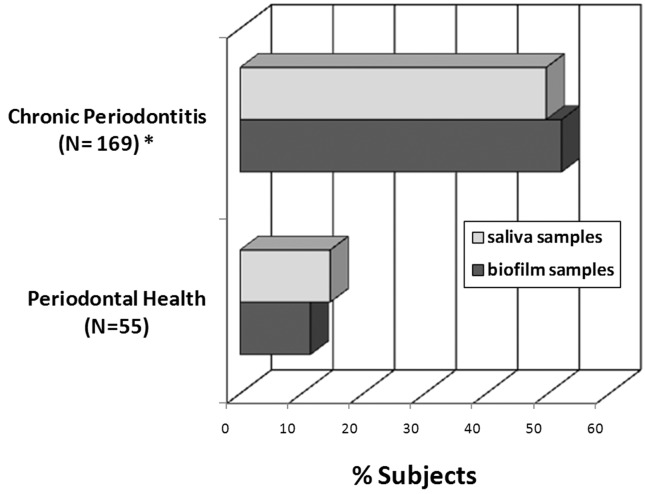
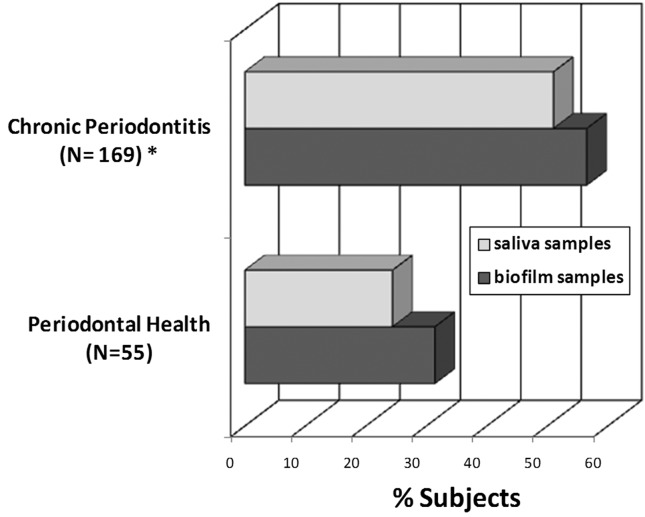
Overall, P. aeruginosa was detected in 40% and Acinetobacter spp. in 45% of all samples. The frequency of P. aeruginosa and Acinetobacter spp. in saliva and biofilm samples from both clinical groups are depicted in Figures 1 and 2. In saliva samples, a significant higher prevalence of P. aeruginosa and Acinetobacter spp. were observed in CP subjects (49.7% and 51%, respectively) compared to PH controls (14.6% and 24.4%, respectively) [p < 0.01; Chi-square test]. Likewise, P. aeruginosa and Acinetobacter spp. were detected significantly more often in subgingival biofilm samples from CP (52.2% and 56.5%, respectively) than PH subjects (11.4% and 31.4%, respectively) [p < 0.01; Chi-square test]. Due to significant differences in smoking status and age between CP and PH, the prevalence of P. aeruginosa and Acinetobacter spp. were evaluated only in non-smokers and subjects with ≤ 35 years of age. Both pathogens were more frequently detected in CP than PH subjects (p < 0.05; Chi-square test, data not shown). Moreover, there were no significant differences in the distribution of P. aeruginosa and Acinetobacter spp. between men and women, subgingival biofilm and saliva samples, patients ≤ 35 and > 35 years of age, and smokers and non-smokers regardless periodontal status (p > 0.05, Chi-square test, Table 2). Associations between P. aeruginosa and Acinetobacter spp. and clinical parameters of disease were also examined (Table 2). For this analysis, only subgingival biofilm samples were considered. Patients with large proportions of visible supragingival biofilm, generalized BOP and periodontal destruction (PD and CAL > 4 mm) presented higher frequencies of P. aeruginosa and Acinetobacter spp. compared to individuals with less signs of disease (p < 0.01, Chi-square test, Table 2). In addition, a strong association between the detection of both species was found. Of the samples positive for P. aeruginosa, 76.4% were also positive for Acinetobacter spp., whereas of the samples negative for P. aeruginosa, 75.2% were negative for Acinetobacter spp. (p < 0.001, Chi-square test, data not shown).
