Abstract
The capture and utilization of light is an exquisitely evolved process. The single-component microbial opsins, although more limited than multicomponent cascades in processing, display unparalleled compactness and speed. Recent advances in understanding microbial opsins have been driven by molecular engineering for optogenetics and by comparative genomics. Here we provide a Primer on these light-activated ion channels and pumps, describe a group of opsins bridging prior categories, and explore the convergence of molecular engineering and genomic discovery for the utilization and understanding of these remarkable molecular machines.
Introduction
Diverse and elegant mechanisms have evolved to enable organisms to harvest light for a variety of survival functions, including energy generation and the identification of suitable environments. A major class of light-sensitive protein consists of 7-transmembrane (TM) rhodopsins that can be found across all kingdoms of life and serve a diverse range of functions (Figure 1). Many prokaryotes employ these proteins to control proton gradients and to maintain membrane potential and ionic homeostasis, and many motile microorganisms have evolved opsin-based photoreceptors to modulate flagellar beating or flagellar motor rotation and thereby direct phototaxis toward environments with optimal light intensities for photosynthesis.
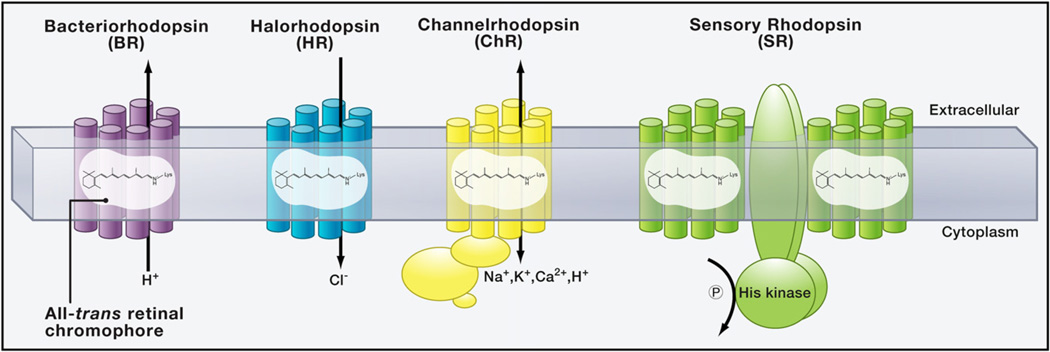
Figure 1. Type I Microbial Rhodopsins.
BR (and PRs) pump protons from the cytoplasm to the extracellular medium, and HRs pump chloride into the cytoplasm; all three hyperpolarize the cell. SRs lack TM ion transport in the presence of the His kinase transducer protein Htr; and algal ChRs conduct cations across the membrane in both directions but always along the electrochemical gradient of the transported ions. In SRs and ChRs, proton translocation within the protein is linked to efficient photocycle progression, but these protons are not necessarily exchanged between the intra- and extracellular spaces.
Owing to their structural simplicity (both light-sensing and effector domains are encoded within a single gene) and fast kinetics, microbial opsins can be treated as precise and modular photosensitization components for introduction into non-light-sensitive cells to enable rapid optical control of specific cellular processes. In recent years, the development of cellular perturbation tools based on these and other light-sensitive proteins has resulted in a technology called optogenetics (Deisseroth, 2011; Deisseroth et al., 2006), which refers to the integration of genetic and optical control to achieve gain or loss of function of precisely defined events within specified cells of living tissue. Details of practical application for neuroscience have been recently summarized elsewhere (Yizhar et al., 2011a; Zhang et al., 2010).
The experimental potential of optogenetics has triggered a surge of genome prospecting and molecular engineering to expand the repertoire of tools and generate new classes of functionality, all of which have catalyzed further mechanistic studies of microbial proteins such as channelrhodopsins (ChRs). Here we provide a Primer on the structural and functional diversity of the microbial opsins, introduce an array of new sequences that inform mechanistic understanding of function, and explore resulting inferences into the principles of operation of this widespread and remarkably evolved class of proteins.
