SUMMARY
Cyclic di-adenosine monophosphate (c-di-AMP) is a broadly conserved second messenger required for bacterial growth and infection. However, the molecular mechanisms of c-di-AMP signaling are still poorly understood. Using a chemical proteomics screen for c-di-AMP interacting proteins in the pathogen Listeria monocytogenes, we identified several broadly conserved protein receptors, including the central metabolic enzyme pyruvate carboxylase (LmPC). Biochemical and crystallographic studies of the LmPC-c-di-AMP interaction revealed a previously unrecognized allosteric regulatory site 25 Å from the active site. Mutations in this site disrupted c-di-AMP binding and affected enzyme catalysis of LmPC as well as PC from pathogenic Enterococcus faecalis. C-di-AMP depletion resulted in altered metabolic activity in L. monocytogenes. Correction of this metabolic imbalance rescued bacterial growth, reduced bacterial lysis, and resulted in enhanced bacterial burdens during infection. These findings greatly expand the c-di-AMP signaling repertoire and reveal a central metabolic regulatory role for a cyclic di-nucleotide.
INTRODUCTION
The colonization of diverse environmental and host niches by bacteria depends on the ability to sense and respond to external stimuli. Signal transduction pathways dependent upon nucleotide-based second messengers are ubiquitous among bacteria (Kalia et al., 2012) and mediate cellular development, group behavior and metabolism as a result of changing environmental conditions. Among such molecules, cyclic di-adenosine monophosphate (c-di-AMP, cdA) has recently emerged as a broadly conserved second messenger of fundamental importance for microbial growth and physiology, with effects on stress responses, antibiotic resistance, cellular morphology, bacterial growth, and virulence (Campos et al., 2014; Corrigan et al., 2011; Corrigan et al., 2013; Luo and Helmann, 2012; Mehne et al., 2013; Oppenheimer-Shaanan et al., 2011; Pozzi et al., 2012; Smith et al., 2012; Witte et al., 2013; Zhang and He, 2013). Moreover, c-di-AMP secreted by L. monocytogenes, M. tuberculosis and C. trachomatis during infection results in an IFN-β-mediated host immune response (Barker et al., 2013; Woodward et al., 2010; Yang et al., 2014). Together these reports have established c-di-AMP as a crucial intracellular signaling molecule and an important component of innate immune detection during infection. Despite this central importance, the molecular mechanisms of c-di-AMP-mediated signal transduction in bacteria are just being elucidated.
Components of second messenger signaling systems include mechanisms to generate, degrade, and relay the signal. C-di-AMP is synthesized from two molecules of ATP (Witte et al., 2008), and in some cases ADP (Bai et al., 2012), through the catalytic activity of di-adenylate cyclases (DACs). The signal is subsequently degraded to pApA or AMP by DHH/DHHA1 domain containing phosphodiesterases (Bai et al., 2013; Rao et al., 2010). Second messengers initiate signal transduction by binding to and altering protein and nucleic acid function, resulting in transcriptional, translational, and post-translational alterations within a cell. Recent reports have identified several mechanisms of c-di-AMP regulation within these contexts, including riboswitches and protein receptors (Bai et al., 2014; Corrigan et al., 2013; Nelson et al., 2013; Zhang et al., 2013). However, c-di-AMP riboswitches are absent in some organisms and protein receptor homologs are not well conserved. Similarly, the functional roles of identified protein receptors cannot explain the breadth of phenotypes associated with altered c-di-AMP levels. Together these observations imply that additional c-di-AMP signaling pathways likely exist.
Listeria monocytogenes is a gram-positive intracellular bacterial pathogen that provides an attractive model to interrogate the molecular details of protein-mediated c-di-AMP signaling. C-di-AMP produced from a single DAC containing protein is a central regulator of numerous fundamental processes within this organism, including growth, cell wall homeostasis, stress responses, and the establishment of infection (Witte et al., 2013). In this study we developed a chemical proteomics approach to identify the c-di-AMP interacting proteins. We identified several c-di-AMP binding proteins that are broadly conserved in numerous bacterial species. Among these proteins, we structurally and biochemically characterized the interaction with pyruvate carboxylase (PC) and characterized the consequences of c-di-AMP regulation of central metabolic activity on L. monocytogenes survival within the host environment. Our findings provide insight into the central biological function of c-di-AMP among the diverse bacteria known to generate this second messenger.
RESULTS
Chemical proteomics to define the c-di-AMP interactome of L. monocytogenes
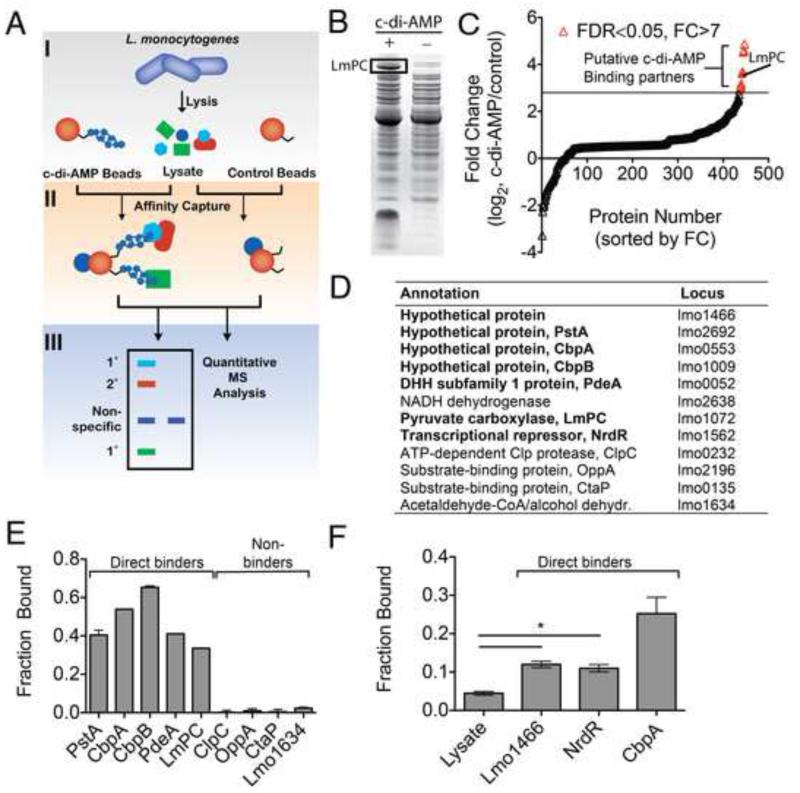
We utilized a proteomics approach to identify the c-di-AMP interactome of L. monocytogenes (Fig. 1A). The nucleotide was covalently conjugated to epoxy-activated sepharose beads (Fig. S1). Nucleotide-bound and control beads were used in parallel for affinity purification of proteins from L. monocytogenes 10403S. Initial binding results were visually analyzed by SDS-PAGE (Fig. 1B) and subsequently using gel-free quantitative shotgun proteomics (Fig. 1C). We identified twelve proteins that were statistically significant among replicates (false discovery rate <0.05) and enriched >7-fold by spectral count ratio (>2 standard deviations) with c-di-AMP versus control sepharose (Fig 1D). The four proteins with highest enrichment are of conserved hypothetical annotation, with homologs found in a large number of bacteria. Among these, the homolog of Lmo2692 in S. aureus was recently identified as a c-di-AMP binding protein (Corrigan et al., 2013). In addition, the known c-di-AMP phosphodiesterase PdeA (Lmo0052) is the fifth protein on our list (Witte et al., 2013). The successful identification of known c-di-AMP binding proteins validated our approach.
Figure 1. Identification of c-di-AMP-interacting proteins from L. monocytogenes.
(A) Schematic diagram of chemical proteomics used to identify c-di-AMP interacting proteins. 1° and 2° represent direct and indirect specific binding proteins. (B) Representative pull-down from bacterial lysates with c-di-AMP (+) and control sepharose (-). The remaining lanes from the SDS-PAGE gel have been removed for clarity. (C) Quantitative shotgun proteomics of c-di-AMP binding proteins. Data are sorted based upon fold change (FC) in spectral count ratio (c-di-AMP sepharose/control sepharose). Data points in red represent proteins with FC >7 (horizontal black line) and a false discovery rate (FDR) < 0.05. (D) List of top hits identified by chemical proteomics. Studies performed in L. monocytogenes strain 10403S. Locus numbers based on strain EGD-e (Accession number: NC_003210.1). Proteins in bold confirmed for direct c-di-AMP binding with (E) purified recombinant proteins or (F) crude cell lysate for proteins identified by chemical proteomics. Data are mean ± SEM (N=2). *P<0.05 (Students t-test, two-tailed). See also Figure S1-S3.