Figure 1.
Frequency of detection of P. aeruginosa in saliva and subgingival biofilm samples from periodontally healthy (N = 55) and chronic periodontitis subjects (N = 169). * Significant difference between groups for saliva and biofilm samples (p < 0.05; χ2 test).
Figure 2.
Frequency of detection of Acinetobacter spp. in saliva and subgingival biofilm samples from periodontally healthy (N = 55) and chronic periodontitis subjects (N = 169). * Significant difference between groups for saliva and biofilm samples (p < 0.05; χ2 test).
Table 2.
Association between the frequency (%) of P. aeruginosa and Acinetobacter spp. and demographic and periodontal clinical parameters in all subjects.
| Demographic and clinical parameters | % P. aeruginosa | % Acinetobacter spp. |
|---|---|---|
| Gender | ||
| males | 37 | 44 |
| females | 42 | 46 |
| Smoking status | ||
| non-smokers | 39 | 43 |
| smokers | 46 | 58 |
| Age | ||
| ≤ 35 years | 35 | 43 |
| > 35 years | 44 | 47 |
| Type of sample | ||
| Saliva | 42 | 45 |
| Subgingival biofilm | 35 | 46 |
| Probing depth * | ||
| ≤ 4 mm | 16 | 28.4 |
| > 4 mm | 55 | 65 |
| Clinical attachment level * | ||
| ≤ 4 mm | 16.3 | 29 |
| > 4 mm | 50 | 57 |
| Bleeding on probing * | ||
| ≤ 30% of sites | 15 | 29 |
| > 30% of sites | 54 | 56 |
| Supragingival biofilm * | ||
| ≤ 30% of sites | 14 | 27 |
| > 30% of sites | 50 | 56 |
Refers to p < 0.01, Chi-square test.
Stepwise logistic regression was performed in order to examine associations between demographic, clinical and bacterial parameters and periodontal status. Table 3 shows the predictor variables entered in the final model to distinguish between periodontal health and chronic periodontitis. Smokers (OR 10.22, CI 2.61–40.0, p < 0.01) presenting P. aeruginosa (OR 7.68, CI 3.13–18.85, p < 0.001) and high frequencies of supragingival plaque (OR 3.77, CI 1.12–12.62, p < 0.05) were more likely to present chronic periodontitis than periodontal health.
Table 3.
Final model of the stepwise logistic regression analysis including demographic, clinical and bacterial parameters as predictor variables for chronic periodontal disease.
| Variables ** | β | OR* | Lower 95% CI | Upper 95% CI | p |
|---|---|---|---|---|---|
| Constant | −1.35 | 0.26 | < 0.001 | ||
| Smoker | 2.32 | 10.22 | 2.61 | 40.0 | < 0.01 |
| Pseudomonas aeruginosa | 2.04 | 7.68 | 3.13 | 18.85 | < 0.001 |
| Supragingival biofilm | 1.33 | 3.77 | 1.12 | 12.62 | < 0.05 |
Reference: Periodontally healthy;
Variables entered on step 1: gender (male 1/female 0), smoking (smokers 1/non-smokers 0), age (> 35 years 1/≤ 35 years 0), PD and CAL (> 4 mm 1/≤ 4 mm 0), supragingival biofilm and BOP (> 30% of sites 1/≤ 30% of sites 0), P. aeruginosa and Acinetobacter spp. (positive 1/negative 0).
