Abstract
Expansions of more than 200 CGG repeats (full mutation) in the FMR1 gene give rise to fragile X syndrome (FXS) through a process that generally involves hypermethylation of the FMR1 promoter region and gene silencing, resulting in absence of expression of the encoded protein, FMRP. However, mosaicism with alleles differing in size and extent of methylation often exist within or between tissues of individuals with FXS. In the current work, CGG-repeat lengths and methylation status were assessed for eighteen individuals with FXS, including 13 mosaics, for which peripheral blood cells (PBMCs) and primary fibroblast cells were available. Our results show that for both PBMCs and fibroblasts, FMR1 mRNA and FMRP expression are directly correlated with the percent of methylation of the FMR1 allele. In addition, Full Scale IQ scores were inversely correlated with the percent methylation and positively correlated with higher FMRP expression. These latter results point toward a positive impact on cognition for full mutation mosaics with lower methylation compared to individuals with fully methylated, full mutation alleles. However, we did not observe a significant reduction in the number of seizures, nor in the severity of hyperactivity or autism spectrum disorder, among individuals with mosaic genotypes in the presentation of FXS. These observations suggest that low, but non-zero expression of FMRP may be sufficient to positively impact cognitive function in individuals with FXS, with methylation mosaicism (lowered methylation fraction) contributing to a more positive clinical outcome.
Keywords: mosaicism, fragile X, FMR1, FMRP, methylation, premutation
Introduction
Fragile X syndrome (FXS) is the most common heritable form of intellectual disability and is caused by an expansion of a CGG trinucleotide repeat tract in the 5' UTR of the FMR1 gene on the X chromosome. CGG-repeat expansion to greater than 200 repeats generally leads to DNA methylation(Sutcliffe et al., 1992; Alisch et al., 2013), aberrant heterochromatinization (Coffee et al., 2002; Tabolacci et al., 2008), subsequent silencing of the FMR1 gene (El-Osta, 2002), and consequent loss of the corresponding gene product, FMRP (Pieretti et al., 1991; Godler et al., 2010). FMRP is an RNA binding protein that functions as a translational repressor at synapses (Antar and Bassell, 2003; Bagni and Oostra, 2013; Darnell and Klann, 2013; Sidorov et al., 2013). FMRP is important for learning and memory, and its absence is associated with the characteristic features of FXS, including intellectual disability, cognitive impairments and behavioral problems, autism spectrum disorders (ASD), Attention Deficit Hyperactivity Disorder (ADHD), seizure, in addition to hyper-responsiveness to sensory stimuli, hyperactivity, impulsive behavior, gaze aversion and shyness (Hull and Hagerman, 1993; Hagerman, 2002; Smith et al., 2012; Schneider et al., 2013; Ballinger et al., 2014; Machalicek et al., 2014; Maurin et al., 2014; Thurman et al., 2014).
Since the mapping of the FMR1 gene to the X chromosome in 1991 (Verkerk et al., 1991), reports have emerged on mosaicism in both the FMR1 allele size and methylation status within lymphocytes as well as between tissue types (Tarleton et al., 1992; Willems et al., 1992; Wohrle et al., 1992; Hagerman et al., 1994; Nolin et al., 1994; Dobkin et al., 1996; Taylor et al., 1999; Tassone et al., 2000a; Han et al., 2006; Govaerts et al., 2007; Hantash et al., 2010; Pretto et al., 2014). The CGG repeats within the FMR1 gene predispose it to instability and the occurrence of size mosaicism, in which individuals present with different CGG repeat allele sizes such that some cells carry a full mutation allele while others carry a premutation allele; a situation that is common among individuals with FXS (Loesch et al., 2004; Lokanga et al., 2013). Notably, Nolin et al. (1994) analyzed a group of affected males with FXS by Southern Blotting and found 41% to be size mosaic. In addition to “size” mosaicism, some individuals exhibit “methylation” mosaicism, in which some cells have fully methylated full mutation alleles while other cells possess unmethylated full mutation alleles (Nolin et al., 1994; Tassone et al., 1999b; Genc et al., 2000). Methylation of the full mutation CGG repeats occurs early in embryonic development and may play a role in stabilization of the expanded repeats (Devys et al., 1992; Malter et al., 1997; Eiges et al., 2007). The hypermethylated state of the fragile X full mutation is found associated locally with histone deacetylation and chromatin remodeling (Coffee et al., 1999, 2002; El-Osta, 2002; Alpatov et al., 2014), and with transcriptional silencing of the gene (Pieretti et al., 1991; Pietrobono et al., 2005; Tabolacci et al., 2008).
Unmethylated alleles from the premutation to full mutation range are virtually transcriptionally active; specifically 2–10 fold overexpressed (Tassone et al., 2000a; Kenneson et al., 2001; Peprah et al., 2010). Overexpression of expanded CGG-repeat alleles is currently believed to lead to “toxicity” of the FMR1 mRNA (Hagerman, 2013; Pretto et al., 2014). However, because expanded CGG-repeat alleles are translated with reduced efficiency (Kenneson et al., 2001; Primerano et al., 2002; Brouwer et al., 2007; Ludwig et al., 2011) FMRP expression is negatively correlated with the CGG repeat number particularly in the upper premutation and full size range (Ludwig et al., 2014; Pretto et al., 2014). Thus, FMR1 and FMRP expression in a mosaic background could add complexity to the clinical presentation and widen the spectrum of involvement in FXS mosaics. This expectation stems from observations of inter- and intra-tissue FMR1 methylation and size mosaicism in premutation carriers and a correlation between methylation and the number of clinical symptoms in a group of premutation carriers with alleles partially methylated (Allingham-Hawkins et al., 1996; Tassone et al., 1999b; Pretto et al., 2014). In addition, while somatic size or methylation mosaicism is not considered as part of a prognostic evaluation, several reports suggest that mosaicism can impact the penetrance of the disorder and that the methylation status of full mutation mosaics affects cognitive functioning (McConkie-Rosell et al., 1993; Hagerman et al., 1994; Schmucker et al., 1996; Wohrle et al., 1998; Helderman-van den Enden et al., 1999). Specifically, McConkie-Rosell et al. (1993), studied a fragile X family with 6 brothers and found that the degree of phenotypic expression of FXS correlates with the degree of FMR1 methylation. The effect of the methylation status was likely due to the overall FMRP levels, since there is a correlation between methylation status and FMRP production (de Vries et al., 1996; Tassone et al., 1999b). A large study of 318 families (2253 individuals) reported that 12% of full mutation males and 6% of females exhibited mosaicism (Rousseau et al., 1994). Notably, a study of 46 males with FXS under 20 years of age was assessed for development of communication, self-care, socialization and motor skills as a function of presence or absence of mosaicism. They reported that adaptive skills development was 2–4 times greater in mosaic cases than in cases with a full mutation suggesting that phenotypic severity can be influenced by the presence of mosaicism (Cohen et al., 1996).
Additional cases of methylation mosaicism have also been reported and support the notion that cognitive function negatively correlate with both the length of CGG repeats and the methylation status (Merenstein et al., 1994; Mueller et al., 1995; Smeets et al., 1995; Schmucker et al., 1996; Wohrle et al., 1998; Helderman-van den Enden et al., 1999; Han et al., 2006; Loesch et al., 2012). Methylation mosaicism has also been associated with higher IQ scores and lower phenotypic presentation in post-pubescent males when compared to males with fully methylated full mutations (Merenstein et al., 1996) and a direct correlation between methylation status and FMRP levels has been demonstrated before (de Vries et al., 1996; Tassone et al., 1999a; Godler et al., 2010; Loesch et al., 2012; Pretto et al., 2014).
