Abstract
Oxacillin/methicillin-resistance is related to the mecA and its regulatory genes mecR1 and mecI. Its origin is still unknown, although evidences support that it is related to CNS, once mecA and a homologue gene, pbpD, were both detected in Staphylococcus sciuri species group. The present work evaluated 210 samples of skin and ear swabs from rodents and 60 nasal swabs from equines of Army Biologic Institute, Rio de Janeiro. Pheno- and genotypic characterization provided 59.52% (25/42) and 78.57% (11/14) S. lentus and S. sciuri, respectively. It was observed that although all S. sciuri isolates tested positive for pbpD, there was no correlation with oxacillin-resistance. On the other hand, isolates tested positive for mecA gene also presented phenotypic oxacillin-resistance in at least one assay. The alignment of the mecA gene showed that the nucleotide sequences were sorted into 2 different groups, one comprising the bovine strains and the other containing human and equine strains.
Keywords: Staphylococcus sciuri, mecA, pbpD, antimicrobial resistance
Antimicrobial resistance is a critical health issue around the world. Oxacillin/methicillin-resistant Staphylococcus aureus represents an ever-increasing global threat especially considering vulnerable people (Souza et al., 2012). Oxacillin/methicillin-resistance is related to the integration of a Staphylococcal cassette chromosome (SSCmec) in the bacterial chromosome made up of a mec gene complex, containing methicillin-resistance determinant mecA and its regulatory genes mecR1 and mecI, and also the ccr gene complex, encoding site-specific recombinase responsible for the movement of the element. The mecA gene codifies a variant of penicillin-binding protein PBP2, termed PBP2a or PBP2’, whose affinity with the beta-lactamic antibiotics is very low (Ito et al., 2004). SCC mechas been identified not only in S. aureusbut also in other coagulase-positive and coagulase-negative staphylococci. Its origin is still unknown, although evidences support that it is related to coagulase-negative staphylococci, once mecA and a homologue gene, pbpD, were both detected in Staphylococcus sciuri species group (Couto et al., 2010, Tsubakishita et al., 2010). S. sciuri species group comprises S. sciuri, S. vitulinus, S. lentus, and S. fleurettii. A recent report considered S. fleurettii and S. sciuri as the highly probable origin of the mecA gene (Tsubakishita et al., 2010). The present work evaluated the oxacillin phenotypic antimicrobial resistance pattern and detected mecA and pbpD genes in S. sciurispecies group from rodents and equines of Army Biologic Institute, Rio de Janeiro. Also, mecA sequences from sciuri group strains were compared to others species in order to evaluate the degree of similarity among them.
A total of 210 samples from the skin and ear swabs of rodents and 60 from nasal swabs from the equines were analyzed and provided 56 S. sciurispecies group isolates. All 270 samples were aseptically collected and transported to the laboratory in coolers with ice (4–8 °C) for the bacteriological analyses. These samples were plated in duplicate in sheep blood agar (Oxoid), and incubated in aerobic conditions for 24 h at 37 °C. The colonies were identified according to routine microbiological diagnostics, including cultural properties, catalase and coagulase production. Then CNS isolates were submitted to phenotypic assays carried out as described by Stepanovic et al. (2005) considering the following tests: oxidase, novobiocin susceptibility, acid production from rafinose, mannose, arabinose, maltose, celobiose, galactose, lactose and salicin. All 56 samples phenotipically identified as Sciuri group were submitted to genotypic assays for S. sciuri and S. lentus were performed according to Yasuda et al. (2002) using the following primers: 16S rRNA Staphylococcus spp. (756 bp) 5′-AAC TCT GTT ATT AGG GAA GAA CA - 3′ and 5′-CCA CCT TCC TCC GGT TTG TCA CC - 3′, 16S rRNA S. sciuri (872 bp) 5′-GAA CCG CAT GGT TCA ATA G - 3′ and 5′-GAC TCT ATC TCT AGA GCG G - 3′, 16S rRNA S. lentus (872 bp) 5′-GAA CCG CAT GGT TCA ATG T - 3′ and 5′-AAC TCT ATC TCT AGA GCG A - 3′. Methicillin-resistance detection was performed according to the recommendations of the Clinical Laboratory Standard Institute (CLSI, 2012) through oxacillin and cefoxitin disk-diffusion test considering all strains presenting an inhibition zone diameter [#LWEQ#] 21 mm as resistant. Polymerase Chain Reactions (PCRs) for the detection of of mecA complex and pbpD genes was carried out as following: DNA was extracted according to the procedures described for the NucliSens® mini MAG System (BioMérieux). PCR for mecA gene (513 bp) was carried out using the primers outlined by Murakami et al. (1991) and methodology described by Coelho et al. (2007). PCRs for regulatory mecI and mecRI