Fundamentals
Each opsin protein requires the incorporation of retinal, a vitamin A-related organic photon-absorbing cofactor, to enable light sensitivity; this opsin-retinal complex is referred to as rhodopsin. The retinal molecule is covalently fixed in the binding pocket within the 7-TM helices and forms a protonated retinal Schiff base (RSBH+; Figure 2A) with a conserved lysine residue located on TM helix seven (TM7). The ionic environment of the RSBH+, heavily influenced by the residues lining the binding pocket, dictates the spectral characteristics of each individual protein; upon absorption of a photon, the retinal chromophore isomerizes and triggers a series of structural changes leading to ion transport, channel opening, or interaction with signaling transducer proteins (discussed below).
Figure 2. Photoreaction Mechanism.
(A) Light-mediated isomerization of the retinal Schiff base (RSB). Top: retinal in the all-trans state, as found in the dark-adapted state of microbial rhodopsins and in the light-activated forms of type II rhodopsins of higher eukaryotes. The absorption of a photon converts the retinal from the all-trans to the 11-cis configuration. Bottom: 11-cis retinal is found only in type II rhodopsins, where it binds to the opsin in the dark state before isomerizing to the all-trans position after photon absorption.
(B) The photocycle of BR is initiated from the dark state where photon absorption activates a sequence of photochemical reactions and structural changes represented by the indicated photointermediates. Also shown is the configuration of the RSB in each step (in red) and the wavelength at which each intermediate maximally absorbs light (in blue).
(C) Summary of proton transport reactions during the BR photocycle. Photon absorption (1) initiates the conformational switch in the RSB, leading to transfer of a proton to Asp85 (2), release of a proton from the proton release complex (PRC, 3), reprotonation of the RSB by Asp96 (4), uptake of a proton from the cytoplasm to reprotonate Asp96 (5), and the reprotonation of the PRC from Asp85 (6), followed by a final proton transfer from D85 to R82 (7).
(D) Light-induced switching of dipole orientation in response to photon absorption in BR, ChR, and HR. In BR and the ChRs, the configuration switch triggers the transfer of the RSB proton to Asp85/Glu123 (for BR/ChR2, respectively). In HR, dipole switching facilitates the transfer of a Cl− ion from the cavity formed between the RSB and Thr143 to a Cl− binding site cytoplasmic to the RSB, enabling the key transport steps of these transporters. Curved arrows indicate isomerization (top row) and ion movement (bottom row).
Opsin genes are divided into two distinct superfamilies: microbial opsins (type I) and animal opsins (type II) (Spudich et al., 2000). Although both opsin families encode 7-TM structures (Luecke et al., 1999; Palczewski et al., 2000), sequence homology between the two families is practically nonexistent; homology within families, however, is high (25%–80% sequence similarity; Man et al., 2003). Type I opsin genes are found in prokaryotes, algae, and fungi and control diverse functions including phototaxis, energy storage, development, and retinal biosynthesis (Spudich, 2006). Type I opsins utilize the all-trans isomer of retinal, which isomerizes to the 13-cis configuration upon photon absorption (Figure 2A, top). The activated retinal molecule in type I rhodopsins remains associated with its opsin protein partner and thermally reverts to the all-trans state while maintaining a covalent bond to its protein partner (Haupts et al., 1997). This reversible reaction occurs rapidly and is critical for allowing microbial rhodopsins to modulate neuronal activity at high frequencies when used as optogenetic tools (Boyden et al., 2005; Zhang et al., 2006, 2007a; Ishizuka et al., 2006); fortuitously, mammalian brains, and indeed the vertebrate tissues thus far examined, contain sufficient levels of retinal so that additional retinal does not need to be supplemented to achieve optical control (Zhang et al., 2006).