In this study we have investigated the expression of FMR1 and FMRP in peripheral blood mononuclear cells (PBMCs) and fibroblast cells derived from FMR1 full mutation and mosaic subjects. We report on inter- and intra-tissue mosaicism in eighteen individuals with FXS, 13 of whom were methylation and/or size mosaics, for which peripheral blood cells (PBMCs) and primary fibroblast cells were available. Using simple linear regression analysis we also investigated the genotype/phenotype relationship including clinical measures such as IQ, seizures, ASD, and ADHD.
Materials and methods
Subjects
Individuals were recruited through the MIND Institute Fragile X Research and Treatment Center. Participants provided informed consent according to protocols approved by the UC Davis Institutional Review Board. Eighteen participants with the FXS mutation, belonging to three mutational categories, were included in this study: fully methylated, full mutation males (n = 2) and females (n = 3); males (n = 12) and females (n = 1) with methylation and size mosaicism. Individuals who were size mosaics and/or methylation mosaics were combined for the purpose of molecular analysis in a mosaic group. Ages ranged from 13 to 73 years (mean ± SD = 31 ± 18 years).
Clinical measures
Clinical assessment of participants included the domains of FSIQ, ASD, ADHD, perseveration, tantrums, anxiety, and seizures. Cognitive testing was carried out with standardized IQ measures as indicated in Table 1 (Mullen and American Guidance Service, 1995; Wechsler, 1997, 2009; Psychological Corporation, 2002; Wechsler et al., 2008); ASD was assessed with the Autism Diagnostic Observation Scale (ADOS) using a module that was developmentally appropriate (Derogatis, 1994; Lord, 2002); ADHD was determined by clinical assessment (Swanson et al., 2001) along with a history of perseverations, tantrums and anxiety (Derogatis, 1994).
Table 1.
Table shows demographic information, mutation category, and molecular outcome measures for the cases included in this study.
| Case | Gender | Age | Category | IQ | IQ test | # Clinical measures | PBMCs | Fibroblasts | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CGG size | % Meth | AR | FMR1 mRNA (Std. Err) | FMRP | CGG size | % Meth | AR | FMR1 mRNA (Std. Err) | FMRP | |||||||
| 1 | F | 13 | FM | 72 | Binet | 3 | 29, 570, 710, 1050 | 0.68 | 0.61 (0.10) | 0.031 | 29, 570, 710, 1050 | 0.3 | 0.16 (0.01) | 0.025 | ||
| 2 | F | 15 | FM | 83 | SB-5 | N/A | 30, 450, 650 | 0.82 | 1.06 (0.09) | 0.080 | 30, 450, 650 | 0.52 | 0.30 (0.01) | 0.300 | ||
| 3 | F | 61 | FM | 69 | SB-5 | N/A | 31, 430, 670 | 0.85 | 1.86 (0.08) | 0.075 | 21, 680 | 0.4 | 0.28 (0.01) | 0.645 | ||
| 4 | F | 26 | FM/ MM | 80 | WISC | 1 | 27, 320–850 (UM smear) | >95 | 0.46 | 5.75 (0.01) | N/A | 27, 720, 940, 1060, 1400 (140) | 54 | 0.55 | N/A | N/A |
| 5 | M | 30 | FM | 55 | WAIS-III | 5 | 370 | 100 | 0.04 (0.00) | 0.0 | 370 | 100 | 0.002 (0.00) | 0.010 | ||
| 6 | M | 26 | FM/ MM | 51 | WAIS-III | 5 | 270, 390, 720 (UM smear) | >95 | 0.20 (0.01) | 0.002 | 290, 540, 790 (UM smear) | >95 | 0.06 (0.01) | 0.040 | ||
| 7 | M | 26 | FM | 54 | SB-5 | N/A | 490, 680, 960 | 100 | 0.03 (0.01) | 0.0 | 500, 820, 960 | 100 | 0.00 | 0.00 | ||
| 8 | M | 32 | FM/ SM | 63 | WAIS-III | N/A | 510, 590, 670, 810 (66) | 76 | 0.45 (0.09) | 0.027 | 510, 590, 670, 810 | 100 | N/A | N/A | ||
| 9 | M | 61 | FM/ MM | 64 | WAIS-III | 1 | 250 (UM smear) | >95 | 1.24 (0.08) | 0.003 | 250 (UM smear) | >95 | 0.29 (1.00) | 0.070 | ||
| 10 | M | 73 | FM/ SM | 40 | SB-5 | N/A | 290 (80–245) | 40 | 2.03 (0.06) | 0.024 | 290 (200–250) | 78 | 0.28 (0.02) | 0.141 | ||
| 11 | M | 32 | FM/ MM | 45 | Weschler | N/A | 610 (260–450) | 74 | 0.39 (0.03) | 0.003 | 425, 580, 680 (210–280) | 79 | 0.58 (0.06) | 0.199 | ||
| 12 | M | 25 | FM/ MM | 55 | WASI | 2 | 345, 450, 650 (215) | 68 | 1.97 (0.08) | 0.009 | 235, 350, 650 (UM smear) | 82 | 0.17 (0.02) | 0.067 | ||
| 13 | M | 16 | FM/ SM | 73 | WPPSI-III | 4 | 500, 820, 980, 1100 (150–230) | 90 | 1.12 (0.28) | N/A | 390, 490, 950, 1140 (160–230) | 94 | 0.16 (0.01) | 0.147 | ||
| 14 | M | 58 | FM/ SM | 64 | WAIS-III | 3 | 180 (30–260) | 37 | 2.19 (0.01) | 0.018 | 200 (250–760) | 21 | 0.29 (0.06) | 0.224 | ||
| 15 | M | 15 | FM/ SM | 63 | SB | 5 | 230 (30–570) | 7 | 2.93 (0.11) | 0.025 | 255 (260–380) | 12 | 1.83 (0.22) | 0.404 | ||
| 16 | M | 16 | FM/ SM | 72 | SB | 3 | 230, 330 (30–270) | 14 | 2.57 (0.14) | 0.010 | 230, 330 (30–200) | 18 | 0.96 (0.02) | 0.394 | ||
| 17 | M | 25 | FM/ SM | 79 | WAIS-III | 4 | 440 (65) | <5 | 2.29 (0.08) | 0.075 | 190, 220, 400, 500, 800 (65) | 49 | 0.77 (0.04) | 0.453 | ||
| 18 | M | 29 | FM/ MM | 117 | WAIS-III | 3 | 180 (210–420) | 7 | 3.85 (0.43) | 0.019 | 180 (200–270) | 16 | 1.32 (0.02) | 0.474 | ||
FM, full mutation; FM/MM, full mutation methylation mosaic; FM/SM, full mutation size mosaic; UM, unmethylated; N/A, non available.
PBMC isolation
Whole blood was collected in Cell Preparation Tube (CPT) vacutainers with sodium citrate (Becton Dickinson) and centrifuged according the manufacturer's recommendations to separate mononuclear cells from whole blood. PBMCs were washed with Dulbecco's phosphate buffered saline (PBS) and frozen in RPMI 1640 media with 10% fetal bovine serum and 10% dimethyl sulfoxide.