Discussion
Over the last few years, studies have indicated that the presence of non-oral species in the oral microbiota is not a transitory event or a result of contamination during sampling, and that the oral cavity may be a reservoir for medically important pathogens (Barbosa et al., 2001; Botero et al., 2007; Colombo et al., 2002; Da Silva-Boghossian et al., 2011; Fourrier et al., 1998; Fritschi et al., 2008; Gonçalves et al., 2009; Slots et al., 1988, 1990; Souto and Colombo, 2008; Souto et al., 2006). It is possible that in addition to putative periodontal pathogens, species such as Acinetobacter spp. and P. aeruginosa, either associated with oral pathogens or not, may also play a role in the etiopathogenesis of periodontal diseases. The current investigation examined the subgingival biofilm and saliva from patients with chronic periodontitis and periodontal health for the presence of P. aeruginosa and Acinetobacter spp. Data correlating periodontal infection and P. aeruginosa or Acinetobacter spp. colonization have indicated that, in general, the prevalence of these species in the oral cavity gets greater proportions in individuals with periodontitis (Barbosa et al., 2001; Botero et al., 2007; Colombo et al., 1998, 2002, 2009; Da Silva-Boghossian et al., 2011; Fourrier et al., 1998; Fritschi et al., 2008; Gonçalves et al., 2009; Persson et al., 2008; Slots et al., 1988, 1990; Souto and Colombo, 2008; Souto et al., 2006). Likewise, our results showed a significantly higher frequency of P. aeruginosa and Acinetobacter spp. in saliva and subgingival biofilm samples from periodontitis patients compared to periodontally healthy control. These species have also been associated with treatment failure in patients with refractory periodontitis treated with mechanical and antimicrobial therapy (Colombo et al., 1998, 2009). Conceivably, the high rates of resistance to multiple prescribed broad spectrum antibiotics, as well as the ability to produce biofilms may result in the persistence of these pathogens in the periodontal pocket after therapy (Pihl et al., 2010; Smith et al., 2003; Woods et al., 1982). Associations between the presence of P. aeruginosa and Acinetobacter spp. and demographic and periodontal clinical parameters were also examined. Both species were detected in significantly higher frequencies in subjects with larger proportions of visible supragingival biofilm, BOP, PD and CAL, reinforcing the relationship of these pathogens with inflammation, tissue destruction and poor oral hygiene (Abe et al., 2001; Colombo et al., 2002, 2009; Da Silva-Boghossian et al., 2011; Paju and Scannapieco, 2007; Scannapieco et al., 1998; Souto and Colombo, 2008; Souto et al., 2006). Indeed, these microorganisms produce virulence factors of relevance to the pathogenesis of periodontitis. P. aeruginosa is tissue invasive, elaborates extracellular leukotoxins, suppresses lymphocyte proliferation, inactivate complement components, degrades basement membrane laminin and releases potent enterotoxins and endotoxins (Pihl et al., 2010; Smith et al., 2003; Woods et al., 1982). Acinetobacter spp. are considered major nosocomial pathogens (Bergogne-Berezin et al., 1996) accounting for about 80% of reported infections. Members of the genus seem to have a remarkable ability to develop resistance to even the most potent antimicrobial agents (Towner, 1997). Obana (1986) demonstrated that some clinical isolates had slime-producing ability, and that the slime also enhanced the virulence of other species, indicating the potential role of Acinetobacter in the enhancement of virulence in mixed infections.
In structured communities, coaggregation of different bacterial species seems to have evolved as an efficient strategy to optimize local opportunities. Studies have indicated that species of Acinetobacter and Pseudomonas, when inserted in highly organized mixed communities, may cooperatively interact with each other, exhibiting direct metabolic communications and frequent genetic exchanges of multi-drug-resistant genes due to the close evolutionary proximity (Fournier et al., 2006; Hansen et al., 2007). We found that over 70% of sites colonized by P. aeruginosa were colonized by Acinetobacter spp. In a previous study with hospitalized individuals, our group also demonstrated a strong association between the oral colonization of these microorganisms (Zuanazzi et al., 2010). Furthermore, P. aeruginosa and Acinetobacter spp. may interact with other species such as periodontal pathogens. Da Silva-Boghossian et al. (2011) demonstrated that Aggregatibacter actinomycetemcomitans, A. baumannii and the red complex (Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola) associated with P. aeruginosa in the subgingival microbiota increased significantly the likelihood of a subject having aggressive periodontitis. P. aeruginosa also seemed to have a synergism with A. actinomycetemcomitans increasing the risk for periodontal disease. Likewise, Persson et al. (2008) showed that P. aeruginosa along with T. forsythia were independent predictors for periodontal disease. However, the exact mechanisms involved in these complex interactions are not elucidated yet.