In contrast, type II opsin genes are present only in higher eukaryotes and are mainly responsible for vision (Sakmar, 2002). A small fraction of type II opsins also play roles in circadian rhythm and pigment regulation (Sakmar, 2002; Shichida and Yamashita, 2003). Type II opsins primarily function as G protein-coupled receptors (GPCRs) and appear to all use the 11-cis isomer of retinal (or derivatives) for photon absorption (Figure 2A, bottom). Upon illumination, 11-cis retinal isomerizes into the all-trans configuration and initiates protein-protein interactions (not ion flux) that trigger the visual phototransduction second messenger cascade. Unlike the situation in type I rhodopsins, here the retinal dissociates from its opsin partner after isomerization into the all-trans configuration, and a new 11-cis retinal must be recruited. Due to these chromophore turnover reactions and the requirement for interaction with downstream biochemical signal transduction partners, type II opsins effect cellular changes with slower kinetics compared to type I opsins.
The power of using microbial opsins to modulate neuronal electrical activity has also stimulated strong interest in using light to control biochemical events in cells. Although not the focus here, it is worth noting that structure-function work in type II vertebrate opsins from many laboratories (such as Kim et al., 2005) inspired the design of synthetic opsins for controlling specific biochemical events in freely moving mammals. By replacing the intracellular loops of bovine rhodopsin with the intracellular loops from GPCRs, an expanding family of synthetic rhodopsins called optoXRs has enabled optical control of Gs, Gq, or Gi signaling in neuronal settings (Airan et al., 2009; Oh et al., 2010). It is also noteworthy that several groups have expanded control of intracellular signaling by engineering non-opsin-based light-regulated proteins to modulate general second messengers such as cAMP and cGMP (Schröder-Lang et al., 2007). For example, the photoactivated adenylyl cyclases (PACs) can be so employed and use the ubiquitous FAD as a cofactor for photoactivation, although early efforts using PACs from Euglena gracilis were hampered by a combination of high levels of basal activity in the dark, poor protein solubility, and large (∼3 kbp) transgene size (Schröder-Lang et al., 2007). More recently, a smaller PAC derivative with lower dark activity from the soil bacterium Beggiatoa has been shown in neurons and Drosophila to alter membrane currents and influence behavior (Stierl et al., 2011). Yet the microbial opsins remain remarkable for both (1) unitary encoding of light sensation and final effector capability by a single compact gene and (2) virtually zero dark activity, along with millisecond-scale response to well-tolerated wavelengths and intensities of light. These core properties have provided a foundation for, and motivated, further investigation and engineering.
Light-Activated Ion Pumps: Bacteriorhodopsin, Proteorhodopsin, and Halorhodopsin
Bacteriorhodopsin (BR) was first described as a single-component TM protein capable of translocating protons from the intra-cellular to the extracellular space (Oesterhelt and Stoeckenius, 1971). Haloarchaea express BR at high levels under low-oxygen conditions to maintain a proton gradient across the cellular membrane to drive ATP synthesis and maintain cellular energetics in the absence of respiration (Michel and Oesterhelt, 1976; Racker and Stoeckenius, 1974). During the proton translocation process, BR undergoes a cascade of photointermediate states, and each state can be identified by a distinct spectral signature (Lanyi, 2004).
Photon absorption by BR first initiates the isomerization of the bound retinal from the all-trans to the 13-cis configuration (Figures 2A and 2B), thereby triggering a series of proton-transfer reactions that constitute the proton translocation mechanism (Figure 2C). This proton transport process, like chloride transport in halorhodopsins, is elegantly evolved to be (necessarily) spatially discontinuous to prevent passive back-diffusion of the ion down the gradient. Internal proton translocation begins when retinal isomerization triggers a conformational change in the protein and shifts the dipole of the RSBH+. This dipole shift raises the pKa of the RSB, thereby resulting in the release of the proton to its nearby acceptor D85 (Figure 2D), and proton movement triggers additional changes in the protein. In the BR pump, the proton is released to the extracellular milieu via a proton release site defined by two surface glutamates. The RSB then indirectly absorbs a second proton from the cytoplasm, such that the photocycle can repeat with absorption of another photon. In sensory rhodopsins (SRs) the internal proton transfer and subsequent structural changes trigger conformational changes in the transducer molecule (Htr) interacting with the rhodopsin. Certain aspects of the internal proton translocation process are conserved across many type I opsins; for example, locations of the carboxylate Schiff base proton acceptor and donor on the third TM helix are conserved across type I proton pumps.