To confirm these interactions, proteins were probed for direct c-di-AMP binding by differential radial capillary action of ligand assay (DRaCALA) (Roelofs et al., 2011). 32P-labeled c-di-AMP was synthesized (Fig. S2) and used to quantify binding. Those proteins predicted to be soluble by in silico analysis (Bernsel et al., 2009) were expressed and purified to homogeneity from E. coli. The three soluble proteins of conserved hypothetical annotation (renamed here PstA, CbpA, and CbpB) as well as PdeA and LmPC exhibited binding in purified form (Fig. 1E). No binding was observed for purified recombinant ClpC, OppA, CtaP, or Lmo1634, which were likely identified because they interact with a direct c-di-AMP interacting protein (labeled 2° in Fig. 1A) or contain post-translational modifications that facilitate c-di-AMP binding. NrdR exhibited significant instability in purified form, while Lmo1466 and NADH dehydrogenase are predicted to be membrane anchored. As such, these proteins were characterized in whole cell lysates. E. coli lysates containing either an empty vector or encoding Lmo0553 were used as negative and positive controls, respectively (Fig. 1F). A significant level of binding was observed for Lmo1466 and NrdR-expressing lysates, consistent with a direct interaction with c-di-AMP. No interaction was observed for lysates derived from E. coli expressing NADH dehydrogenase. However, overexpression of NADH dehydrogenase was not observed and the direct interaction with c-di-AMP was therefore inconclusive.
The top three proteins that displayed positive binding and could be purified were investigated in further detail. CbpA, CbpB, and PstA demonstrated c-di-AMP binding constants <10 μM (Fig. S3), consistent with levels of c-di-AMP reported in bacteria (Corrigan et al., 2011). Specificity of the c-di-AMP interaction was studied by competitive binding with cold nucleotides. For the proteins CbpA, CbpB, and PstA, only excess c-di-AMP could abrogate binding when present at 400 μM of unlabeled competing nucleotide (Fig. S3). These observations support a specific interaction with c-di-AMP.
C-di-AMP is an allosteric inhibitor of LmPC
Among those newly identified c-di-AMP-binding proteins, only LmPC (120 kDa) has a known biological function. In fact, LmPC is essential for L. monocytogenes growth (Schar et al., 2010). Therefore, we focused our initial studies on the detailed characterization of this molecular interaction and its biological consequences. PC is a multi-domain enzyme that catalyzes the ATP-dependent carboxylation of pyruvate to oxaloacetate. During catalysis, the biotin carboxyl carrier protein (BCCP) domain is carboxylated in an ATP-dependent manner by the biotin carboxylase (BC) domain. Subsequently, the carboxyltransferase (CT) domain mediates transfer of the carboxyl from biotin to the substrate pyruvate. The PC tetramerization (PT) domain is important for the quaternary structure of the enzyme (St Maurice et al., 2007; Xiang and Tong, 2008) and together with the BC domain defines the binding site of the allosteric activator Ac-CoA (St Maurice et al., 2007; Yu et al., 2009).
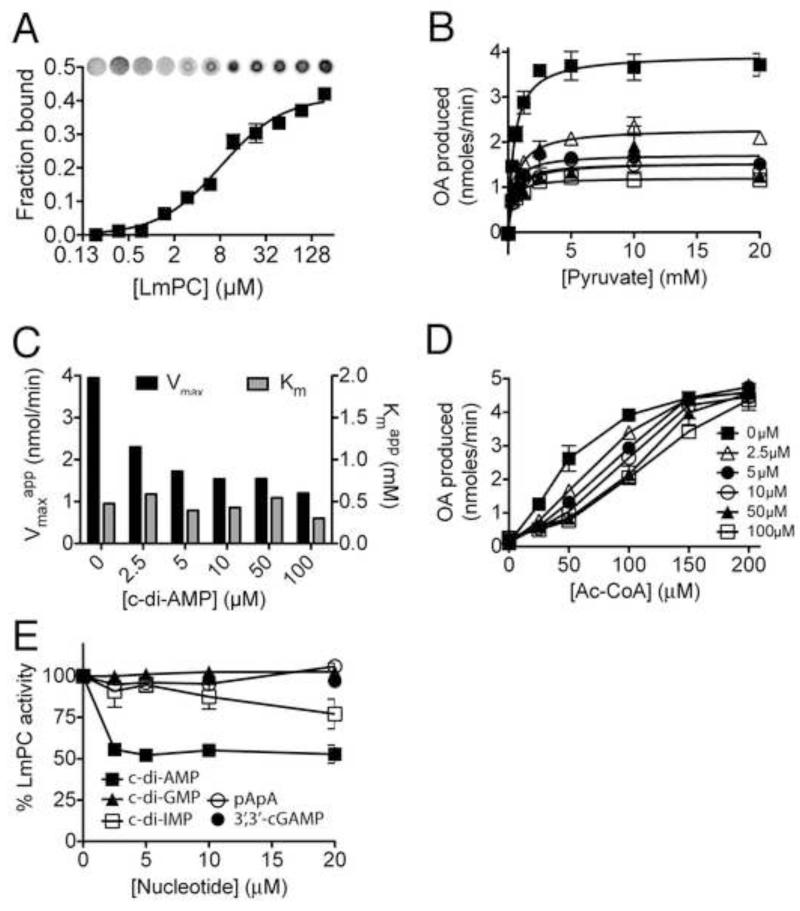
Binding studies indicated that LmPC interacts with c-di-AMP with a Kd of 8 ± 0.2 μM (Fig. 2A). We observed diminished PC acitivity in the presence of increasing concentrations of c-di-AMP (Ki ~3 μM, Fig. 2B). The inhibitory effect was at the level of the apparent kcat with slight lowering of the apparent Km (Fig. 2C), indicating that c-di-AMP does not compete with the pyruvate substrate. We also found that the inhibition by c-di-AMP could be overcome by high concentrations of Ac-CoA (Fig. 2D). Finally, the inhibition was specific to c-di-AMP as most of structurally related di-nucleotides (Fig. S4), including c-di-GMP, cGAMP (cyclic[Gp(3′,5′)Ap(3′,5′)]), or pApA, the breakdown product of c-di-AMP, showed no effect on LmPC activity, while c-di-IMP did show weak inhibitory activity (Fig. 2E). Overall, these biochemical studies define c-di-AMP as an allosteric inhibitor of LmPC.
Figure 2. LmPC specifically binds and is inhibited by c-di-AMP.
(A) Binding titration of c-di-AMP and LmPC using 32P-c-di-AMP. (B) Enzymatic activity of LmPC in the presence of Acetyl-CoA (100 μM), varied pyruvate, and c-di-AMP concentrations as indicated. (C) Rate constants of pyruvate turnover in the presence of c-di-AMP. (D) Enzymatic activity of PC in the presence of pyruvate (20 mM), varied Acetyl-CoA (allosteric activator) and c-di-AMP concentrations as indicated. (E) PC activity in the presence of nucleotide analogues. Data reported as percent activity relative to that without added nucleotide. All data reported as the mean +/− SEM. See also Figure S4.
Binding mode of c-di-AMP in LmPC
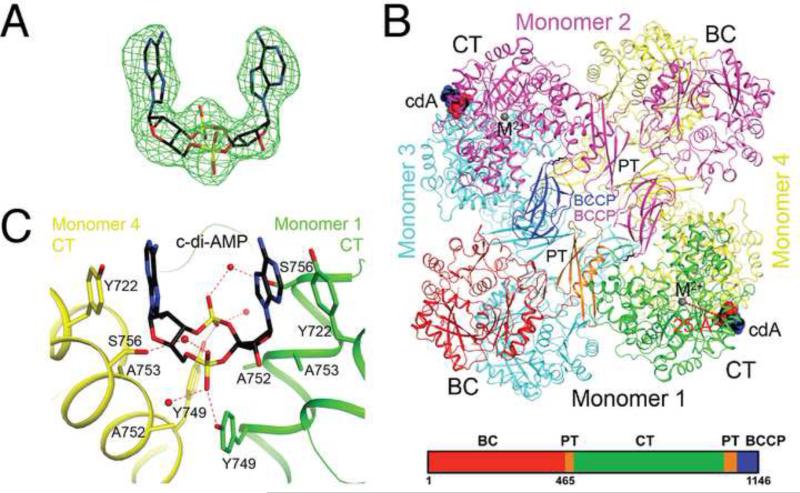
To understand the molecular basis for the enzyme regulation, we determined the structure of the LmPC-c-di-AMP complex at 2.5 Å resolution (Table S1). Clear electron density was observed for the c-di-AMP ligand based on the crystallographic analysis (Fig. 3A). A citrate molecule was found in the acetyl-CoA binding site, although our kinetic studies showed that citrate had only small effects on the catalytic activity of LmPC. Its presence in the structure was likely due to the high concentration in the crystallization solution. Unexpectedly, we observed another c-di-AMP that bridges two c-di-AMP molecules bound to neighboring LmPC in this crystal (Fig. S5A). The bridging c-di-AMP inserts one of its adenine bases between the two bases of the bound c-di-AMP, forming π-stacking interactions, but it has no direct contacts with the protein (Fig. S5A). We observed a second crystal form of the complex from the same crystallization condition, and determined its structure at 2.7 Å resolution (Table S1). The bridging c-di-AMP molecule is absent in this crystal (Fig. S5B), but the structure of the complex is essentially identical to the other crystal form (Fig. S5C). Therefore, the bridging c-di-AMP is most likely an artifact in that crystal form and will not be discussed further.