In summary, our findings provide additional evidence for the oral cavity as a reservoir for P. aeruginosa and Acinetobacter spp., as well as of the association of these microorganisms with the presence of periodontal infection. Considering that eradication of this species from subgingival biofilm in deep periodontal pockets may be limited, close attention should be given to these patients in order to reduce the risk for periodontal breakdown and/or development of systemic diseases caused by these species in other areas of the body.
Acknowledgments
This work was supported in part by National Council for Scientific and Technological Development (CNPq), Coordination of Improvement of Higher Education Personnel (CAPES), and Foundation for Research Support of the State of Rio de Janeiro (FAPERJ), Brazil.
References
- Abe S, Ishihara K, Okuda K. Prevalence of potential respiratory pathogens in the mouths of elderly patients and effects of professional oral care. Arch Gerontol Geriatr. 2001;32:45–55. doi: 10.1016/s0167-4943(00)00091-1. [DOI] [PubMed] [Google Scholar]
- Agodi A, Barchitta M, Cipresso R, Giaquinta L, Romeo MA, Denaro C. Pseudomonas aeruginosa carriage, colonization, and infection in ICU patients. Intensive Care Med. 2007;33:1155–1161. doi: 10.1007/s00134-007-0671-6. [DOI] [PubMed] [Google Scholar]
- Ali RWC, Bakken V, Nilsen R, Skaug N. Comparative detection frequency of 6 putative periodontal pathogens in Sudanese and Norwegian adult periodontitis patients. J Periodontol. 1994;65:1046–1052. doi: 10.1902/jop.1994.65.11.1046. [DOI] [PubMed] [Google Scholar]
- Ali RWC, Velcescu MC, Jivanescu B, Lophtus B, Skaug N. Prevalence of 6 putative periodontal pathogens in subgingival plaque samples from Romanian adult periodontitis patients. J Clin Periodonto. 1996;18:411–415. doi: 10.1111/j.1600-051x.1996.tb00546.x. [DOI] [PubMed] [Google Scholar]
- Barbosa FC, Mayer MP, Saba-Chujfi E, Cai S. Subgingival occurrence and antimicrobial susceptibility of enteric rods and pseudomonads from Brazilian periodontitis patients. Oral Microbiol Immunol. 2001;16:306–310. doi: 10.1034/j.1399-302x.2001.016005306.x. [DOI] [PubMed] [Google Scholar]
- Bergogne-Berezin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: Microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9:148–165. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botero JE, Contreras A, Lafaurie G, Jaramillo A, Betancourt M, Arce RM. Occurrence of periodontopathic and superinfecting bacteria in chronic and aggressive periodontitis subjects in a Colombian population. J Periodontol. 2007;78:696–704. doi: 10.1902/jop.2007.060129. [DOI] [PubMed] [Google Scholar]
- Broderick NA, Raffa KF, Goodman RM, Handelsman J. Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl Environ Microbiol. 2004;70:293–300. doi: 10.1128/AEM.70.1.293-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastre J, Fagon J. Ventilator-associated pneumonia. Am J Respir Crit Care Med. 2002;165:867–903. doi: 10.1164/ajrccm.165.7.2105078. [DOI] [PubMed] [Google Scholar]
- Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo AP, Haffajee AD, Dewhirst FE, Paster BJ, Smith CM, Cugini MA, et al. Clinical and microbiological features of refractory periodontitis subjects. J Clin Periodontol. 1998;25:169–180. doi: 10.1111/j.1600-051x.1998.tb02424.x. [DOI] [PubMed] [Google Scholar]
- Colombo AP, Teles RP, Torres MC, Souto R, Rosalem WJ, Mendes MC, et al. Subgingival microbiota of Brazilian subjects with untreated chronic periodontitis. J Periodontol. 2002;73:360–369. doi: 10.1902/jop.2002.73.4.360. [DOI] [PubMed] [Google Scholar]