Fibroblast cell lines
Explants of ~3-mm dermal biopsies were minced and placed in a 100-mm TC-treated tissue culture dish (Corning Life Science, USA) with a small drop of Fibroblast medium [Gibco AmnioMAX-C100 Basal Medium supplemented with 15% AmnioMAX-C100 Supplement (Invitrogen, Carlsbad, CA, USA). Dishes were incubated at 37°C in a humidified 5% CO2 atmosphere and media was replaced every 3–4 days. Fibroblast outgrowths were harvested by trypsinization, transferred into a new dish with a modified Fibroblast Medium (1:1 solution of Gibco AmnioMAX-C100 supplemented with 15% AmnioMAX-C100 Supplement (Invitrogen, Carlsbad, CA, USA) and RPMI-1640 Basal Medium supplemented with 10% fetal bovine serum (Invitrogen), 1X Primocin (Invitrogen), 1% non-essential amino acids (Invitrogen), with media exchange every 3–4 days and allowed to reach 90% confluence prior to splitting. Fibroblast cultures were passaged no more than 3 times prior to collection for DNA, RNA, or protein extracts isolation or cryopreservation.
CGG repeat length
DNA from PBMCs or primary fibroblast cell lines was isolated using Gentra Puregene Blood Kit (Qiagen, Valencia, CA). DNA was analyzed by Southern Blot as previously described to establish CGG repeat length, percentage of CpG methylated FMR1 alleles, and, in females, activation ratio was also determined (Tassone et al., 1999a, 2008). CGG repeat length was also measured by PCR amplification followed by capillary electrophoresis, as previously described (Chen et al., 2010; Filipovic-Sadic et al., 2010).
FMR1 mRNA expression levels
Whole blood was collected in Tempus vacutainers (Applied Biosystems, Foster City, CA) and processed to isolate total RNA. Fibroblast cell total RNA was isolated using Trizol (Qiagen, Valencia, CA). Total RNA was reverse transcribed into cDNA, quantitative real time PCR (qRT-PCR) was performed on cDNA template to determine FMR1 mRNA levels. FMR1 mRNA levels were reported in relative abundance compared to reference gene β-glucoronidase (GUS). Details of the qRT-PCR methodology are as previously described (Tassone et al., 2000b).
FMRP expression levels by western blot analysis
PBMCs and fibroblast cells were thawed on ice and gently pelleted at 5000 rpm for 5 min. Cell pellets were lysed with rapid shaking at 70°C using a thermomixer (Eppendorf) for 20 min. Lysis buffer contained 0.125 M Tris HCL (pH 6.8), 2% SDS, 10% Glycerol, and 5% BME. The samples were cooled to room temperature and centrifuged at 13,000 rpm in a benchtop centrifuge for 10 min after which the protein extracts were transferred to clean microcentrifuge tubes. Protein concentrations were measured using the Detergent Compatible Protein Assay RC/DC (Bio-Rad Laboratories Inc., United States). Proteins (10 μg) were separated by electrophoresis in Any KD Criterion TGX Gels (Bio-Rad Laboratories Inc., United States) in 1X Tris/Glycine/SDS buffer and transferred overnight at 4°C, 30 volts, to 0.2 μm nitrocellulose membranes (Bio-Rad Laboratories Inc., Germany) in 1X Tris/Glycine/SDS buffer containing 10% methanol. The membranes were blocked using Licor blocking buffer (Licor) with 50% 1X PBS for 1 h and hybridized overnight at 4°C with 1:10,000 chicken anti-GAPDH (Millipore) and 1:5000 mouse anti-FMRP (Chemicon, Millipore). Membranes were washed with 1X PBST and hybridized with secondary antibodies for 1 h at room temperature (1:25,000 goat anti-chicken IRDye 800CW and 1:20,000 goat anti-mouse IRDye 680LT; Licor). Membranes were washed in 1X PBST and finally 1X PBS prior to being imaged using Licor Odyssey System with Image Studio Version 2.1 (Licor).
Statistical analysis
FMR1 expression was compared between groups, adjusting for percent methylation, using a multiple regression model including group and percent methylation. Percent methylation values indicating >95% were converted to 95% for use in analysis. FMRP expression was compared between groups using one-way analysis of variance. FMR1 expression, FMRP expression, IQ, numbers of clinical features, and percent methylation were correlated with each other using simple linear regression. FMRP expression was correlated with FMR1 expression, adjusting for percent methylation, using multiple linear regression analysis. FMR1 data and FMRP expression were log-transformed prior to analysis whenever they were used as the response variable in a linear model. Analyses were conducted using R, version 3.0.3 (Team, 2014).
Results
Clinical history of two representative cases
The group of patients examined in this study consists of 18 subjects that presented with clinical characteristics of FXS and molecularly with size or methylation mosaicism. The clinical history of two of the 18 cases is described in detail below.
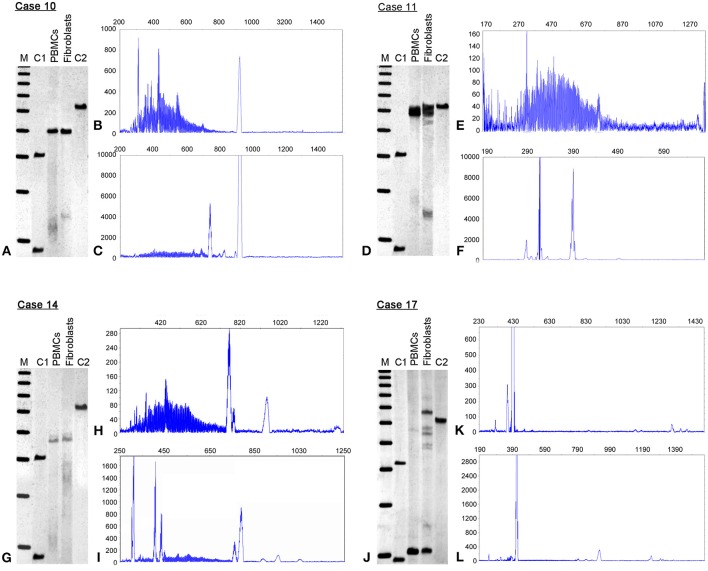
Case 14 is a 58 year old male with a history of learning problems and shyness in childhood. He was dyslexic, and had difficulty reading and writing. On evaluation at age 55, he demonstrated psychotic symptoms on the Structured Clinical Interview for DSM IV (SCID) with both visual and auditory delusions. He was also subthreshold for agorophobia. He has developed a neuropathy in the last year with pain, numbness, and tingling in both his hands and in his feet, with burning pain in his feet particularly evident during the past few months. He trips frequently, but has not reported ataxia or tremor by history or by examination. His family history includes a mother who was a premutation carrier with dementia, tremor, and ataxia. His brother has FXTAS, his sister has FXS, and his daughter is a premutation carrier with a son affected with FXS. On molecular testing, he displays substantial somatic instability (size mosaicism) as evidenced by a broad range of CGG repeat alleles more so in PBMCs compared to fibroblasts (Figures 1H,I). He also has methylation mosaicism in both PBMCs and fibroblasts (Figure 1G) with most of the cells carrying unmethylated alleles in both PBMCs (63%) and fibroblasts (79%), although the CGG repeat sizes were larger in the fibroblasts (Table 1). FMR1 mRNA levels were over two-fold higher than normal (2.19 ± 0.01; mean FMR1 mRNA in controls = 1.42 ± 0.26; Tassone et al., 2000b) in PBMCs, and but slightly lower compared to normals in fibroblasts (0.29 ± 0.06; mean in FMR1 mRNA in controls = 0.40 ± 0.08; Garcia-Arocena, 2010). His FMRP expression levels were low; approximately 10% of normal in PBMCs and in fibroblasts, likely due to inefficient translation of large expanded alleles.
Figure 1.