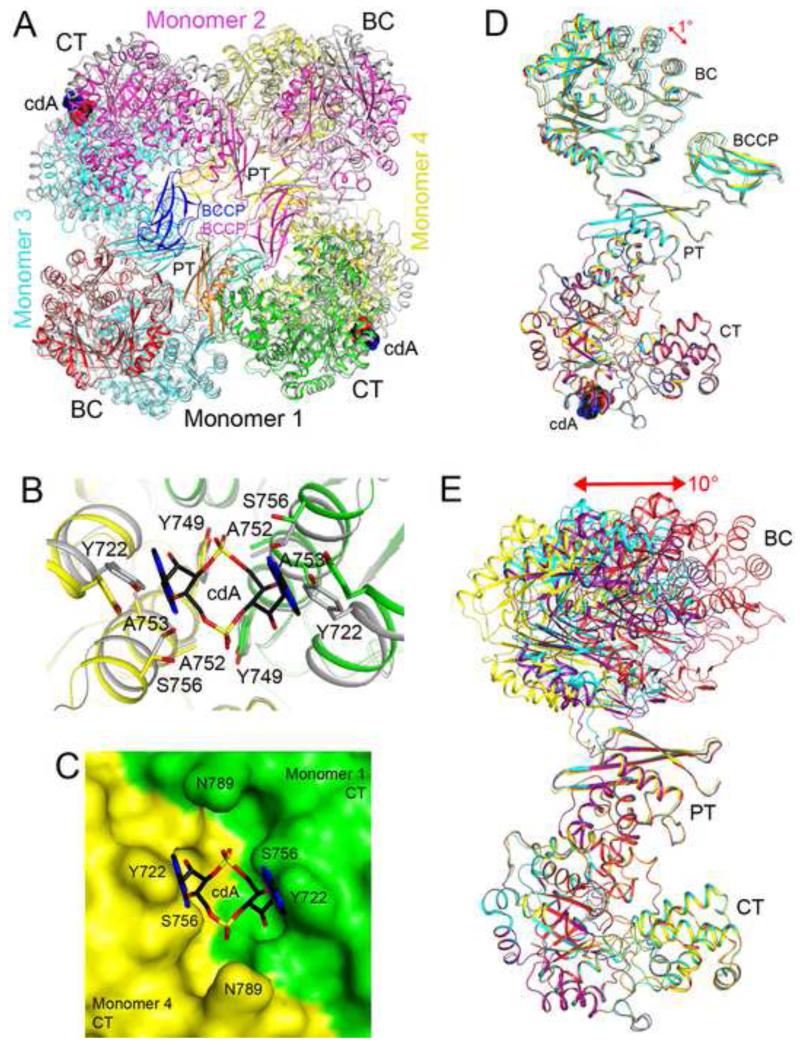
Figure 3. Crystal structure of LmPC in complex with c-di-AMP.
(A). Omit Fo-Fc electron density map for c-di-AMP at 2.5 Å resolution, contoured at 3′ (B). Schematic drawing of the structure of LmPC tetramer in complex with two c-di-AMP molecules. The domains of monomer 1 are colored separately, according to the diagram at the bottom of the panel. c-di-AMP is shown as a sphere model and labeled cdA (carbon atoms in black). The metal ion in the active site of the CT domain is shown as a gray sphere and labeled M2+. The distance from the CT active site and c-di-AMP binding site is indicated with the red line. (C). Detailed interactions between c-di-AMP and LmPC. Hydrogen-bonding interactions are indicated with dashed lines (in red). Water molecules are shown as red spheres. All structure figures were produced with PyMOL (www.pymol.org). See also Table S1 and Figure S5.
LmPC is a tetramer in the crystal structure (Figs. 3B, S5C), consistent with solution studies as well as earlier structures of R. etli PC (RePC), S. aureus (SaPC) and H. sapiens (HsPC) (St Maurice et al., 2007; Xiang and Tong, 2008). The tetramer is composed of two layers, with two monomers in each layer. There are extensive interactions between the two layers, mediated through the BC and CT domain dimers, which are located at opposing corners of the diamond-shaped tetramer. In contrast, there are few direct contacts between the two monomers in the same layer. All four BCCP domains are observed, but the linkers between the PT and the BCCP domains (residues 1060-1066) are disordered. As a result, each BCCP domain is assigned to a monomer based on the earlier structure of SaPC (Fig. 3B), although the actual PT-BCCP connectivity is ambiguous in LmPC. The position of BCCP is similar to that in the exo site of the SaPC structure (Xiang and Tong, 2008), but its orientation is very different (Fig. S6A). Consequently, the biotin group in the current structure is projected into the solvent and disordered. Besides the PT-BCCP linker, the B domain of BC (residues 135-205) is also disordered in the crystals.
The c-di-AMP nucleotide is bound at the dimer interface of two CT domains (Figs 3B), with the two-fold symmetry axis of the compound aligned with that of the CT domains (Fig. 3C). Therefore, each LmPC tetramer binds only two c-di-AMP molecules (Fig. 3B). The binding site is formed by two helices in the TIM barrel fold of the CT domain (Fig. 3C), but it is located ~25 Å away and on the opposite face from the CT active site (Figs. 3B). This region of PC has not been known previously to have any regulatory or structural significance.
The c-di-AMP ligand adopts a U-shaped conformation (Fig. 3A) and interacts with LmPC through hydrophobic and hydrogen-bonding interactions (Fig. 3C). The aromatic ring of Tyr722 has a direct face-to-face π-stacking arrangement with an adenine base. One of the terminal oxygen atoms of the phosphate group of c-di-AMP forms a hydrogen bond with the hydroxyl group of Tyr749, while the other terminal oxygen interacts with the Ser756 side chain through a water molecule. The ribose is located near Ala753, which together with Ala752 may provide the space necessary for the ligand to bind.
The U-shaped conformation of c-di-AMP is similar to the binding mode of c-di-GMP in some proteins, such as the bacterial PilZ domain (Benach et al., 2007) and the human STING protein (Huang et al., 2012; Ouyang et al., 2012; Shang et al., 2012; Shu et al., 2012; Yin et al., 2012). In addition, c-di-GMP is also aligned with the two-fold symmetry axis of a STING dimer, -stacked with a Tyr side chain. Together these observations identify a binding site of c-di-AMP to LmPC and confirm kinetic studies that the site is unique from known substrates or allosteric regulators of this family of enzymes.