- Da Silva-Boghossian CM, Souto R, Luiz RR, Colombo APV. Association of red complex, A. actinomycetemcomitans and non-oral bacteria with periodontal diaseases. Arch Oral Biol. 2011;56:899–906. doi: 10.1016/j.archoralbio.2011.02.009. [DOI] [PubMed] [Google Scholar]
- De Vos D, Lim A, Jr, Pirnay JP, Struelens M, Vandenvelde C, Duinslaeger L, et al. Direct detection and identification of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectorations by multiplex PCR based on two outer membrane lipoprotein genes, oprI and oprL. J Clin Microbiol. 1997;35:1295–1299. doi: 10.1128/jcm.35.6.1295-1299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourrier F, Duvivier B, Boutigny H, Roussel-Delvallez M, Chopin C. Colonization of dental plaque: A source of nosocomial infections in intensive care unit patients. Crit Care Me. 1998;26:301–308. doi: 10.1097/00003246-199802000-00032. [DOI] [PubMed] [Google Scholar]
- Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, et al. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2006;10:2–7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritschi BZ, Albert-Kiszely A, Persson GR. Staphylococcus aureus and other bacteria in untreated periodontitis. J Dent Re. 2008;87:589–593. doi: 10.1177/154405910808700605. [DOI] [PubMed] [Google Scholar]
- Hansen SK, Haagensen JA, Gjermansen M, Jorgensen TM, Tolker-Nielsen T, Molin S. Characterization of a Pseudomonas putida rough variant evolved in a mixed-species biofilm with Acinetobacter sp. strain C6. J Bacteriol. 2007;189:4932–4493. doi: 10.1128/JB.00041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Goncalves LS, Souto R, Colombo AP. Detection of Helicobacter pylori, Enterococcus faecalis, and Pseudomonas aeruginosa in the subgingival biofilm of HIV-infected subjects undergoing HAART with chronic periodontitis. Eur J Clin Microbiol Infect Dis. 2009;28:1335–1342. doi: 10.1007/s10096-009-0786-5. [DOI] [PubMed] [Google Scholar]
- Karlowsky JA, Draghi DC, Jones ME, Thornsberry C, Friedland IR, Sahm DF. Surveillance for antimicrobial susceptibility among clinical isolates of Pseudomonas aeruginosa and Acinetobacter baumannii from hospitalized patients in the United States, 1998 to 2001. Antimicrob Agents Chemother. 2003;47:1681–1688. doi: 10.1128/AAC.47.5.1681-1688.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine MJ, Farre MA, Crusius BA, Pena S. The mouthwash: A non- invasive sampling method to study cytokine gene polymorphisms. J Periodontol. 2000;71:1315–1318. doi: 10.1902/jop.2000.71.8.1315. [DOI] [PubMed] [Google Scholar]
- Luna CM, Aruj PK. Nosocomial Acinetobacter pneumonia. Respirology. 2007;12:787–791. doi: 10.1111/j.1440-1843.2007.01147.x. [DOI] [PubMed] [Google Scholar]
- Obana Y. Pathogenic significance of Acinetobacter calcoaceticus: Analysis of experimental infection in mice. Microbiol Immunol. 1986;30:645–657. doi: 10.1111/j.1348-0421.1986.tb02991.x. [DOI] [PubMed] [Google Scholar]
- Page RC, Kornman KS. The pathogenesis of human periodontitis: An introduction. Periodontol 2000. 1997;14:9–11. doi: 10.1111/j.1600-0757.1997.tb00189.x. [DOI] [PubMed] [Google Scholar]
- Paju P, Scannapieco FA. Oral biofilms, periodontitis, and pulmonary infections. Oral Diseases. 2007;13:508–512. doi: 10.1111/j.1601-0825.2007.1410a.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- Persson GR, Hitti J, Paul K, Hirschi R, Weibel M, Rothen M, et al. Tannerella forsythia and Pseudomonas aeruginosa in subgingival bacterial samples from parous women. J Periodontol. 2008;79:508–516. doi: 10.1902/jop.2008.070350. [DOI] [PubMed] [Google Scholar]