BR-type proton-pumping opsins have been found across other kingdoms of life; for example, proteorhodopsins (PRs) have been found in marine proteobacteria with photocycles similar to that of BR (Váró et al., 2003). Because marine PRs share a high degree of sequence similarity across species and have action spectra that are tuned according to the ocean depth and latitude of their origin, several groups have explored genomic approaches to understand opsin spectral tuning (Man et al., 2003). Interestingly, absorption variance between blue and green wavelengths can depend on a single amino acid residue (Béjà et al., 2001; Man et al., 2003), but attempts to transfer mutations conferring spectral tuning from PR to other microbial opsins have met with limited success (Yoshitsugu et al., 2009). More extensive high-resolution crystal structures and molecular dynamics/molecular modeling of PRs, fungal opsins, and BRs may provide an opportunity to deepen understanding and extend functionality.
A BR-type proton pump called Archaerhodopsin-3 (initially identified by Ihara et al., 1999) has been shown to allow detection of voltage transients in neurons through generation of a voltage-dependent optical signal (Kralj et al., 2011); although it remains to be seen whether this functionality will be of utility in vivo, this class of experiment represents a potentially interesting value for the microbial opsins in neuroscience. The same protein (Archaerhodopsin-3) is also capable of generating hyperpolarizing currents that can be used to inhibit neural activity (Chow et al., 2010), as with other BR-type proton pumps (and indeed BR itself, optimized for mammalian expression; Gradinaru et al., 2010); however, the efflux of protons elicited by all of these proton pumps under typical steady-illumination experimental conditions will result in decreased extracellular pH. A distinct class of outward current-generating archaeal opsins known as halorhodopsins (HRs) (Matsuno-Yagi and Mukohata, 1977) instead use chloride as the charge carrier. HRs control gradients across the cell membrane by transporting chloride ions from the extracellular medium into the cell (Bamberg et al., 1984; Schobert and Lanyi, 1982). The primary photocycle, although qualitatively similar to that of BR, does not show RSBH+ deprotonation (Essen, 2002; Oesterhelt et al., 1985) due to a single amino acid substitution of the Asp acceptor with Thr. Therefore after the light-induced retinal isomerization and RSBH+ dipole switch, the proton cannot be released due to the absence of an appropriate acceptor. Instead, a Cl− ion already present in the HR protein is transported from the external side of the RSBH+ chromophore to the internal side (Figure 2D) and is subsequently released into the intracellular space (Kolbe et al., 2000).
An experimental screen (Zhang et al., 2007b) revealed that the best known HR (from Halobacterium salinarum) failed to maintain stable photocurrents when expressed heterologously, whereas the HR from the less halophilic Egyptian Natronomonas pharaonis (NpHR) (initially identified by Lanyi et al., 1990; Scharf and Engelhard, 1994) was capable of blocking animal (C. elegans) behavior by hyperpolarizing neurons with electrogenic inward Cl− currents. Pump desensitization is modest, allowing stable, step-like currents over many tens of minutes in response to steady yellow light, but due to the stoichiometry of only one transported ion per photocycle (true for all light-driven pumps), robust expression and fast photocycles are required. Increased heterologous membrane expression can be achieved with addition of trafficking signals from mammalian membrane proteins (Gradinaru et al., 2008), and ultimately this version allowed the first optogenetic inhibition of behavior in mammals (Witten et al., 2010; reviewed in Yizhar et al., 2011a). Moreover, by significantly increasing the number of HR molecules on the neuronal membrane, NpHR (in this case, eNpHR3.0)-expressing neurons can even be inhibited by 680 nm far-red light, which is far from the action spectrum peak (Gradinaru et al., 2010).