The c-di-AMP binding site affects PC catalytic activity
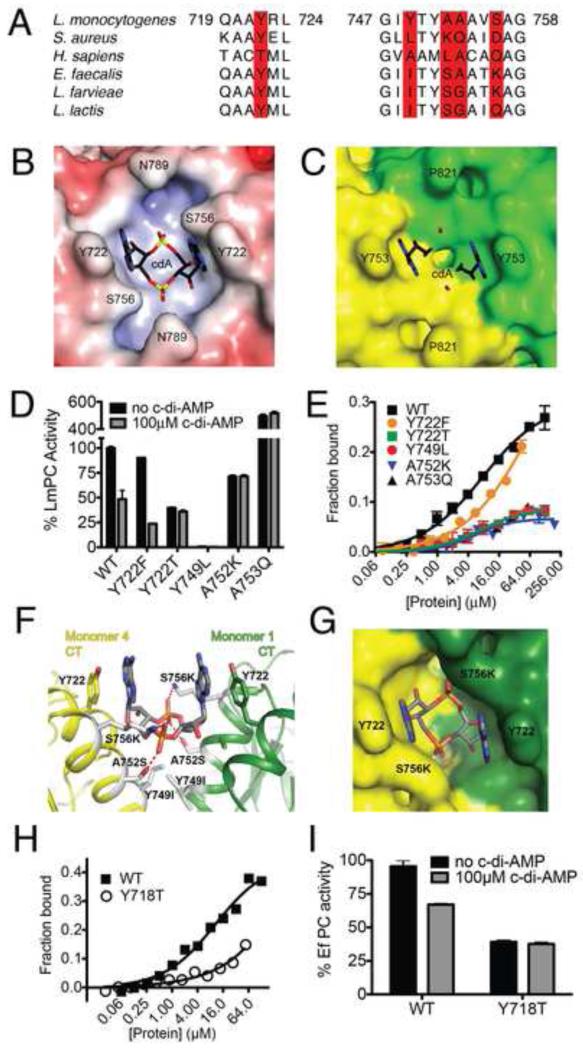
To assess the functional importance of the binding site, we introduced amino acid mutations into LmPC. We noted conservation of a π-stacking aromatic amino acid among many cyclic di-nucleotide binding proteins, including STING, VpsT, and the EAL domain protein YahA (Huang et al., 2012; Krasteva et al., 2010; Ouyang et al., 2012; Shang et al., 2012; Sundriyal et al., 2014; Yin et al., 2012), implicating Y722 as a critical residue for nucleotide recognition. Two amino acid substitutions (Y722F and Y722T) were introduced into this site. The Y722F mutant was chosen to assess the importance of the hydroxyl group interacting with the N6 of the adenine base. The Y722T mutant was chosen due to the presence of this amino acid in the corresponding site of HsPC (Fig. 4A). Comparison of the LmPC and SaPC structures revealed that the analogous binding pocket does not apparently exist within the latter enzyme (Figs 4B-C and S6A-B). This can be attributed to the presence of a Lys and Gln substitution in SaPC corresponding to A752 and A753 of LmPC, respectively, which introduce significant steric bulk into the pocket. Similarly, the Y749 residue required for hydrogen bonding to the terminal oxygen of the phosphate at the bottom of this pocket has been lost. These observations were used to guide amino acid substitutions in the LmPC protein.
Figure 4. Functional analysis of the c-di-AMP binding site of PC.
(A). Conservation of residues in the c-di-AMP binding site. The five residues in the binding site are highlighted in red. (B). Molecular surface of the LmPC binding site for c-di-AMP (labeled cdA), colored by the electrostatic potential (red: negative, blue: positive). (C). Molecular surface of SaPC at the CT dimer interface. The c-di-AMP molecule is shown as a reference in stick models. (D). Effects of c-di-AMP on the catalytic activity of wild-type and mutant LmPC. (E). Titration curve showing varied sensitivity of wild-type and mutant LmPC to c-di-AMP. (F) LmPC c-di-AMP binding pocket with corresponding EfPC amino acid variations. (G) Surface view of modeled EfPC. (H) C-di-AMP binding to WT and Y718T EfPC. (I) Activity of WT and Y718T EfPC in the presence and absence of c-di-AMP. Data are mean ± SEM (D, E, G, and H). See also Figure S6.
The mutants were expressed and purified following the same protocol as was used for the wild-type protein. All amino acid mutants, except Y749L, were catalytically active. However, the Y722T, A752K, and A753Q variants lost sensitivity to inhibition by c-di-AMP even at 100 μM concentration (Fig. 4D). Interestingly, the enzyme activity in the absence of c-di-AMP was substantially different from WT, with both increased and reduced activity being observed for the mutants, suggesting that this region of the enzyme has a functionally relevant effect on PC catalysis. The Y722F mutant behaved similar to the wild-type enzyme in terms of its catalytic activity and responsiveness to c-di-AMP, establishing that the hydroxyl group of Y722 is dispensable for nucleotide recognition. In agreement with the kinetic data, binding to c-di-AMP was only observed for Y722F and none of the other amino acid variants (Fig. 4E).
We noted that although several of the residues required for c-di-AMP binding by LmPC are absent in other bacterial enzymes, the required chemical features of the binding pocket might be conserved. Specifically, when comparing LmPC to PC from lactic acid bacteria (Fig 4A), such as PC from the opportunistic pathogen Enterococcus faecalis (EfPC), structural modeling predicted that the A752S and S756K variants could establish favorable interactions with the c-di-AMP phosphate group and compensate for the loss of the Y749 residue (Fig. 4F-G). Indeed, recombinant EfPC binds c-di-AMP and its catalytic activity is inhibited by the nucleotide (Fig. 4G-H). Consistent with a conserved binding site between the enzymes, mutation of the base stacking tyrosine results in loss of c-di-AMP binding and regulation. Similarly, SaPC is insensitive to c-di-AMP. Overall, the results from these studies confirm the c-di-AMP binding site identified by the structural analysis and identify a previously unrecognized region important for PC regulation in L. monocytogenes and other bacteria.
C-di-AMP binding induces large conformational changes in LmPC
We have also determined the structure of apo LmPC (Table S1), in the absence of c-di-AMP. The best quality X-ray diffraction data set we could collect after screening through a large number of crystals was at 3.3 Å resolution. There are two tetramers in the asymmetric unit that have essentially the same conformation, with root mean square (rms) distance of 0.1 Å among crystal had pseudo C2 space-group symmetry). Therefore, only one of these tetramers will be used for further analysis. All four BCCP domains and residues 850-950 in the CT domain of one of the monomers had poor electron density and were not modeled. The B domain of BC was partially ordered in the four monomers and there was weak density for an ATP molecule.
A comparison of the structure of apo LmPC and that bound to c-di-AMP showed dramatic rearrangements in the tetramer upon ligand binding (Fig. 5A). Near the c-di-AMP binding site, there are significant changes in the orientation and position of the two CT domains of the dimer, corresponding to a rotation of ~4.5° for each CT domain (Fig. 5B). As a consequence, the side chains of both Tyr722 and Ser756 in the apo enzyme would clash with the c-di-AMP molecule in the complex. These changes greatly reduce the size of the pocket in this region, such that it can no longer accommodate c-di-AMP (Fig. 5C).
Figure 5. Large conformational differences in the structure of apo LmPC.
(A). Overlay of the structure of LmPC in complex with c-di-AMP (in color) with that of apo LmPC (in gray). The superposition was based on monomer 1. Large differences in the positions of the other monomers are visible. (B). Conformational differences in the c-di-AMP binding site in the structure of apo LmPC. (C). Molecular surface of apo LmPC at the CT dimer interface. The c-di-AMP molecule would clash with the enzyme in this conformation. (D). Overlay of the four monomers of the LmPC tetramer in the complex with c-di-AMP. The superposition is based on the CT domain only. A small conformational difference is seen for the BC domain. (E). Overlay of the four monomers of the apo LmPC tetramer. Large differences are observed in the positions and orientations of the BC domains.
The BC dimer in apo LmPC has roughly the same structure as that in the complex with c-di-AMP (Fig. S5D). However, the position and orientation of the BC domain relative to the CT domain are different between the apo enzyme and the c-di-AMP complex. In addition, the four LmPC monomers in the c-di-AMP complex have essentially the same conformation, while the monomers in the apo enzyme have large structural differences. This is best visualized by overlaying the CT domains of the four monomers in each structure. For the c-di-AMP complex, overlaying the CT domains essentially brought the BC and PT domains into overlay as well, with a difference of only ~1° in their orientations (Fig. 5D). In comparison, the four monomers differ by ~10° in the orientations of their BC domains after the CT domains are overlaid for the apo enzyme, and the hinge rotation is located near the boundary between the BC and PT domains (Fig. 5E). In comparison, the overlay of the CT domains brought the PT domains into superposition. The PT domains of SaPC and HsPC have important roles in mediating contacts in the tetramer (Xiang and Tong, 2008). The PT domains in the c-di-AMP complex of LmPC and apo LmPC are positioned further away from each other and do not make direct contacts (Fig. 5A).