genes were carried out using the primers and methodology outlined by Rosato et al. PCR screening for pbpD gene (1020 bp) was carried out using the primers and methodology outlined by Couto et al. (2010): 5′-ATC CAT CAA TAT TGA ACC A -3′ and 5′-TAT ATC TTC ACC AAC ACC -3′. Amplicons were detected by 1.5% agarose gel, stained with SYBR Green (Invitrogen) and examined under UV transilluminator (UvTrans). Also, three mecA gene sequences obtained from S. sciuri and S. lentus isolated from equines at the present study were included in a further step to evaluate the degree of similarity among mecA from sciuri group strains and others species. For this study, distinct primer pairs were designed to amplify different but overlapping segments of the whole mecA gene based on the nucleotide sequence of Staphylococcus aureus (HE681097) and Staphylococcus sciuri (AY820253) (Melo, 2013). The products of mecA gene PCR product were purified using Exo-Sap (USB Corporation) according to the manufacturer’s recommended protocol. To assure fidelity, sequencing of both strands was performed at Helixxa Company, Brazil. Nucleotides sequences were edited using the software Bioedit version 7.0.9.0 (Hall, 1999) and assembled using the Mobyle@Pasteur program – (http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::merger) (Rice et al., 2000). Dendogram was performed by a neighbor joining (NJ) algorithm method using p-distance model with MEGA version 4.0 (Tamura et al., 2007). The robustness of each branch was determined using the non-parametric bootstrap test (Felsenstein, 1985) with 1000 replicates.
Phenotypic identification provided 42 S. lentus and 14 S. sciuri isolates. After genotypic characterization, this profile had changed for 25 S. lentus (44.64%), being 24 from rodents and just one from equine, and 11 S. sciuri (19.64%), 8 from equine and 3 from rodents. Pheno- and genotypic results were concordant for 59.52% (25/42) and 78.57% (11/14) S. lentus and S. sciuri isolates, respectively. The remaining 20 isolates (35.72%) did not amplify the specific genes and from now on will be considered for this study as sciurispecies group. It is important to emphasize that this difference in species identification due to atypical biochemical patterns and that most routine diagnostic laboratories only perform phenotypic identification which can lead to misidentification. Genotypic characterization assays are more reliable, otherwise, it is still too expensive once equipments and skilled staff needs make it difficult to be used routinely. For such a long time, CNS Staphylococcus has been considered harmless and minor important species. Nowadays, the advances in molecular taxonomy and systematic allow researchers to a better comprehension of their role. Besides a direct clinical impact in diseases etiology, they are been investigated as reservoirs of resistance genes and consequently as part of the threat to the public represented by antimicrobial resistance. The sciuri group species are rarely associated to human or animal infections. Otherwise, similarities of mecA and pbpD genes suggest a closely related phylogeny (Couto et al., 2010). In our study, sciuri species group were evaluated for oxacillin-resistance by means of oxacillin and cefoxitin disk diffusion and mecA and pbpD genes detection. It was detected a 14.29% oxacillin- and 8.93% cefoxitin-resistance pattern by disk diffusion assay. Stepanovic et al. (2005) detected oxacillin-resistant S. sciuri strains from human clinical samples. Bagcigil et al. (2007) isolated oxacillin-resistant CNSs from nasal cavities of equines exposed to antimicrobials, including β-lactams. PCRs for pbpD, mecA, mecI and mecRI genes revealed that 21.42% (12/56) isolates tested positive for pbpD, 10.71% (6/56) for mecA and 5.35% (3/56) for both mecI and mecRI. Although 12 isolates tested positive for pbpD, no correlation was observed with antimicrobial resistance for isolates that presented only mecA homologue (Table 1). These results agree to the reports of its relation to essential functions as growth and cell wall synthesis (Antignac and Tomasz, 2008). On the other hand, isolates tested positive for mecA gene also presented phenotypic resistance in at least one assay.
Table 1.
Distribution of sciuri group Staphylococcus patterns.
| Pattern (n*) | Disk diffusion | Genes | Species | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Oxacillin | Cefoxitin | pbpD | mecA | mecI | mecRI | ||
| 1 (2) | R** | R | + | + | + | + | S. sciuri |
| 2 (1) | R | R | − | + | + | + | S. lentus |
| 3 (1) | S* | S | + | − | − | − | S. sciuri group |
| 4 (2) | R | R | − | + | − | − | S. sciuri group |
| 5(1) | R | S | − | + | − | − | S. sciuri group |
| 6(1) | R | S | − | − | − | − | S. lentus |
| 7 (9) | S | S | + | − | − | − | S. sciuri |
n: number of isolates;
R: resistant; S: sensible.