Methylation status and CGG size instability. Southern blots (SB) and electrophoregrams in four representative cases of full mutation mosaic males, cases 10, 11, 14 and 17 are illustrated. The SB analysis (left, A,D,G,J) demonstrates differences in FMR1 percent of methylation when comparing peripheral blood lymphocytes (PBMCs) to primary cultured fibroblasts (Fibroblasts). See also Table 1. M = 1 Kb DNA size ladder marker; C1, normal female, negative control and C2, full mutation male, positive control. The electrophoregrams show CGG instability as illustrated by the presence of serial peaks, each representing single distinct alleles, with differences between PBMCs (B,E,H,K) and fibroblasts (C,F,I,L). The X axis marks the size of the alleles in base pairs. The Y-axis marks the fluorescence intensity of each allele.
Case 17 presented at 20 years of age with a history of a normal pregnancy; but with delayed development, hypotonia, ADHD, shyness, and social anxiety during early childhood. He also had seizures documented by EEG during his first 2 years, and night terrors during his first 5 years. He has characteristic FXS behaviors including intermittent poor eye contact, tactile defensiveness, social anxiety, sensitivity to stimuli, and perseveration. However, he does not have tantrums or aggression. He has had a joint dislocation of his toe, and right exotropia. He has a large head circumference (>99%) with mild obesity. His ears are large and prominent with cupping bilaterally, his fingers are hyperextensible and his skin is soft and velvet-like, although striae are present over his lateral chest and abdomen. He has flat feet with a moderate degree of pronation. He has a high arched palate, and macroorchidism with a testicular volume of 50 mL bilaterally. He was able to tandem walk without difficulty, which is unusual for a man with FXS. He is higher functioning than most adult males with FXS since his FSIQ is in the borderline to low normal range: WAIS-III FSIQ is 79; performance IQ 78; and verbal IQ is 83, with a verbal comprehension score of 93, perceptional organization of 70, working memory of 69, and perceptional speed of 79. At the molecular level, he showed size mosaicism in PBMCs and fibroblasts with a premutation allele of 65 CGG repeats and full mutation alleles present in a small percent of PBMC cells (<5%), but much higher than in fibroblast cells (49%) (Figures 1J,K; Table 1). FMR1 mRNA levels were over 2-fold higher than normal both in PBMCs and in fibroblasts (Table 1). Within the mosaic group, he showed a higher FMRP expression in both tissues likely due to the premutation allele of 65 CGG repeat present in over 50% of the cells (Table 1).
Differences in methylation and size mosaicism occur between PBMCs and fibroblasts
Southern Blot and PCR analyses were used to determine the methylation status and the CGG repeat size of the FMR1 allele in both PBMC and fibroblast samples from individuals with FXS (males, n = 14 and females, n = 4) (Table 1). Among the FXS cases, three females had both a normal allele and a full mutation allele in both tissues (Cases 1–3, Table 1) while one female had size and methylation mosaicism that differed between PBMCs and fibroblasts (Case 4, Table 1). Two males with FXS showed fully methylated full mutation alleles (Cases 5 and 7), while the remaining 12 (Table 1) were methylation or size mosaics in both PBMCs and/or fibroblasts. The percent of methylation and the CGG size varied between PBMCs and fibroblast cells in the majority of cases (Table 1). Variations in CGG size and methylation status, as observed by Southern Blot and PCR assay, for 4 representative cases are shown in Figure 1.
In the methylation mosaic males, the CGG size of the unmethylated alleles spanned the full mutation range in all individuals; in some cases alleles within the premutation and normal range were present, particularly in the PBMCs (Table 1). However, it was not possible to estimate the molecular contribution and the consequent potential clinical outcome due to any single allele present in the sample. Although the observed differences in methylation were variable between individuals and involved both differences in percent of methylation and allele size, the percent of methylation was consistently higher in the majority of the fibroblast cells. The activation ratio, expressing the percent of cells carrying the normal allele on the active X chromosome, was higher in PBMCs than fibroblast cells for three of the four female cases (Cases 1–3) while the remaining female (Case 4) had approximately the same activation ratio between cell types (0.46 in PBMCs, 0.55 in fibroblast cells) (Supplementary Figure 1). The higher activation ratio in Cases 1–3, and thus the higher proportion of normal alleles on the active X chromosome, likely reflects the FMR1 mRNA levels observed in the PBMCs of these individuals closer to the levels observed in normal controls (mean FMR1mRNA in controls = 1.42 ± 0.26; Tassone et al., 2000b). However, the FMRP levels were less than 50% of the control levels for both PBMCs and fibroblasts (mean FMRP in control PBMCs = 0.17 ± 0.06; mean FMRP in control fibroblasts = 1.2 ± 0.02; data not shown) (Table 1).
FMR1 mRNA and FMRP expression levels are higher in individuals with mosaicism
FMR1 mRNA levels adjusted for percent methylation were compared between the three mutational groups, namely males with full mutations, females with full mutations and individuals with mosaicism (methylation and size male and female mosaics, n = 13). FMR1 mRNA levels measured by qRT-PCR varied between 0 and 5.75 (mean 1.79). As previously reported (Tassone et al., 2000a), FMR1 mRNA levels were higher in both tissues in mosaics than in full mutation males (Table 1; Figures 2A,B). A correlation between FMR1 mRNA expression and percent of methylation in PBMCs (Figure 2C) and fibroblasts (Figure 2D) showed a decrease in FMR1 expression with increased methylation using simple linear regression. FMRP expression between the three mutational categories showed a 5.1 fold-greater FMRP expression in the mosaic group compared to the full mutation males in PBMCs (Figure 3A) (although this was not statistically significant, P = 0.199) and significantly higher in the female group compared to full mutations (p < 0.001). FMRP was 37.0-fold greater in mosaic males and females than in full mutation males (P = 0.001) in fibroblast cells (Figure 3B). FMRP expression in PBMCs and fibroblasts decreased significantly with increasing percent methylation (P = 0.001 and p = 0.02 respectively; Figures 3C,D).
Figure 2.
FMR1 gene expression. Boxplots show FMR1 expression in PBMCs (A) and fibroblasts (B) for full mutation females, full mutation males and mosaic individuals as measured by qRT-PCR relative to a reference gene (Gus). The line across each box represents the group median. Outliers are shown as circles. Scatterplots of FMR1 mRNA expression levels (y-axis) as a function of percent methylation (x-axis) show single data points demonstrating decrease FMR1 mRNA expression with increased methylation in PBMC's (C) and in fibroblasts (D). Circles represent observed data. The solid line shows the linear regression fit.
Figure 3.
FMRP expression. Boxplots show semi-quantitative measures of FMRP expression in PBMCs (A) and fibroblasts (B) for control individuals, full mutation females, full mutation males, and mosaic individuals as detected by Western Blot analysis. The line across each box represents the group median and outliers are shown as circles. Scatterplots show single data points for FMRP expression (y-axis) as a function of percent of methylation in PBMCs (C) and fibroblasts (D), illustrating an inverse association between FMRP levels and percent of methylation and in both cases, a dramatic loss of FMRP expression in highly methylated alleles.
FMRP expression levels correlate with IQ
Participants FSIQ decreased significantly with increased FMR1 methylation in both PBMCs (P = 0.022) and fibroblasts (P = 0.02) (Figures 4A,B) while the IQ increased significantly with increasing FMRP expression in fibroblasts (P = 0.028) (Figure 4D) while a similar trend, although not statistically significant, was observed in PBMCs (P = 0.118) (Figure 4C). Notably, the plots in Figures 4A,B show a wide scatter likely due to the large range of unmethylated CGG allele sizes, particularly in individuals with mosaicism (Table 1). Both CGG allele size and the methylation status affect the FMRP expression levels, which ultimately can influence the severity of the phenotype.
Figure 4.