Additional retinal-binding proteins have been identified from Halobacterium salinarum as behaviorally relevant photosensors (Hildebrand and Dencher, 1975; Takahashi et al., 1985), such as sensory rhodopsins SRI (Bogomolni and Spudich, 1982; Hildebrand and Dencher, 1975; Takahashi et al., 1985) and SRII, initially termed phoborhodopsin (Tomioka et al., 1986) or P480 (Marwan and Oesterhelt, 1987). The photocycle of SRs is similar to that of BR with analogous internal proton movements (Spudich, 1998; Spudich and Bogomolni, 1984), except that light-initiated conformational changes of the opsin are used to activate a closely associated transducer molecule Htr (Figure 1D) (Büldt et al., 1998; Chen and Spudich, 2002). When activated, Htr initiates a phosphorylation cascade that controls the directionality of the flagellar motor and directs phototaxis toward green and yellow light (SRI, peak absorption 587 nm) and away from blue light (SRII, peak absorption 487 nm) (Spudich, 2006; Spudich and Bogomolni, 1984). Given that prokaryotic kinase cascades are fundamentally different from eukaryotic second messengers (Scharf, 2010), opportunities to translate SR function to heterologous systems may be more complicated than with the ion pumps; such an effort could require reconstitution of the entire signal transduction cascade.
Light-Gated Ion Channels
ChRs are 7-TM proteins capable of conducting passive nonselective cation flow across the cellular membrane upon illumination and, in algae, also mediate intracellular signaling via a long C-terminal extension (Figure 1); for optogenetic applications, only the 7-TM ChR fragments are used, but in the native environment, the intracellular signaling activates a limited number of secondary ion channels that are seen as photocurrents at low light. In contrast, at high light intensities, the intrinsic conductances seen as short-delay currents are dominant and contribute up to 80% of the total current (Berthold et al., 2008; Ehlenbeck et al., 2002; Sineshchekov and Govorunova, 1999; Sineshche-kov et al., 2002). A structural alignment of the 7-TM fragment (based solely on homology models) is shown in Figure 3A (BR and ChR2); note the polar residues that may contribute to the conducting pore. The recently discovered ChR1 of Mesostigma viride lacks two of the five anionic residues of this group, which might explain the small currents of this ChR in human embryonic kidney cells (Govorunova et al., 2011).
Figure 3. Structural and Functional Homology between BR and ChR.
(A) Homology model-based structural alignment of ChR showing the 7-TM helices, next to the BR structural representation. For ChR2, the sequence of the illustrated residues may create a polar environment for water molecules and cation permeation. In BR, R82 functions as a connector between counterion and proton release complex, and D85 is the counterion to which the RSB proton is transferred. To emphasize the spatial discontinuity involved in pumping, only the proton transfer steps after photon absorption and proton transfer to D85 are shown.
(B) Simplified model for the photocycle of ChRs. The D470 dark state is converted by a light-induced isomerization of retinal via the early intermediate P500 and the transient P390 intermediate to the conducting-state P520. The recovery of the D470 dark state proceeds either thermally via the nonconducting P480 intermediate or photochemically via possible short-lived intermediates (green arrow). The late or desensitized P480 state can also be activated (blue arrow) to yield the early intermediate P500. Additional parallel cycles may be present (Yizhar et al., 2011b).
(C) Sample photocurrents show the key kinetic properties that govern function, including inactivation (inact), deactivation (deact), and recovery (rec).
(D) Homology near the retinal-binding pocket between BR and ChR2. The BR retinal-binding pocket is shown based on structure 1KGB (Facciotti et al., 2001) with key amino acids that are involved in the proton transfer reaction. The ChR residues are shown based on sequence homology in the relevant positions.
The first known and described ChR, channelrhodopsin-1 (ChR1), was identified as a light-gated ion channel in Chlamydomonas reinhardtii, a green unicellular alga from temperate freshwater environments (Nagel et al., 2002). Although originally considered proton selective (Nagel et al., 2002), later studies have found that ChR1 has broad cation conductance, including for Na+, K+, and even Ca2+ ions (Lin et al., 2009; Tsunoda and Hegemann, 2009). A second ChR, channelrhodopsin-2 (ChR2), was later characterized from the same organism (Figure 3B). Similar to ChR1, ChR2 also conducts cations (Nagel et al., 2003; Tsunoda and Hegemann, 2009), and both ChRs exhibit fast on and off kinetics. When introduced into neurons, ChRs can insert into the plasma membrane and mediate membrane potential changes in response to blue light (Boyden et al., 2005; Ishizuka et al., 2006; Li et al., 2005). Although ChR2 expresses at higher levels than ChR1 in mammalian systems, a chimera of ChR1 with a ChR from another algal species, Volvox carteri (VChR1, described below), contains no ChR2 sequence elements yet generates significantly greater photocurrents than ChR2 (Yizhar et al., 2011b).