Metabolic balance mediated by c-di-AMP affects intracellular growth of L. monocytogenes
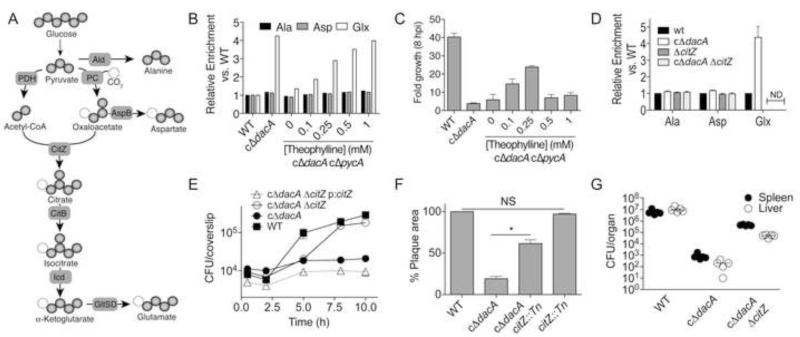
Structural and biochemical characterization of the interaction between LmPC and c-di-AMP implicates the nucleotide as a metabolic regulator in L. monocytogenes. Listeria species have an incomplete citric acid cycle and the oxaloacetate (OA) generated by LmPC is used as a precursor for aspartate and glutamate biosynthesis (Fig. 6A) (Kim et al., 2006). To study the effects of c-di-AMP levels on metabolism in L. monocytogenes, we measured incorporation of isotope label from U-13C-glucose into amino acids alanine (Ala), aspartate (Asp), and glutamate/glutamine (Glx) in both WT and a conditional depletion strain of DacA (cΔdacA), which exhibits diminished levels of c-di-AMP (Witte et al., 2013). Bacteria with depleted c-di-AMP exhibited substantially increased 13C incorporation into Glx relative to bacteria with normal levels of the nucleotide (Fig. 6B). De novo synthesis of Glx is entirely dependent on LmPC activity in L. monocytogenes (Schar et al., 2010) and flux through the oxidative branch of the citric acid cycle. To determine if these observations are a result of PC activity directly, we generated a double conditional depletion strain, cΔdacA cΔpycA, with IPTG inducible expression of DacA and theophylline inducible expression of PC (Fig. S7A). Indeed, increased levels of theophylline in the absence of IPTG led to a direct increase in Glx synthesis (Fig. 6B). Thus, c-di-AMP, through its action on LmPC and possibly other downstream targets that affect flux through this pathway, negatively affects de novo Glx biosynthesis. In comparison, reduced c-di-AMP levels did not affect Asp synthesis, indicating that LmPC is not the limiting activity for this pathway.
Figure 6. Metabolic imbalance specifically alters L. monocytogenes intracellular growth.
(A) De novo biosynthesis of amino acids and labeling patterns from 13C-glucose by L. monocytogenes. Dark spheres, 13C. Light spheres, natural abundance carbon. (B) 13C enrichment into Ala, Asp, and Glx from mid-exponential L. monocyotogenes strains. (C) Magnitude of growth of L. monocytogenes strains in iBMMs. Data reported as the ratio of CFU recovered at 8 hpi relative to 0.5 hpi. (D) 13C enrichment into Ala, Asp, and Glx from mid-exponential L. monocyotogenes strains. (E) Immortalized murine bone marrow derived macrophages were infected with L. monocytogenes and CFU were enumerated at the indicated times. (F) Plaque area from mouse fibroblasts (L2 cells) infected with indicated strains and normalized to WT. (G) Bacterial burden in organs 48 h post infection with the median indicated by a horizontal bar. (C-F) Data are mean ± SEM. (B-G) Data are representative of at least two independent experiments.. (D) *P<0.001 (Students t-test, two-tailed). See also Figure S7.
We hypothesized that this metabolic imbalance may be responsible for the defects in bacterial growth and host colonization associated with low levels of c-di-AMP. To test this possibility we infected macrophages with the cΔdacA cΔpycA strain grown in the presence of varying concentrations of theophylline. Increased cytosolic growth was observed as theophylline levels were increased until a critical threshold was achieved, after which the strain exhibited limited growth comparable to cΔdacA (Fig. 6C). These observations are consistent with previous reports where low PC activity results in diminished bacterial growth in the cytosol. However, our findings also show that enhanced PC activity is detrimental in the absence of c-di-AMP and are consistent with the requirement for balanced metabolic activity to establish infection in the host.
It has not been established why PC is essential for L. monocytogenes, though disruption of aspB resulted in diminished bacterial growth (data not shown), consistent with this being the essential branch emanating from the precursor metabolite oxaloacetate. Based on these observations, we hypothesized that the metabolic imbalance associated with diminished c-di-AMP levels was a consequence of elevated flux to Glx. To segregate the detrimental effects of hypo and hyper-active PC we deleted citrate synthase (CitZ, Fig. 6A), the first committed step to Glx biosynthesis. Incorporation of 13C into Glx was abolished in the WT and cΔdacA strains containing the citZ mutation (Fig. 6D), while levels of Asp and Ala incorporation remained unchanged. Thus, disruption of citZ prevents unregulated glutamate/glutamine biosynthesis from glucose under conditions of low c-di-AMP.
Having corrected the elevated synthesis of Glx, we detailed the effects of metabolic imbalance associated with altered c-di-AMP signaling on L. monocytogenes growth and infection. Although disruption of citZ had no discernable effect on broth growth in the presence or absence of c-di-AMP production (Fig. S7B), a significant increase in intracellular growth in the absence of c-di-AMP was observed within immortalized bone marrow derived macrophages (iBMM) and mouse fibroblasts (Fig. 6E-F). No change in growth was observed for citZ disruption in the WT background (Fig. 6F and S7C). Analysis of bacterial growth revealed that the doubling time was not significantly altered by loss of citZ, consistent with observations in rich media. However, the ability of L. monocytogenes to maintain intracellular growth between five and eight hours was rescued by disruption of citZ, resulting in nearly WT levels of bacterial burden at later time points. Expression of the citrate synthase gene in trans attenuated intracellular growth of the cΔdacA ΔcitZ strain back to cΔdacA levels (Fig. 6E).
To further characterize the effects of metabolic imbalance associated with c-di-AMP deficiency, a murine model of acute listeriosis was characterized. As previously reported (Witte et al., 2013), the cΔdacA strain exhibited 3.8 and 4.6-log defects in both the spleen and liver, respectively (Fig. 6G). When citZ was disrupted in the cΔdacA strain, 2.7 and 2.4-logs of growth were rescued in the spleen and liver, respectively. Together with ex vivo analysis of intracellular growth, these findings support a model whereby metabolic imbalance associated with altered c-di-AMP levels specifically affects the magnitude of L. monocytogenes growth within the host cell cytosol with significant consequences on bacterial virulence.
Metabolic imbalance in L. monocytogenes causes bacteriolysis within the host cytosol
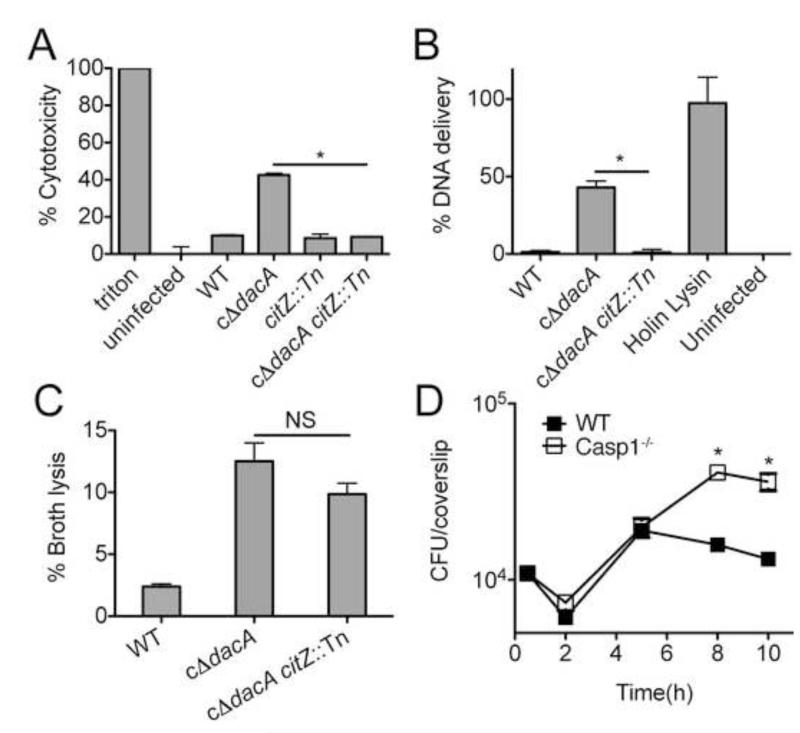
To further elucidate the basis for the intracellular growth defect resulting from altered bacterial metabolic activity, we characterized the host response to infection. DNA released by L. monocytogenes leads to activation of Caspase 1 through the AIM2 inflammasome, resulting in pyroptosis during L. monocytogenes infection (Sauer et al., 2010). We found that bacteria with diminished levels of c-di-AMP production induce increased cell death during infection (Witte et al., 2013), while those with corrected metabolic activity through citZ disruption exhibited significantly decreased levels of host cell death (Fig. 7A). Consistent with these observations, bacteria deficient in c-di-AMP production exhibit increased DNA delivery during infection, while the metabolic correction of this strain results in significantly reduced intracellular bacteriolysis (Fig. 7B). Interestingly, no significant change in bacteriolysis is observed during growth in broth media (Fig. 7C).