FMR1 methylation and FMRP are associated with FSIQ scores as measured by Stanford Binet-5, WISC, WAIS-III, Weschler, and WPPSI-III Standardized tests. Scatterplots show IQ scores (y-axis) as a function of percent methylation (x-axis) in PBMCs (A) and fibroblasts (B), demonstrating a significant correlation between lower IQ and greater percent of methylation in both cell types. Circles represent observed data and the solid line shows the linear regression fit. Similarly, scatterplots of IQ scores (y-axis) as a function of FMRP expression show that lower IQs significantly correlate with reduced FMRP expression in fibroblasts (D) and a similar trend can be observed in PBMCs (C). Circles represent observed data, the solid line shows the linear regression fit.
Molecular measures do not correlate with the number of clinical features present
In order to determine whether the percent of methylation in FMR1 correlated with clinical presentation we examined the relationship between methylation and number of clinical features that were diagnosed by their physician as reported in their medical records. Scores for severity of symptoms were not considered in our evaluation; only the presence or absence of each feature was taken into consideration. Clinical diagnoses used in this study included ASD, ADHD, perseveration, tantrums, anxiety, and seizures. A minimum of one (n = 2) and a maximum of five (n = 4) clinical features out of six considered were observed in the 18 participants for which this clinical information was available. Out of the 18 individuals with FXS, 50% (n = 9) presented with ASD. The number of clinical features was not significantly associated with percent methylation in either PMBCs (P = 0.926) or fibroblasts (P = 0.803). In addition, FMRP expression did not significantly correlate with the number of clinical features.
Discussion
Reports of methylation and size mosaicism have been observed in a large proportion of individuals with a full mutation (McConkie-Rosell et al., 1993; Nolin et al., 1994; Genc et al., 2000; Tassone et al., 2000a; Loesch et al., 2012; Pretto et al., 2014). However, despite the fact that mosaics can produce some FMRP, depending on the methylation and the CGG allele size, they usually present with developmental delay. This is likely due to the limited amount of FMRP present, due both to lowered gene expression and the difficulty in translating residual mRNA with the expanded CGG repeat (Primerano et al., 2002; Ludwig et al., 2011, 2014). Alternatively, because DNA testing is normally performed in PBMCs, the results may not accurately reflect the mutation pattern in other tissues. Mosaicism in different tissues has been investigated and reported in several studies (Genc et al., 2000; Bonarrigo et al., 2014; Pretto et al., 2014), which have reported similarities across tissues in some cases and extreme difference in mosaicism in others. Thus, it is difficult to predict on an individual basis whether mosaicism observed in blood will reflect the pattern in other tissues, particularly brain, and therefore the patterns of clinical involvement/severity. However, the studies in general show association between mosaicism and prognosis (Merenstein et al., 1994; Mueller et al., 1995; Smeets et al., 1995; Dobkin et al., 1996; Schmucker et al., 1996; Wohrle et al., 1998; Helderman-van den Enden et al., 1999; Genc et al., 2000; Han et al., 2006; Govaerts et al., 2007; Loesch et al., 2012; Pretto et al., 2014).
In the current study the impact of mosaicism was assessed in 18 individuals with a full mutation by comparing molecular measures in both PBMC and fibroblast cells. In several cases CGG allele sizes and their corresponding FMR1 and FMRP expression was similar between PBMC and fibroblast samples, however, noticeable differences in CGG size allele distribution were observed in the majority of cases (Table 1) likely contributing to the variations observed in the phenotypic presentation of FXS. Percent of methylation was in general lower in the PBMCs than in fibroblasts. Notably, the mRNA expression was higher in PBMCs suggesting that the unmethylated alleles even in the full mutation range are actively transcribed (Tassone et al., 2000). Although an excess in transcription, producing higher than normal levels of mRNA was observed for many of the subjects included in this study, these are long expanded alleles, which are inefficiently translated to FMRP (Primerano et al., 2002; Loesch et al., 2004; Peprah et al., 2010). Thus, it is not surprising that some individuals exhibited elevated FMR1 mRNA but not lower FMRP levels.
The fully-methylated, full mutation males and those mosaics with a higher percent of cells carrying a methylated allele had an IQ rating under 70, within the range of intellectual disability, as is common in FXS. Higher IQ scores were observed in mosaics with less methylation (Table 1). This data suggest that low FMRP levels are biologically critical, and small increases in its expression may have a significant impact on cognitive function. Alternatively, it is also possible that different methylation and consequently different FMRP levels may exist in neural tissues. We found no significant correlation between percent of unmethylated alleles and the number of clinical phenotypes present, suggesting the importance of the genetic background, in addition to the presence of a full mutation in the complexity of the clinical phenotype.
As expected, the IQ was in the borderline range for females as the FXS phenotype in females is normally milder than that of FXS in males due the normal, active allele expressing FMRP at normal levels. The activation ratio, which reflects random X inactivation, was measured in the full mutation females in both PMBCs and fibroblasts, revealing inter-tissue difference that can have a biological and ultimately a clinical impact on a full mutation female (Table 1). Higher activation ratios are favorable, as they indicate more normal alleles being expressed. Because activation ratios vary between tissue types, measurements from PBMC or fibroblast samples may not reflect what may be present in clinically relevant tissue types such as brain.
In all cases the presence of full mutation alleles contributed to a lower IQ and perhaps to the higher incidence of ASD (50%) observed in these individuals.
Although mosaic individuals can have milder cognitive involvement, they can be at risk for developing FXTAS if their FMR1 mRNA levels are elevated as suggested from cases previously reported in the literature (Loesch et al., 2012; Pretto et al., 2013, 2014; Santa Maria et al., 2013). In addition, individuals with both lowered FMRP and elevated FMR1 mRNA can also be at greater risk for psychotic symptoms (Schneider et al., 2013).
One limitation of this study is represented by the small number of subjects (n = 18), particularly due to the wide range of CGG size and methylation, for which both PBMCs and primary fibroblasts were available. The results presented in this study, may have clinical relevance, as a detailed molecular diagnosis (including information about methylation status, FMR1mRNA and FMRP levels) could provide additional information and guide to clinicians and expectations for the family of a patient with FXS. However, it also underlines the complexity of the disorders with many molecular facets, including CGG repeat size, methylation, FMRP expression, and intra-tissue mosaicism that can lead to the broad spectrum of clinical involvement in FXS, particularly in those with mosaicism.