Both ChR1 and ChR2 bear sequence similarity to BR and other type I opsins, with strong homology in residues corresponding to the retinal-binding pocket and proton-conducting network (Hegemann et al., 2001; Nagel et al., 2002, 2003; Sineshchekov et al., 2002; Suzuki et al., 2003), suggesting that ChRs may share partially related ion conduction mechanisms with other microbial opsins. Indeed, the photocycle of ChR2 (Figure 3B) (Yizhar et al., 2011b) is similar to that of BR except with different spectral characteristics, likely due to variations in the ionic environment near the retinal Schiff base, larger conformational changes within the protein (Radu et al., 2009; Ritter et al., 2008), and possibly higher water content within the proton network. In ChR2, a dark-adapted state absorbing at 470 nm (D470) converts rapidly upon illumination to the conducting state P520, via the shortlived photointermediates P500 and P390. Illumination of the open channel at this step with green light terminates the photocurrent (Bamann et al., 2008; Berndt et al., 2009) by photochemically shifting the channel back into a closed state, which may be the dark-adapted state D470 or the light-adapted state P480 (Stehfest and Hegemann, 2010), effectively resetting the photocycle. This photocycle-shortcut pathway may be relevant only at very high light intensities with wild-type ChR2 but acquires a special utility with certain molecularly engineered mutants known as step-function opsin genes (SFOs) with mutation in C128 (Berndt et al., 2009), which can essentially eliminate the inactivation and deactivation photocycle processes (Figures 3B–3D) and create bistable photocurrents.
Indeed, as with the light-activated ion pumps, molecular engineering and genomic strategies have begun to bear fruit for enhancing function of the light-activated ion channels and for understanding the mechanistic differences between rhodopsins that function as active pumps and passive channels. Molecular engineering has largely depended on molecular models derived from the available 3D structures of other microbial rhodopsins such as BR, HR, and SRII. The TM3 of ChRs contains residues important (Figure 3D) for controlling channel gating and the lifetime of the conducting state P520 (residues E123 and C128) and ion selectivity and competition (residues L132 and H134). The roles these residues play in ChR function are discussed in detail next.
Mechanistic Considerations
The most fundamental biophysical properties that influence the performance of opsins at the single-molecule level are efficiency of light absorption, which is dependent on both extinction coefficient (εmax typically between 50,000 and 70,000 M−1 cm−1) and quantum efficiency (Φ, typically between 0.3 and 0.7), and the turnover time of the photocycle. The latter is a critical parameter both for native function and for neuroscience application. For most light-driven pumps (HR and BR), the photocycle turnover time is ∼10–20 ms, thereby capping the temporal precision at tens of milliseconds. Other opsin pumps such as blue PRs have much slower turnover time (∼80–100 ms) (Wang et al., 2003) and therefore have limited applications for neuroscience. It is important to note that for most transporters, these values are determined at neutral membrane potential (0 mV), but the photocycle turnover time can be dramatically slower at hyperpolarized membrane potentials, extending from ∼10–20 ms to ∼100–400 ms (Geibel et al., 2001). Thus reductions in membrane potential slow down the pump, and more light is needed to achieve the same amount of hyperpolarization at more negative membrane potentials. In general, pump direction remains unchanged under all physiologically relevant conditions; the reported “pump inversion” phenomenon observed in PR is due to a leakiness of the pump under extremely negative voltages plus strong pH gradients (Lörinczi et al., 2009).