Figure 7. C-di-AMP-induced metabolic imbalance provokes intracellular bacteriolysis and host cell pyroptosis.
(A) Cytotoxicity induced in primary BMMs by the indicated strains following infection was measured by LDH release. (B) Intracellular lysis of bacterial strains in iBMMs as measure by reported plasmid delivery. Percent lysis was determined by normalizing to Holin-Lysin and uninfected controls. (C) Lysis of L. monocytogenes strains grown in BHI. (D) Growth of cΔdacA L. monocytogenes in WT (closed symbols) and Caspase 1−/− (open symbols) BMMs. Data are mean ± SEM and are representative of at least two independent experiments. *P<0.05 (Students t-test, two-tailed); NS, not significant.
Pyroptosis is a protective host response to L. monocytogenes infection (Sauer et al., 2011). As such, we predicted that removal of this inflammatory response might rescue the growth of c-di-AMP deficient bacteria that exhibit c-di-AMP-related metabolic imbalance. Consistent with this hypothesis, intracellular growth of bacteria deficient in c-di-AMP production significantly improved in the absence of pyroptosis in primary bone marrow derived macrophages (Figure 7D). These observations suggest that the metabolic imbalance associated with diminished c-di-AMP signaling leads in part to diminished bacterial growth as a consequence of an enhanced restrictive host response. Together with our biochemical studies we conclude that these effects are most likely due to c-di-AMP mediated regulation of LmPC and perhaps other metabolic factors.
DISCUSSION
Chemical proteomics provides a powerful approach to interrogate small molecule-protein interactions (Rix and Superti-Furga, 2009). The nucleotide interactome revealed here substantially broadens the known c-di-AMP protein-based signaling repertoire; identifying five previously unrecognized c-di-AMP interacting proteins in L. monocytogenes, in addition to LmPC. The large number of effectors, their domain diversity, and seemingly disparate functional roles (Grinberg et al., 2006; Leigh and Dodsworth, 2007; Liu et al., 2006; Scott et al., 2004) are consistent with the pleiotropic effects of the dacA gene in L. monocytogenes. Our studies genetically segregate the effects of c-di-AMP on the magnitude of intracellular growth and intracellular bacterial lysis from other known phenotypes associated with loss of c-di-AMP production. We anticipate that the identification of several other protein receptors for c-di-AMP will guide future studies to define the molecular details governing changes in other c-di-AMP related phenotypes, including bacterial cell stability in broth, susceptibility to cell-wall targeting antibiotics, and bacterial growth rate in and out of the host.
Beyond broadening the known c-di-AMP effectors, our studies provide a detailed molecular characterization of the effects of c-di-AMP on PC activity. Our structural studies of apo and c-di-AMP bound LmPC in comparison to known PC structures provide mechanistic insight into an unexpected mode of enzyme regulation. The nearly identical structure of the four LmPC monomers in the c-di-AMP complex is rather unusual among known PC structures. For SaPC, the differences in the orientations of the BC domains vary between 6 and 18° after their CT domains are superimposed (Xiang and Tong, 2008; Yu et al., 2009), and the differences are even larger for RePC (St Maurice et al., 2007). The structural differences among the four monomers in apo LmPC, as well as in SaPC and RePC, indicate that domain movements are necessary during PC catalysis, which is supported by recent EM studies on SaPC (Lasso et al., 2014). Based on these observations we favor a mechanism whereby c-di-AMP ‘freezes’ the enzyme into a single (symmetrical) conformation, which is incompatible with catalysis.
Structure-function studies also clearly establish the location and strict specificity of the c-di-AMP binding pocket. However, it is currently not clear how nucleotide specificity is achieved, as there are no hydrogen-bonding interactions to the base. Mutations of the sole base-interacting amino acid, Tyr722, do not extend inhibition activity to related nucleotides, and therefore other residues may contribute to this specificity. Beyond conferring specificity, changes in this pocket have significant consequences on enzyme function, even in the absence of ligand binding. These observations may be indicative of a previously unrecognized allosteric site for PC enzymes in response to c-di-AMP and possibly other stimuli. Significant variability is observed within the regulatory site identified by our studies. The EfPC-c-di-AMP interaction illustrates this mechanism of regulation extends beyond L. monocytogenes and that in the absence of amino acid conservation, divergent amino acids within the c-di-AMP binding site can compensate to maintain nucleotide-mediated enzyme regulation. Given the central function of PC, proper integration of this enzyme into existing metabolic networks is required to maintain metabolic balance. As such, we postulate that the diversity of the sequence at this novel functional site may reflect divergent mechanisms of regulation, either through allosteric ligands or altered enzyme dynamics, which fine tune activity such that PC can properly integrate into the unique metabolic capacity of the diverse organisms that utilize this enzyme.
Bacteria have the capacity to thrive within a variety of environmental and host niches. This adaptability is largely reliant on the capacity to utilize diverse nutrients encountered in these disparate environments. The phosphoenolpyruvate-pyruvate-oxaloacetate node of central metabolism shapes the distribution of carbon metabolites among anabolic, catabolic and energy-producing pathways (Jitrapakdee et al., 2008; Jitrapakdee and Wallace, 1999; Sauer and Eikmanns, 2005; Schar et al., 2010). Given this central function, aberrant metabolic activity as a consequence of altered c-di-AMP signaling may disrupt any one or a combination of the pathways required for bacterial adaptation to accessible nutrients. During infection, intracellular L. monocytogenes undergo a major metabolic transition from utilization of glucose to alternative carbon sources (Joseph et al., 2006), a process that affects activation of the master virulence regulator PrfA (Joseph et al., 2008; Stoll et al., 2008). Consequently, changes in PEP levels through increased pyruvate consumption may diminish PrfA activation and directly link c-di-AMP-related metabolic activity to virulence capacity. However, our observation that bacterial lysis is a downstream consequence of altered c-di-AMP-dependent metabolism suggests the answer partially relates to the integrity of the bacterial cell. The metabolic burden generated by unregulated flux through the citric acid cycle may cause insufficiency in cell wall biosynthesis intermediates, leading to bacteriolysis. No apparent deficiency in de novo Asp, Ala, or Glx synthesis, each of which contributes to biosynthesis of the glycan and peptide components of the cell wall, suggests that limitations in these metabolites are not responsible. As the critical compound utilized by L. monocytogenes to combat acid stress and the primary counter ion to osmotic stabilizing potassium (Booth and Higgins, 1990; Cotter et al., 2005; Yan et al., 1996), synthesis of glutamate must be tightly regulated to appropriately respond to changing conditions. As such, increased production of this and metabolites in this pathway may have toxic consequences on bacterial stability in the cytosol. Finally, hyperactivity of LmPC and subsequent turnover of OA by CitZ could lead to defects in lipid synthesis by consuming Ac-CoA, a major precursor for the acyl-chain required to generate branched chain fatty acids. It is not known if L. monocytogenes strains deficient in these lipids exhibit enhanced intracellular lysis, however, the synthesis of these membrane components is required for L. monocytogenes survival within the cytosol of the host cell (Sun and O’Riordan, 2010). Clearly the effects of PC regulation extend beyond this intracellular pathogen. The evidence supporting PC regulation among lactic acid bacteria suggests that both extracellular pathogenic (i.e. E. faecalis) as well as non-pathogenic commensal Lactobacilli spp. utilize a similar mechanism of metabolic regulation, though the effects of this regulation remain an open question. Future studies to detail the functional consequences of c-di-AMP mediated metabolic regulation through PC and likely other protein receptors will provide important insight into the physiology of these medically important host associated organisms.
In summary, the c-di-AMP binding proteins reported here are widespread and conserved among environmental and pathogenic microbes alike, and broadly expand the protein targets of c-di-AMP in bacteria. Furthermore, our biochemical and structural studies may provide important insight into a new molecular mechanism of central metabolic regulation through the enzyme PC. Finally, our studies demonstrate that c-di-AMP signaling regulates bacterial central metabolism and survival within the host environment. These findings lead us to propose that c-di-AMP serves a conserved and central role in the regulation of fundamental metabolic processes with important physiological implications in the unique environments occupied by the bacteria that utilize this secondary signaling molecule. A detailed understanding of bacterial signaling and metabolic regulation has profound practical and therapeutic applications. As a central element of these processes, the possibility of targeting c-di-AMP signaling pathways in this regard is an attractive option that remains to be explored.