- Pihl MLEC, Schimidtchen A, Svensater G, Davies JR. Effects of clinical isolates of Pseudomonas aeruginosa on Staphylococcus epidermidis biofilm formation. FEMS Immunol Med Microbiol. 2010;59:504–512. doi: 10.1111/j.1574-695X.2010.00707.x. [DOI] [PubMed] [Google Scholar]
- Scannapieco FA. Position paper of The American Academy of Periodontology: Periodontal disease as a potential risk factor for systemic diseases. J Periodontol. 1998;69:841–850. [PubMed] [Google Scholar]
- Scannapieco FA, Papandonatos GD, Dunford RG. Associations between oral conditions and respiratory disease in a national sample survey population. Ann Periodontol. 1998;3:251–256. doi: 10.1902/annals.1998.3.1.251. [DOI] [PubMed] [Google Scholar]
- Slots J, Feik D, Rams TE. Prevalence and antimicrobial susceptibility of Enterobacteriacea, Psedomonadaceaea and Acinetobacter in human periodontitis. Oral Microbiol Immunol. 1990;5:149–154. doi: 10.1111/j.1399-302x.1990.tb00413.x. [DOI] [PubMed] [Google Scholar]
- Slots J, Rams TE, Litsgarten MA. Yeasts, enteric rods and pseudomonads in the subgingival flora of severe adult periodontitis. Oral Microb Immunol. 1988;3:47–52. doi: 10.1111/j.1399-302x.1988.tb00080.x. [DOI] [PubMed] [Google Scholar]
- Slots J, Rams TE, Feik D, Taveras HD, Gillespie GM. Subgingival microflora of advanced periodontitis in the Dominican Republic. Oral Microbiol Immunol. 1991;62:543–547. doi: 10.1902/jop.1991.62.9.543. [DOI] [PubMed] [Google Scholar]
- Smith RS, Iglewski BH. P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol. 2003;6:56–60. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Souto R, Colombo AP. Detection of Helicobacter pylori by polymerase chain reaction in the subgingival biofilm and saliva of non-dyspeptic periodontal patients. J Periodontol. 2008;79:97–103. doi: 10.1902/jop.2008.070241. [DOI] [PubMed] [Google Scholar]
- Souto R, de Andrade AFB, Uzeda M, Colombo APV. Prevalence of “non-oral” pathogenic bacteria in subgingival biofilm of subjects with chronic periodontitis. Braz J Microbiol. 2006;37:208–215. [Google Scholar]
- Towner KJ. Clinical importance and antibiotic resistance of Acinetobacter spp. J Med Microbiol. 1997;46:721–746. doi: 10.1099/00222615-46-9-721. [DOI] [PubMed] [Google Scholar]
- Vanbroekhoven K, Ryngaert AK, Wattiau P, Mot R, Springael D. Acinetobacter diversity in environmental samples assessed by 16S rRNA gene PCR-DGGE fingerprinting. FEMS Microbiol Ecol. 2004;50:37–50. doi: 10.1016/j.femsec.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Wagner VE, Iglewski BH. P. aeruginosa biofilms in CF infection. Clin Rev Allerg Immul. 2008;35:124–134. doi: 10.1007/s12016-008-8079-9. [DOI] [PubMed] [Google Scholar]
- Willis SG, Smith KS, Dunn VL, Gapter LA, Riviere KH, Riviere GR. Identification of seven Treponema species in health- and disease-associated dental plaque by nested PCR. J Clin Microbiol. 1999;37:867–869. doi: 10.1128/jcm.37.3.867-869.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DE, Cryz SJ, Friedman RL, Iglewski BH. Contribution of toxin A and elastase to virulence of Pseudomonas aeruginosa in chronic lung infections of rats. Infect Immun. 1982;36:1223–1228. doi: 10.1128/iai.36.3.1223-1228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuanazzi D, Souto R, Mattos MB, Zuanazzi MR, Tura BR, Sansone C, Colombo AP. Prevalence of potential bacterial respiratory pathogens in the oral cavity of hospitalised individuals. Arch Oral Biol. 2010;55:21–28. doi: 10.1016/j.archoralbio.2009.10.005. [DOI] [PubMed] [Google Scholar]