Conflict of interest statement
Dr. Randi Hagerman has received funding from Roche, Novartis, Seaside Therapeutics, Forest Curemark and the National Fragile X Foundation for clinical trials in fragile X syndrome and/or autism. She has also consulted with Novartis, Genentech and Roche regarding treatment in fragile X syndrome. Dr. Flora Tassone has consulted with Novartis and Genentech and has received funds from Roche. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by National Institute of Health grants HD02274 and HD040661 and by the National Center for Advancing Translational Sciences, National Institutes of Health, through grant number UL1 TR000002. This work is dedicated to the memory of Matteo.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fgene.2014.00318/abstract
References
- Alisch R. S., Wang T., Chopra P., Visootsak J., Conneely K. N., Warren S. T. (2013). Genome-wide analysis validates aberrant methylation in fragile X syndrome is specific to the FMR1 locus. BMC Med. Genet. 14:18 10.1186/1471-2350-14-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allingham-Hawkins D. J., Brown C. A., Babul R., Chitayat D., Krekewich K., Humphries T., et al. (1996). Tissue-specific methylation differences and cognitive function in fragile X premutation females. Am. J. Med. Genet. 64, 329–333 [DOI] [PubMed] [Google Scholar]
- Alpatov R., Lesch B. J., Nakamoto-Kinoshita M., Blanco A., Chen S., Stutzer A., et al. (2014). A chromatin-dependent role of the Fragile X mental retardation protein FMRP in the DNA damage response. Cell 157, 869–881 10.1016/j.cell.2014.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar L. N., Bassell G. J. (2003). Sunrise at the synapse: the FMRP mRNP shaping the synaptic interface. Neuron 37, 555–558 10.1016/S0896-6273(03)00090-4 [DOI] [PubMed] [Google Scholar]
- Bagni C., Oostra B. A. (2013). Fragile X syndrome: from protein function to therapy. Am. J. Med. Genet. A 161A, 2809–2821 10.1002/ajmg.a.36241 [DOI] [PubMed] [Google Scholar]
- Ballinger E. C., Cordeiro L., Chavez A. D., Hagerman R. J., Hessl D. (2014). Emotion Potentiated Startle in Fragile X Syndrome. J. Autism Dev. Disord. [Epub ahead of print]. 10.1007/s10803-014-2125-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonarrigo F. A., Russo S., Vizziello P., Menni F., Cogliati F., Giorgini V., et al. (2014). Think about it: FMR1 gene mosaicism. J. Child Neurol. 29, NP74–NP77 10.1177/0883073813503187 [DOI] [PubMed] [Google Scholar]
- Brouwer J. R., Mientjes E. J., Bakker C. E., Nieuwenhuizen I. M., Severijnen L. A., Van der Linde H. C., et al. (2007). Elevated Fmr1 mRNA levels and reduced protein expression in a mouse model with an unmethylated Fragile X full mutation. Exp. Cell Res. 313, 244–253 10.1016/j.yexcr.2006.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Hadd A., Sah S., Filipovic-Sadic S., Krosting J., Sekinger E., et al. (2010). An information-rich CGG repeat primed PCR that detects the full range of fragile X expanded alleles and minimizes the need for southern blot analysis. J. Mol. Diagn. 12, 589–600 10.2353/jmoldx.2010.090227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B., Zhang F., Ceman S., Warren S. T., Reines D. (2002). Histone modifications depict an aberrantly heterochromatinized FMR1 gene in fragile x syndrome. Am. J. Hum. Genet. 71, 923–932 10.1086/342931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B., Zhang F., Warren S. T., Reines D. (1999). Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat. Genet. 22, 98–101 10.1038/8807 [DOI] [PubMed] [Google Scholar]
- Cohen I. L., Nolin S. L., Sudhalter V., Ding X. H., Dobkin C. S., Brown W. T. (1996). Mosaicism for the FMR1 gene influences adaptive skills development in fragile X-affected males. Am. J. Med. Genet. 64, 365–369 [DOI] [PubMed] [Google Scholar]
- Darnell J. C., Klann E. (2013). The translation of translational control by FMRP: therapeutic targets for FXS. Nat. Neurosci. 16, 1530–1536 10.1038/nn.3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derogatis L. R. (1994). Symptom Checklist-90-R (SCL-90-R): Administration, Scoring, and Procedures Manual. Minneapolis, MN: National Computer Systems [Google Scholar]
- de Vries B. B., Jansen C. C., Duits A. A., Verheij C., Willemsen R., van Hemel J. O., et al. (1996). Variable FMR1 gene methylation of large expansions leads to variable phenotype in three males from one fragile X family. J. Med. Genet. 33, 1007–1010 10.1136/jmg.33.12.1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devys D., Biancalana V., Rousseau F., Boue J., Mandel J. L., Oberle I. (1992). Analysis of full fragile X mutations in fetal tissues and monozygotic twins indicate that abnormal methylation and somatic heterogeneity are established early in development. Am. J. Med. Genet. 43, 208–216 10.1002/ajmg.1320430134 [DOI] [PubMed] [Google Scholar]
- Dobkin C. S., Nolin S. L., Cohen I., Sudhalter V., Bialer M. G., Ding X. H., et al. (1996). Tissue differences in fragile X mosaics: mosaicism in blood cells may differ greatly from skin. Am. J. Med. Genet. 64, 296–301 [DOI] [PubMed] [Google Scholar]
- Eiges R., Urbach A., Malcov M., Frumkin T., Schwartz T., Amit A., et al. (2007). Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell 1, 568–577 10.1016/j.stem.2007.09.001 [DOI] [PubMed] [Google Scholar]
- El-Osta A. (2002). FMR1 silencing and the signals to chromatin: a unified model of transcriptional regulation. Biochem. Biophys. Res. Commun. 295, 575–581 10.1016/S0006-291X(02)00682-4 [DOI] [PubMed] [Google Scholar]
- Filipovic-Sadic S., Sah S., Chen L., Krosting J., Sekinger E., Zhang W., et al. (2010). A novel FMR1 PCR method for the routine detection of low abundance expanded alleles and full mutations in fragile X syndrome. Clin. Chem. 56, 399–408 10.1373/clinchem.2009.136101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Arocena D. (2010). Fibroblast phenotype in male carriers of FMR1 premutation alleles. Hum. Mol. Genet. 19, 299–312 10.1093/hmg/ddp497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genc B., Muller-Hartmann H., Zeschnigk M., Deissler H., Schmitz B., Majewski F., et al. (2000). Methylation mosaicism of 5'-(CGG)(n)-3' repeats in fragile X, premutation and normal individuals. Nucleic Acids Res. 28, 2141–2152 10.1093/nar/28.10.2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godler D. E., Tassone F., Loesch D. Z., Taylor A. K., Gehling F., Hagerman R. J., et al. (2010). Methylation of novel markers of fragile X alleles is inversely correlated with FMRP expression and FMR1 activation ratio. Hum. Mol. Genet. 19, 1618–1632 10.1093/hmg/ddq037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govaerts L. C., Smit A. E., Saris J. J., VanderWerf F., Willemsen R., Bakker C. E., et al. (2007). Exceptional good cognitive and phenotypic profile in a male carrying a mosaic mutation in the FMR1 gene. Clin. Genet. 72, 138–144 10.1111/j.1399-0004.2007.00829.x [DOI] [PubMed] [Google Scholar]
- Hagerman P. (2013). Fragile X-associated tremor/ataxia syndrome (FXTAS): pathology and mechanisms. Acta Neuropathol. 126, 1–19 10.1007/s00401-013-1138-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman R. J. (2002). Physical and behavioral phenotype, in Fragile X Syndrome: Diagnosis, Treatment and Research, 3rd Edn., eds Hagerman R. J., Hagerman P. J. (Baltimore, MD: The Johns Hopkins University Press; ), 3–109 [Google Scholar]