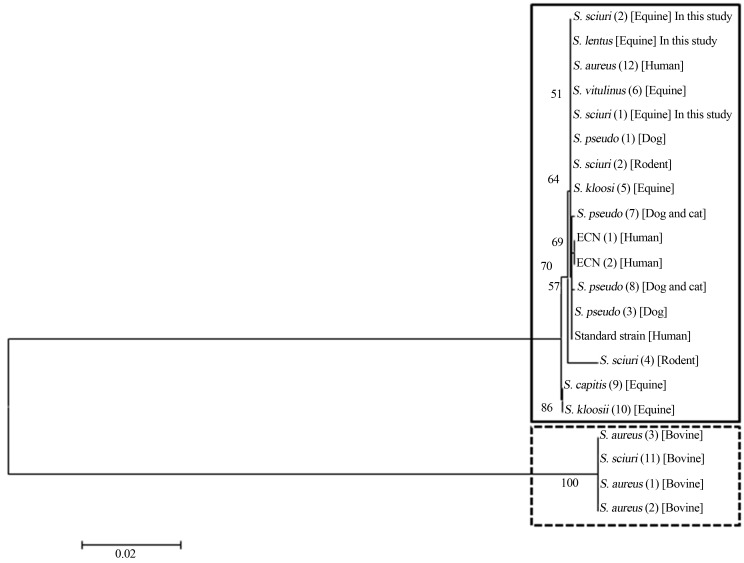
PCR using primers outlined by Melo (2013) based on the nucleotide sequence of S. aureus mecA gene (HE681097) successfully amplified mecA gene fragments from the two S. sciuri and one S. lentus from equine. The resulting mecA gene fragments were sequenced and the overlapping reads were assembled in contigs. Otherwise, newly designed primers based on bovine S. sciuri mecA sequence (AY820253) tested negative for this isolates. Sequences of mecA gene from different hosts provided by our studies and available at NCBI GenBank were used to generate a dendrogram (Figure 1). Genomic divergences between mecA genes originated two different clusters of Staphylococcus spp., one comprising dog, cat, rodent, equine and human isolates and the other just bovine isolates (Figure 1). The puzzling question concerning mecA gene evolutive pathway is being investigated for researches around the world. The idea that it has been generated in or transferred into a broad-host-range Staphylococcus species is well established. Also it is recognized that the mecA region carried by SCC mec might have spread beyond the host animal species (Coelho et al., 2007). Otherwise, there are some blanks to be filled such as SCC mecis generated inS. aureus, which is both a human- and animal-relatedStaphylococcusspecies.
Figure 1.
Dendrogram showing the genetic divergence of nucleotide sequences of bovine (dotted line) and others species (continuous line) Staphylococcal mecA gene. Sequences available of GenBank: S. pseudo (1): S. pseudintermedius: AM904731; S. sciuri (2): S. sciuri: Y13096; S. pseudo (3): S. pseudintermedius: AM904732; S. sciuri (4): S. sciuri: Y13095; S. kloosii (5): S. kloosii: AM048803; S. vitulinus (6): S. vitulinus: AM048802; S. pseudo (7): S. pseudintermedius: EU929082; S. pseudo (8): S. pseudintermedius: EU929081; S. capitis (9): S. capitis: AM048805; S. kloosii (10): S. kloosii: AM048804; S. sciuri (11): S. sciuri: AY820253; S. aureus (12): S. aureus: HE681097. Sequences of Melo (10): Standard Strain: S. aureus; ECN (1) and (2): CNS isolates 1 and 2; S. aureus (1), (2) and (3): S. aureus isolates 1, 2 and 3. Sequences of this study: S. sciuri (1) and (2): S. sciuri isolates 1 and 2; S. lentus: S. lentus.
We have been recently confronted with the rapid spread of community-acquired MRSA and facilitated transmission of animal-derived MRSA to humans (Bagcigil et al., 2007; Coelho et al., 2007; Furowicz, 2010). In the present study, mecA from equine S. sciuri and S. lentus was similar to mecA from human S. aureus whereas divergent to mecA from bovine S. sciuri. These findings outline the hypothesis that the evolution of a resistance gene is more closely related to hosts environment, that bacterial specie. Tenover (2008) in his article “Vancomicin-resistant Staphylococcus aureus: a Perfect but Geographically Limited Storm?” gives us a clue that antibitioc resistance issue is not so simple answer. Science is a creative activity that request exploration and gambling. We have to be open-minded to understand that some evolutionary steps are more successful than others and the pathways to resistance are not so predictable. The biggest challenge is keep researching in order to enhance our knowledge of the mechanisms beyond resistance, the evolutionary pathways of resistance among microorganisms, and selective pressure factors that contribute to the expression of underlying genes (Souza et al., 2012).
Acknowledgments
We are grateful to National Council for Scientific and Technological Development (CNPq, Rio de Janeiro, Brazil – process 472119/2011-7) and Foundation for Research Support in the State of Rio de Janeiro (FAPERJ; process E-26/110.526/2011).