EXPERIMENTAL PROCEDURES
C-di-AMP pull-downs
Soluble lysates from L. monocytogenes grown in 1.5 L BHI broth at 37 °C to mid-exponential phase (OD600 = 0.8) were prepared in 30 mL of buffer (0.1 M Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% v/v Tween-20, 1 mM PMSF). Lysates were incubated with rotation for 2 hours at 4 °C with 100 μL ethanoamine or c-di-AMP conjugated beads. Beads were washed three times with 5 mL PBS, mixed with 100 μL of SDS-PAGE sample loading buffer, incubated at 56 °C for 10 minutes, and the soluble fraction removed. The elution was repeated and each was pooled and processed for quantitative mass spectrometry as detailed in the Supplemental Information. Pulldowns were performed three independent times, one with the ΔpdeA strain and twice with WT L. monocytogenes.
Nucleotide binding assays
The DRaCALA assay were performed with recombinant proteins or E. coli lysates in binding buffer (40mM Tris, 100mM NaCl, 20 mM MgCl2, pH 7.5) and analyzed as described previously (Roelofs et al., 2011). For competition assays, 400 μM unlabeled nucleotide [ATP, GTP, cAMP, cGMP, NAD, NADH, c-di-AMP, c-di-GMP, pApA] was added prior to the addition of 32P-c-di-AMP.
Protein crystallization
LmPC crystals were grown by the sitting-drop vapor diffusion method at 20 °C. For the c-di-AMP complex, the protein was incubated with 2.5 mM c-di-AMP and 2.5 mM ATP for 30 min at 4 °C before setup. The reservoir solution contained 19% (w/v) PEG3350 and 0.2 M ammonium citrate (pH 7.0). The crystals appeared within 1-2 weeks and grew to full-size after a few additional days. The crystals were cryo-protected in the reservoir solution supplemented with 10% (v/v) PEG200 and were flash-frozen in liquid nitrogen for data collection at 100 K. For the apo structure, the protein at 5 mg/ml was incubated with 2.5 mM ATP and 2.5 mM pyruvate for 30 min at 4 °C before setup. The reservoir solution contained 16% (w/v) PEG3350, 0.1 M Bis-Tris (pH 6.5), and 1% (w/v) tacsimate (pH 7.0, Hampton). The crystals appeared within 1 week and grew to full-size within a few more days. They were cryo-protected in the reservoir solution supplemented with 15% (v/v) ethylene glycol and flash-frozen in liquid nitrogen. Data collection and structure determination were as detailed in the Supporting Information.
13C Incorporation studies
Bacterial strains were grown overnight in BHI at 37 °C. The cells were sedimented and washed with PBS. BHI was supplemented with 2.5 mg/ml [U-13C6] glucose and were subsequently inoculated with an aliquot of the cell suspensions to an OD600 of 0.1. The cultures were grown at 37 °C until OD600 = 1.0. Sodium azide was then added to a final concentration of 10 mM. The cells were centrifuged and washed three times with water. Bacterial cells (approximately 20 mg washed wet pellet) were suspended in 0.5 ml of 6 M hydrochloric acid. The mixture was heated at 105 °C for 24 h. The hydrolysate was placed on a column of Dowex 50W × 8 (H+form; 200 to 400 mesh; 5 by 10 mm) previously washed with 4 ml of 7 % formic acid and water. After loading the acid hydrolysate the column was washed with 4 mL of water. Sample was then eluted with 4 mL of 1 N ammonium hydroxide. The samples were air dried under vacuum, and the residue was dissolved in 30 μl of pyridine. A total of 30 μL of-(tert-butyldimethyl-silyl)-methyl-trifluoroacetamide containing 1% tert-butyl-dimethyl-silylchloride (Sigma) was added. The mixture was kept at 90 °C for 60 minutes. Derivatized amino acids were analyzed by GC/MS as detailed in the Supporting Information.
Tissue culture and infection assays
Primary bone marrow derived macrophages were generated as previously described (Jones and Portnoy, 1994). Macrophage growth curves, LDH, and DNA release assays were performed in primary or J2 immortalized BMMs (Sauer et al., 2010), as indicated. Plaque assays we conducted in L2 mouse fibroblasts (Sun et al., 1990). Murine infections were performed as described previously (Witte et al., 2013). All protocols were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Wisconsin, Madison and the University of Washington.
Supplementary Material
HIGHLIGHTS.
Chemical proteomics identifies c-di-AMP protein receptors in L. monocytogenes
C-di-AMP binds to and regulates the essential enzyme pyruvate carboxylase
Structure-function studies identified a novel regulatory site of pyruvate carboxylase
Metabolic regulation imposed by c-di-AMP is required for L. monocytogenes virulence
ACKNOWLEDGEMENTS
We would like to thank Dan Portnoy (UC Berkeley) for generous support and reagents, Brad Cookson and Dan Stetson (University of Washington) for murine bone marrow, Jason Smith and Sam Miller (University of Washington) for instrument access, Jake McKinlay (University of Indiana) for 13C-enrichement protocols, Nicholas Olivarez for technical support, and Joseph Mougous (University of Washington) for feedback on the preparation of the manuscript. We thank Linda Yu for earlier studies on PC expression and purification; Neil Whalen, Rick Jackimowicz and Howard Robinson for access to the X29A beamline at the NSLS. The in-house instrument for X-ray diffraction screening was purchased with an NIH grant to LT (OD012208). PHC was also supported by an NIH Medical Scientist Training Program (GM007367). This work was supported by NIH grants U54AI057141 (to JJW) and R01DK067238 (to LT).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceived and designed the experiments: J.J.W., L.T., K.S. and P.C. Performed the experiments: J.J.W., L.T., K.S., P.C., T.N.H., A.R.J., M.P., M.D., N.P.O, D.P., and J.D.S. Contributed reagent/materials/analysis tools: A.T.I., Y.A.G, and M.S. Wrote the manuscript: J.J.W., L.T., K.S. and P.C.
ACCESSION NUMBERS
The Research Collaboratory for Structural Bioinformatics Protein Data Bank accession numbers for the new structures reported in this paper are 4QSH, 4QSK, and 4QSL. The LC MS/MS proteomics sequencing files have been deposited to Peptide Atlas database with submission number PASS00528.