- Hagerman R. J., Hull C. E., Safanda J. F., Carpenter I., Staley L. W., O'Connor R. A., et al. (1994). High functioning fragile X males: demonstration of an unmethylated fully expanded FMR-1 mutation associated with protein expression. Am. J. Med. Genet. 51, 298–308 10.1002/ajmg.1320510404 [DOI] [PubMed] [Google Scholar]
- Han X. D., Powell B. R., Phalin J. L., Chehab F. F. (2006). Mosaicism for a full mutation, premutation, and deletion of the CGG repeats results in 22% FMRP and elevated FMR1 mRNA levels in a high-functioning fragile X male. Am. J. Med. Genet. A 140, 1463–1471 10.1002/ajmg.a.31291 [DOI] [PubMed] [Google Scholar]
- Hantash F. M., Goos D. G., Tsao D., Quan F., Buller-Burckle A., Peng M., et al. (2010). Qualitative assessment of FMR1 (CGG)n triplet repeat status in normal, intermediate, premutation, full mutation, and mosaic carriers in both sexes: implications for fragile X syndrome carrier and newborn screening. Genet. Med. 12, 162–173 10.1097/GIM.0b013e3181d0d40e [DOI] [PubMed] [Google Scholar]
- Helderman-van den Enden A. T., Maaswinkel-Mooij P. D., Hoogendoorn E., Willemsen R., Maat-Kievit J. A., Losekoot M., et al. (1999). Monozygotic twin brothers with the fragile X syndrome: different CGG repeats and different mental capacities. J. Med. Genet. 36, 253–257 [PMC free article] [PubMed] [Google Scholar]
- Hull C., Hagerman R. J. (1993). A study of the physical, behavioral, and medical phenotype, including anthropometric measures, of females with fragile X syndrome. Am J Dis Child 147, 1236–1241 [DOI] [PubMed] [Google Scholar]
- Kenneson A., Zhang F., Hagedorn C. H., Warren S. T. (2001). Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum. Mol. Genet. 10, 1449–1454 10.1093/hmg/10.14.1449 [DOI] [PubMed] [Google Scholar]
- Loesch D. Z., Huggins R. M., Hagerman R. J. (2004). Phenotypic variation and FMRP levels in fragile X. Ment. Retard. Dev. Disabil. Res. Rev. 10, 31–41 10.1002/mrdd.20006 [DOI] [PubMed] [Google Scholar]
- Loesch D. Z., Sherwell S., Kinsella G., Tassone F., Taylor A., Amor D., et al. (2012). Fragile X-associated tremor/ataxia phenotype in a male carrier of unmethylated full mutation in the FMR1 gene. Clin. Genet. 82, 88–92 10.1111/j.1399-0004.2011.01675.x [DOI] [PubMed] [Google Scholar]
- Lokanga R. A., Entezam A., Kumari D., Yudkin D., Qin M., Smith C. B., et al. (2013). Somatic expansion in mouse and human carriers of fragile X premutation alleles. Hum. Mutat. 34, 157–166 10.1002/humu.22177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C. (2002). Autism Diagnostic Observation Schedule: ADOS. Los Angeles, CA: Western Psychological Services [Google Scholar]
- Ludwig A. L., Espinal G. M., Pretto D. I., Jamal A. L., Arque G., Tassone F., et al. (2014). CNS expression of murine fragile X protein (FMRP) as a function of CGG-repeat size. Hum. Mol. Genet. 23, 3228–3238 10.1093/hmg/ddu032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A. L., Hershey J. W., Hagerman P. J. (2011). Initiation of translation of the FMR1 mRNA Occurs predominantly through 5'-end-dependent ribosomal scanning. J. Mol. Biol. 407, 21–34 10.1016/j.jmb.2011.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machalicek W., McDuffie A., Oakes A., Ma M., Thurman A. J., Rispoli M. J., et al. (2014). Examining the operant function of challenging behavior in young males with fragile X syndrome: a summary of 12 cases. Res. Dev. Disabil. 35, 1694–1704 10.1016/j.ridd.2014.03.014 [DOI] [PubMed] [Google Scholar]
- Malter H. E., Iber J. C., Willemsen R., de Graaff E., Tarleton J. C., Leisti J., et al. (1997). Characterization of the full fragile X syndrome mutation in fetal gametes. Nat. Genet. 15, 165–169 10.1038/ng0297-165 [DOI] [PubMed] [Google Scholar]
- Maurin T., Zongaro S., Bardoni B. (2014). Fragile X syndrome: from molecular pathology to therapy. Neurosci. Biobehav. Rev. [Epub ahead of print]. 10.1016/j.neubiorev.2014.01.006 [DOI] [PubMed] [Google Scholar]
- McConkie-Rosell A., Lachiewicz A. M., Spiridigliozzi G. A., Tarleton J., Schoenwald S., Phelan M. C., et al. (1993). Evidence that methylation of the FMR-I locus is responsible for variable phenotypic expression of the fragile X syndrome. Am. J. Hum. Genet. 53, 800–809 [PMC free article] [PubMed] [Google Scholar]
- Merenstein S. A., Shyu V., Sobesky W. E., Staley L., Berry-Kravis E., Nelson D. L., et al. (1994). Fragile X syndrome in a normal IQ male with learning and emotional problems. J. Am. Acad. Child Adolesc. Psychiatry 33, 1316–1321 10.1097/00004583-199411000-00014 [DOI] [PubMed] [Google Scholar]
- Merenstein S. A., Sobesky W. E., Taylor A. K., Riddle J. E., Tran H. X., Hagerman R. J. (1996). Molecular-clinical correlations in males with an expanded FMR1 mutation. Am. J. Med. Genet. 64, 388–394 [DOI] [PubMed] [Google Scholar]
- Mueller O. T., Hartsfield J. K., Jr., Amar M. J., Gallardo L. A., Kousseff B. G. (1995). Fragile X syndrome: discordant levels of CGG repeat mosaicism in two brothers. Am. J. Med. Genet. 60, 302–306 10.1002/ajmg.1320600408 [DOI] [PubMed] [Google Scholar]
- Mullen E. M., American Guidance Service. (1995). Mullen Scales of Early Learning. Minneapolis, MN: Pearson: 1 item administration book, 25 identical record forms, 21 stimulus book, assorted test materials, 21 manual, numerous toys [Google Scholar]
- Nolin S. L., Glicksman A., Houck G. E., Jr., Brown W. T., Dobkin C. S. (1994). Mosaicism in fragile X affected males. Am. J. Med. Genet. 51, 509–512 10.1002/ajmg.1320510444 [DOI] [PubMed] [Google Scholar]
- Peprah E., He W., Allen E., Oliver T., Boyne A., Sherman S. L. (2010). Examination of FMR1 transcript and protein levels among 74 premutation carriers. J. Hum. Genet. 55, 66–68 10.1038/jhg.2009.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieretti M., Zhang F. P., Fu Y. H., Warren S. T., Oostra B. A., Caskey C. T., et al. (1991). Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 66, 817–822 10.1016/0092-8674(91)90125-I [DOI] [PubMed] [Google Scholar]
- Pietrobono R., Tabolacci E., Zalfa F., Zito I., Terracciano A., Moscato U., et al. (2005). Molecular dissection of the events leading to inactivation of the FMR1 gene. Hum. Mol. Genet. 14, 267–277 10.1093/hmg/ddi024 [DOI] [PubMed] [Google Scholar]
- Pretto D. I., Hunsaker M. R., Cunningham C. L., Greco C. M., Hagerman R. J., Noctor S. C., et al. (2013). Intranuclear inclusions in a fragile X mosaic male. Transl. Neurodegener. 2:10 10.1186/2047-9158-2-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretto D. I., Mendoza-Morales G., Lo J., Cao R., Hadd A., Latham G. J., et al. (2014). CGG allele size somatic mosaicism and methylation in FMR1 premutation alleles. J. Med. Genet. 51, 309–318 10.1136/jmedgenet-2013-102021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primerano B., Tassone F., Hagerman R. J., Hagerman P. J., Amaldi F., Bagni C. (2002). Reduced FMR1 mRNA translation efficiency in Fragile X patients with premutations. RNA 8, 1482–1488 10.1017/S1355838202020642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychological Corporation. (2002). Wais-III, Wechsler Adult Intelligence Scale, Third Edition: WMS-III, Wechsler Memory Scale, 3rd Edn.: Technical Manual. San Antonio, TX: Psychological Corp [Google Scholar]