References
- Antignac A, Tomasz A. Reconstruction of the Phenotypes of Methicilin-Resistant Staphylococcus aureus by Replacement of the Staphylococcal Cassete Chromossome mec with a Plasmid-Borne Copy of the Staphylococcus sciuri pbpD Gene. Antimicrob Agents Chemother. 2008;53:435–441. doi: 10.1128/AAC.01099-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagcigil FA, Moodley A, Baptiste KE, Jensen VF, Guardabassi L. Occurrence, species distribution, antimicrobial resistance and clonality of methicillin- and erythromycin-resistant staphylococci in the nasal cavity of domestic animals. Vet Microbiol. 2007;121:307–315. doi: 10.1016/j.vetmic.2006.12.007. [DOI] [PubMed] [Google Scholar]
- CLSI. 20th Informational Supplement, M100-S20. CLSI. Wayne, PA: Clinical and Laboratory Standards Institute; 2012. Performance Standards for Antimicrobial Susceptibility Testing. [Google Scholar]
- Coelho SMO, Menezes RA, Soares LC, Pereira IA, Gomes LP, Souza MMS. Mapeamento do Perfil de Resistência e Detecção do gene mecA em Staphylococcus aureus e Staphylococcus intermedius oxacilina-resistentes isolados de espécies humanas e animais. Ciênc Rural. 2007;371:195–200. [Google Scholar]
- Couto I, Lencastre H, Severina E, Kloos W, Webster JA, Hubner RJ, Sanches IS, Tomasz A. Ubiquitous Presence of a mecA Homologue in Natural Isolates of Staphylococcus sciuri. Microb Drug Resist. 2010;24:377–391. doi: 10.1089/mdr.1996.2.377. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Mol Biol and Evol. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Ito T, Ma XX, Takeuchi F, Okuma K, Yuzawa H, Hiramatsu K. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccr. Antimicrob Agents Chemother. 2004;48:2637–2651. doi: 10.1128/AAC.48.7.2637-2651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakulska J, Fijalkowski K, Nawrotek P, Pobucewicz A, Poszumski F, Czernomysy-Furowicz D. Identification and methicillin resistance of coagulase-negative Staphylococci isolated from nasal cavity of healthy horses. J Microbiol. 2012;503:444–451. doi: 10.1007/s12275-012-1550-6. [DOI] [PubMed] [Google Scholar]
- Melo DA. M.Sc. Dissertation. Rio de Janeiro, Brasil: Instituto de Veterinária, CPGCV UFRRJ; 2013. Implicações da utilização de parâmetros humanos na detecção do gene mecA em Staphylococcus spp isolados de mastite bovina e seus impactos na predição da resistência aos betalactâmicos em ambiente de produção leiteira; p. 74. [Google Scholar]
- Murakami KW, Minamide K, Wada W, Nakamura E, Teraoka H, Watanbe S. Identification of methicillin resistant strains of staphylococci by Polymerase Chain Reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Soft. Suite Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rosato AE, Kreiswirth BN, Craig WA, Eisner W, Climo M, Archer GL. mecA-blaZ corepressors in clinical Staphylococcus aureus isolates. Antimicrob Agents Chemother. 2003;47:1460–1463. doi: 10.1128/AAC.47.4.1460-1463.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza MMS, Coelho SMO, Pereira IA, Soares LC, Pribul BR, Coelho IS. Antibiotic resistance in Staphylococcus species of animal origin. In: Pana Marina., editor. Antibiotic Resistant Bacteria - A Continuous Challenge in the New Millennium. InTech; Croatia: 2012. pp. 273–302. [Google Scholar]
- Stepanovic S, Dakic I, Morrison D, Hauschild T, Jezek P, Petrás P, Martel A, Vukovic D, Shittu A, Devriese LA. Identification and Characterization of Clinical Isolates of Members of the Staphylococcus sciuri Group. J Clin Microbiol. 2005;432:956–958. doi: 10.1128/JCM.43.2.956-958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tenover FC. Vancomycin-resistant Staphylococcus aureus: a perfect but geographically limited storm? Clin Infect Dis. 2008;46:675–677. doi: 10.1086/527393. [DOI] [PubMed] [Google Scholar]
- Tsubakishita S, Kuwahara-Arai K, Sasaki T, Hiramatsu K. Origin and Molecular Evolution of the Determinant of Methicilin Resistance in Staphylococci. Antimicrob Agents Chemother. 2010;548:4352–4359. doi: 10.1128/AAC.00356-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda R, Kawano J, Matsuo E, Masuda T, Shimizu A, Anzai T, Hashikura S. Distribution of mecA-harboring staphylococci in healthy horses. J Vet Med Sci. 2002;694:821–82. doi: 10.1292/jvms.64.821. [DOI] [PubMed] [Google Scholar]