REFERENCES
- Bai Y, Yang J, Eisele LE, Underwood AJ, Koestler BJ, Waters CM, Metzger DW, Bai G. Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence. J Bacteriol. 2013;195:5123–5132. doi: 10.1128/JB.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Yang J, Zarrella TM, Zhang Y, Metzger DW, Bai G. Cyclic Di-AMP Impairs Potassium Uptake Mediated by a Cyclic Di-AMP Binding Protein in Streptococcus pneumoniae. J Bacteriol. 2014;196:614–623. doi: 10.1128/JB.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Yang J, Zhou X, Ding X, Eisele LE, Bai G. Mycobacterium tuberculosis Rv3586 (DacA) is a diadenylate cyclase that converts ATP or ADP into c-di-AMP. PLoS One. 2012;7:e35206. doi: 10.1371/journal.pone.0035206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker JR, Koestler BJ, Carpenter VK, Burdette DL, Waters CM, Vance RE, Valdivia RH. STING-Dependent Recognition of Cyclic di-AMP Mediates Type I Interferon Responses during Chlamydia trachomatis Infection. MBio. 2013;4 doi: 10.1128/mBio.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, et al. The structural basis of cyclic diguanylate signal tranduction by PilZ domains. EMBO J. 2007;26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsel A, Viklund H, Hennerdal A, Elofsson A. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 2009;37:W465–468. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth IR, Higgins CF. Enteric bacteria and osmotic stress: intracellular potassium glutamate as a secondary signal of osmotic stress? FEMS Microbiol Rev. 1990;6:239–246. doi: 10.1111/j.1574-6968.1990.tb04097.x. [DOI] [PubMed] [Google Scholar]
- Campos SS, Ibarra-Rodriguez JR, Barajas-Ornelas RC, Ramirez-Guadiana FH, Obregon-Herrera A, Setlow P, Pedraza-Reyes M. Interaction of Apurinic/Apyrimidinic Endonucleases Nfo and ExoA with the DNA Integrity Scanning Protein DisA in the Processing of Oxidative DNA Damage during Bacillus subtilis Spore Outgrowth. J Bacteriol. 2014;196:568–578. doi: 10.1128/JB.01259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Grundling A. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci U S A. 2013;110:9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PD, Ryan S, Gahan CG, Hill C. Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl Environ Microbiol. 2005;71:2832–2839. doi: 10.1128/AEM.71.6.2832-2839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinberg I, Shteinberg T, Gorovitz B, Aharonowitz Y, Cohen G, Borovok I. The Streptomyces NrdR transcriptional regulator is a Zn ribbon/ATP cone protein that binds to the promoter regions of class Ia and class II ribonucleotide reductase operons. J Bacteriol. 2006;188:7635–7644. doi: 10.1128/JB.00903-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat Struct Mol Biol. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- Jitrapakdee S, St Maurice M, Rayment I, Cleland WW, Wallace JC, Attwood PV. Structure, mechanism and regulation of pyruvate carboxylase. Biochem J. 2008;413:369–387. doi: 10.1042/BJ20080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitrapakdee S, Wallace JC. Structure, function and regulation of pyruvate carboxylase. Biochem J. 1999;340(Pt 1):1–16. doi: 10.1042/bj3400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Portnoy DA. Characterization of Listeria monocytogenes pathogenesis in a strain expressing perfringolysin O in place of listeriolysin O. Infect Immun. 1994;62:5608–5613. doi: 10.1128/iai.62.12.5608-5613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Mertins S, Stoll R, Schar J, Umesha KR, Luo Q, Muller-Altrock S, Goebel W. Glycerol metabolism and PrfA activity in Listeria monocytogenes. J Bacteriol. 2008;190:5412–5430. doi: 10.1128/JB.00259-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Przybilla K, Stuhler C, Schauer K, Slaghuis J, Fuchs TM, Goebel W. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia D, Merey G, Nakayama S, Zheng Y, Zhou J, Luo Y, Guo M, Roembke BT, Sintim HO. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem Soc Rev. 2012 doi: 10.1039/c2cs35206k. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Mittal M, Sonenshein AL. CcpC-dependent regulation of citB and lmo0847 in Listeria monocytogenes. J Bacteriol. 2006;188:179–190. doi: 10.1128/JB.188.1.179-190.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasso G, Yu LP, Gil D, Lazaro M, Tong L, Valle M. Functional Conformations for Pyruvate Carboxylase during Catalysis Explored by Cryoelectron Microscopy. Structure. 2014 doi: 10.1016/j.str.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JA, Dodsworth JA. Nitrogen regulation in bacteria and archaea. Annual review of microbiology. 2007;61:349–377. doi: 10.1146/annurev.micro.61.080706.093409. [DOI] [PubMed] [Google Scholar]
- Liu S, Bayles DO, Mason TM, Wilkinson BJ. A cold-sensitive Listeria monocytogenes mutant has a transposon insertion in a gene encoding a putative membrane protein and shows altered (p)ppGpp levels. Appl Environ Microbiol. 2006;72:3955–3959. doi: 10.1128/AEM.02607-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Helmann JD. Analysis of the role of Bacillus subtilis sigma(M) in beta-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol. 2012;83:623–639. doi: 10.1111/j.1365-2958.2011.07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehne FM, Gunka K, Eilers H, Herzberg C, Kaever V, Stulke J. Cyclic Di-AMP Homeostasis in Bacillus subtilis: BOTH LACK AND HIGH LEVEL ACCUMULATION OF THE NUCLEOTIDE ARE DETRIMENTAL FOR CELL GROWTH. J Biol Chem. 2013;288:2004–2017. doi: 10.1074/jbc.M112.395491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nature chemical biology. 2013;9:834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011;12:594–601. doi: 10.1038/embor.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi C, Waters EM, Rudkin JK, Schaeffer CR, Lohan AJ, Tong P, Loftus BJ, Pier GB, Fey PD, Massey RC, et al. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 2012;8:e1002626. doi: 10.1371/journal.ppat.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F, See RY, Zhang D, Toh DC, Ji Q, Liang ZX. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem. 2010;285:473–482. doi: 10.1074/jbc.M109.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nature chemical biology. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- Roelofs KG, Wang J, Sintim HO, Lee VT. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc Natl Acad Sci U S A. 2011;108:15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Pereyre S, Archer KA, Burke TP, Hanson B, Lauer P, Portnoy DA. Listeria monocytogenes engineered to activate the Nlrc4 inflammasome are severely attenuated and are poor inducers of protective immunity. Proc Natl Acad Sci U S A. 2011;108:12419–12424. doi: 10.1073/pnas.1019041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Witte CE, Zemansky J, Hanson B, Lauer P, Portnoy DA. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe. 2010;7:412–419. doi: 10.1016/j.chom.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U, Eikmanns BJ. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol Rev. 2005;29:765–794. doi: 10.1016/j.femsre.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Schar J, Stoll R, Schauer K, Loeffler DI, Eylert E, Joseph B, Eisenreich W, Fuchs TM, Goebel W. Pyruvate carboxylase plays a crucial role in carbon metabolism of extra- and intracellularly replicating Listeria monocytogenes. J Bacteriol. 2010;192:1774–1784. doi: 10.1128/JB.01132-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, Liu C, Yu Q, Zhao Y, Xu S, et al. Crystal structure of STING protein reveal basis for recognition of cyclic di-GMP. Nat Struct Mol Biol. 2012;19:725–727. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WM, Pham TH, Lei L, Dou J, Soomro AH, Beatson SA, Dykes GA, Turner MS. High temperature growth induced spontaneous mutation of llmg_1816 (gdpP) results in heat resistance and salt hypersensitivity in Lactococcus lactis. Appl Environ Microbiol. 2012 doi: 10.1128/AEM.02316-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Maurice M, Reinhardt L, Surinya KH, Attwood PV, Wallace JC, Cleland WW, Rayment I. Domain architecture of pyruvate carboxylase, a biotin-dependent multifunctional enzyme. Science. 2007;317:1076–1079. doi: 10.1126/science.1144504. [DOI] [PubMed] [Google Scholar]
- Stoll R, Mertins S, Joseph B, Muller-Altrock S, Goebel W. Modulation of PrfA activity in Listeria monocytogenes upon growth in different culture media. Microbiology. 2008;154:3856–3876. doi: 10.1099/mic.0.2008/018283-0. [DOI] [PubMed] [Google Scholar]
- Sun AN, Camilli A, Portnoy DA. Isolation of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect Immun. 1990;58:3770–3778. doi: 10.1128/iai.58.11.3770-3778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, O’Riordan MX. Branched-chain fatty acids promote Listeria monocytogenes intracellular infection and virulence. Infect Immun. 2010;78:4667–4673. doi: 10.1128/IAI.00546-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundriyal A, Massa C, Samoray D, Zehender F, Sharpe T, Jenal U, Schirmer T. Inherent regulation of EAL domain-catalyzed hydrolysis of second messenger c-di-GMP. J Biol Chem. 2014 doi: 10.1074/jbc.M113.516195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte CE, Whiteley AT, Burke TP, Sauer JD, Portnoy DA, Woodward JJ. Cyclic di-AMP Is Critical for Listeria monocytogenes Growth, Cell Wall Homeostasis, and Establishment of Infection. MBio. 2013;4 doi: 10.1128/mBio.00282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte G, Hartung S, Buttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Tong L. Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction. Nat Struct Mol Biol. 2008;15:295–302. doi: 10.1038/nsmb.1393. [DOI] [PubMed] [Google Scholar]
- Yan D, Ikeda TP, Shauger AE, Kustu S. Glutamate is required to maintain the steady-state potassium pool in Salmonella typhimurium. Proc Natl Acad Sci U S A. 1996;93:6527–6531. doi: 10.1073/pnas.93.13.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Bai Y, Zhang Y, Gabrielle VD, Jin L, Bai G. Deletion of the cyclic di-AMP phosphodiesterase gene (cnpB) in Mycobacterium tuberculosis leads to reduced virulence in a mouse model of infection. Mol Microbiol. 2014 doi: 10.1111/mmi.12641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, TIan Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu LP, Xiang S, Lasso G, Gil D, Valle M, Tong L. A symmetrical tetramer for S. aureus pyruvate carboxylase in complex with coenzyme A. Structure. 2009;17:823–832. doi: 10.1016/j.str.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, He ZG. Radiation-sensitive gene A (RadA) targets DisA, DNA integrity scanning protein A, to negatively affect cyclic Di-AMP synthesis activity in Mycobacterium smegmatis. J Biol Chem. 2013;288:22426–22436. doi: 10.1074/jbc.M113.464883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li W, He ZG. DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J Biol Chem. 2013;288:3085–3096. doi: 10.1074/jbc.M112.428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.