- Rousseau F., Heitz D., Tarleton J., MacPherson J., Malmgren H., Dahl N., et al. (1994). A multicenter study on genotype-phenotype correlations in the fragile X syndrome, using direct diagnosis with probe StB12.3: the first 2,253 cases. Am. J. Hum. Genet. 55, 225–237 [PMC free article] [PubMed] [Google Scholar]
- Santa Maria L., Pugin A., Alliende M., Aliaga S., Curotto B., Aravena T., et al. (2013). FXTAS in an unmethylated mosaic male with fragile X syndrome from Chile. Clin. Genet. [Epub ahead of print]. 10.1111/cge.12278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker B., Ballhausen W. G., Pfeiffer R. A. (1996). Mosaicism of a microdeletion of 486 bp involving the CGG repeat of the FMR1 gene due to misalignment of GTT tandem repeats at chi-like elements flanking both breakpoints and a full mutation. Hum. Genet. 98, 409–414 10.1007/s004390050230 [DOI] [PubMed] [Google Scholar]
- Schneider A., Ligsay A., Hagerman R. J. (2013). Fragile X syndrome: an aging perspective. Dev. Disabil. Res. Rev. 18, 68–74 10.1002/ddrr.1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorov M. S., Auerbach B. D., Bear M. F. (2013). Fragile X mental retardation protein and synaptic plasticity. Mol. Brain 6:15 10.1186/1756-6606-6-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets H. J., Smits A. P., Verheij C. E., Theelen J. P., Willemsen R., van de Burgt I., et al. (1995). Normal phenotype in two brothers with a full FMR1 mutation. Hum. Mol. Genet. 4, 2103–2108 10.1093/hmg/4.11.2103 [DOI] [PubMed] [Google Scholar]
- Smith L. E., Barker E. T., Seltzer M. M., Abbeduto L., Greenberg J. S. (2012). Behavioral phenotype of fragile X syndrome in adolescence and adulthood. Am. J. Intellect. Dev. Disabil. 117, 1–17 10.1352/1944-7558-117.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. S., Nelson D. L., Zhang F., Pieretti M., Caskey C. T., Saxe D., et al. (1992). DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum. Mol. Genet. 1, 397–400 10.1093/hmg/1.6.397 [DOI] [PubMed] [Google Scholar]
- Swanson J. M., Kraemer H. C., Hinshaw S. P., Arnold L. E., Conners C. K., Abikoff H. B., et al. (2001). Clinical relevance of the primary findings of the MTA: success rates based on severity of ADHD and ODD symptoms at the end of treatment. J. Am. Acad. Child Adolesc. Psychiatry 40, 168–179 10.1097/00004583-200102000-00011 [DOI] [PubMed] [Google Scholar]
- Tabolacci E., Moscato U., Zalfa F., Bagni C., Chiurazzi P., Neri G. (2008). Epigenetic analysis reveals a euchromatic configuration in the FMR1 unmethylated full mutations. Eur. J. Hum. Genet. 16, 1487–1498 10.1038/ejhg.2008.130 [DOI] [PubMed] [Google Scholar]
- Tarleton J., Wong S., Schwartz C. (1992). Direct analysis of the FMR-1 gene provides an explanation for an exceptional case of a fragile X negative, mentally retarded male in a fragile X family. J. Med. Genet. 29, 919–920 10.1136/jmg.29.12.919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone F., Hagerman R. J., Ikle D. N., Dyer P. N., Lampe M., Willemsen R., et al. (1999a). FMRP expression as a potential prognostic indicator in fragile X syndrome. Am. J. Med. Genet. 84, 250–261 [DOI] [PubMed] [Google Scholar]
- Tassone F., Hagerman R. J., Loesch D. Z., Lachiewicz A., Taylor A. K., Hagerman P. J. (2000a). Fragile X males with unmethylated, full mutation trinucleotide repeat expansions have elevated levels of FMR1 messenger RNA. Am. J. Med. Genet. 94, 232–236 [DOI] [PubMed] [Google Scholar]
- Tassone F., Hagerman R. J., Taylor A. K., Gane L. W., Godfrey T. E., Hagerman P. J. (2000b). Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet. 66, 6–15 10.1086/302720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone F., Longshore J., Zunich J., Steinbach P., Salat U., Taylor A. K. (1999b). Tissue-specific methylation differences in a fragile X premutation carrier. Clin. Genet. 55, 346–351 10.1034/j.1399-0004.1999.550508.x [DOI] [PubMed] [Google Scholar]
- Tassone F., Pan R., Amiri K., Taylor A. K., Hagerman P. J. (2008). A rapid polymerase chain reaction-based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J. Mol. Diagn. 10, 43–49 10.2353/jmoldx.2008.070073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A. K., Tassone F., Dyer P. N., Hersch S. M., Harris J. B., Greenough W. T., et al. (1999). Tissue heterogeneity of the FMR1 mutation in a high-functioning male with fragile X syndrome. Am. J. Med. Genet. 84, 233–239 [DOI] [PubMed] [Google Scholar]
- Team R. C. (2014). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing [Google Scholar]
- Thurman A. J., McDuffie A., Hagerman R., Abbeduto L. (2014). Psychiatric symptoms in boys with fragile X syndrome: a comparison with nonsyndromic autism spectrum disorder. Res. Dev. Disabil. 35, 1072–1086 10.1016/j.ridd.2014.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk A. J., Pieretti M., Sutcliffe J. S., Fu Y. H., Kuhl D. P., Pizzuti A., et al. (1991). Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65, 905–914 10.1016/0092-8674(91)90397-H [DOI] [PubMed] [Google Scholar]
- Wechsler D. (1997). WMS-III Wechsler Memory Scale. San Antonio, TX: Psychological Corp.: 1 plastic 3 dimensional board with 10 blocks, 12 stimulus booklets, 11 technical manual, 25 visual reproduction response booklets, 25 record forms, 11 manual [Google Scholar]
- Wechsler D. (2009). WMS-IV Wechsler Memory Scale-fourth Edition. San Antonio, TX: Pearson: 2 books, 2 manuals, scoring template, response booklet, record forms, memory grid, cards [Google Scholar]
- Wechsler D., Psychological Corporation, Pearson Education Inc. (2008). WAIS-IV Wechsler Adult Intelligence Scale. San Antonio, TX: Psychological Corp.: 1 administration and scoring manual (258 p. 224 cm.), 251 technical and interpretive manual (218 p. 228 cm.), 251 stimulus book 251 (228 cm.), 251 stimulus book 252 (228 cm.), 225 test record forms (228 cm.), 225 response booklets #251 (ill. 228 cm.), 225 response booklets #252 (ill. 228 cm.), 251 coding scoring template (251 sheet 228 cm.), 252 symbol search scoring keys (252 sheets 228 cm., A-B, C), 251 cancellation scoring template (244 x 228 cm., folded to 228 x 222 cm.), 259 design blocks in box [Google Scholar]
- Willems P. J., Van Roy B., De Boulle K., Vits L., Reyniers E., Beck O., et al. (1992). Segregation of the fragile X mutation from an affected male to his normal daughter. Hum. Mol. Genet. 1, 511–515 10.1093/hmg/1.7.511 [DOI] [PubMed] [Google Scholar]
- Wohrle D., Hirst M. C., Kennerknecht I., Davies K. E., Steinbach P. (1992). Genotype mosaicism in fragile X fetal tissues. Hum. Genet. 89, 114–116 10.1007/BF00207057 [DOI] [PubMed] [Google Scholar]
- Wohrle D., Salat U., Glaser D., Mucke J., Meisel-Stosiek M., Schindler D., et al. (1998). Unusual mutations in high functioning fragile X males: apparent instability of expanded unmethylated CGG repeats. J. Med. Genet. 35, 103–111 10.1136/jmg.35.2.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.