Unlike opsin-derived pumps, the kinetic parameters related to ChRs are determined by the efficiency of light absorption and the lifetime of the resulting conducting state, given that ion transport (down the gradients) is coupled to occupancy of the conducting state. The TM3 of ChRs contains key residues governing channel gating and the lifetime of the conducting state. The H134R point mutation in ChR2 (Figure 3D), homologous to the proton donor Asp96 in BR, increases the sodium conductance of ChR2 by ∼2-fold; however temporal precision is significantly reduced due to slower deactivation kinetics (Gradinaru et al., 2007; Nagel et al., 2005). Another variant, ChR2-L132C (CatCH), has been shown to exhibit 1.6-fold more Ca2+ influx as well as reduced inactivation, which result together in 3-fold higher Ca2+ influx compared with wild-type ChR2 (Kleinlogel et al., 2011). And as noted above, modification of the C128 residue (C128S, C128A, C128T) in ChR2, alone or in combination with D156 (Berndt et al., 2009; Bamann et al., 2010; Yizhar et al., 2011b) (Figure 3D), extends the lifetime of the open state by several hundred- or even several thousand-fold and enables long-acting (step function-like) inward current in response to single pulses of light. This property leads to useful bistable behavior in the form of sustained subthreshold activation of neurons lasting up to many minutes (Berndt et al., 2009). Moreover, these variants in principle can achieve maximal current magnitudes similar to those in wild-type ChR2 but using much lower levels of light (Berndt et al., 2009; Schoenenberger et al., 2009; Yizhar et al., 2011b).
In addition to conducting-state lifetime, the unitary conductance of individual ChR molecules also affects performance. The single-channel conductance of ChR2 is estimated to be in the femtosiemens range; for order-of-magnitude estimation, in Chlamydomonas the integrated current in response to a half-saturating flash is carried by 1 × 106 ions (Harz et al., 1992), and with 10,000–120,000 ChRs per eyespot (Harz et al., 1992; Berthold et al., 2008; Govorunova et al., 2004) this would correspond to 10–100 ions per ChR and a single-channel conductance between 30 and 300 fS (Harz et al., 1992), a range that has been confirmed for ChR2 by noise analysis (Feldbauer et al., 2009; Lin et al., 2009). In this order-of-magnitude estimation we do not consider minority components from secondarily induced ionic currents that could be recruited, either in the native alga or in neurons under heterologous expression. We also note that it is not yet clear whether any ChR variants, identified by genomics or created by molecular engineering, have altered unitary conductance magnitude.
In addition to these single-molecule or intrinsic factors, population or extrinsic factors will also affect the functionality of microbial opsins, either in the native environment or when expressed in heterologous systems. Indeed, in order to achieve the highest levels of rhodopsin performance for experimental use, transcription, translation, folding, trafficking, and membrane targeting need to be optimized through a combination of codon optimization and addition of trafficking and targeting signals from the heterologous cellular host. TM helix shuffling may also prove influential; chimeras of ChR containing a combination of TMs from ChR1 and ChR2 (C1C2) (Lin et al., 2009; Tsunoda and Hegemann, 2009; Wang et al., 2009) or ChR1 and VChR1 (C1V1) (Yizhar et al., 2011a, 2011b) show reduced inactivation (as with ChR1) and also improved expression in mammalian cells.
In some cases, mutagenesis to achieve improved heterologous expression performance has also illuminated basic mechanisms of ChR function. For example, wild-type ChR2 exhibits a deactivation time constant after light-off of ∼10 ms and therefore can be used to drive neuronal firing at frequencies up to 40 to 50 Hz; however, achieving higher frequencies of neuronal firing requires faster formation and decay of the conducting state (P520 photocycle intermediate). Removal of the putative proton acceptor in the E123 position (“ChETA” mutation E123T or E123A; Figure 3D), at the residue that is homologous to D85 in BR, leads to more rapid channel closing and enables ultrafast optical control of spiking (at least up to 200 Hz; Gunaydin et al., 2010). The ChETA photocurrent is slightly smaller than wild-type ChR2 due to shorter open time, but this effect can be compensated for with more intense or longer flashes (for instance 2 ms instead of 1 ms) (Gunaydin et al., 2010). The E123A mutant therefore demonstrates that this residue, normally considered to be the proton acceptor, is not needed for any proton or other ion flux that would be an essential aspect of ChR function.
Outlook: Genome Mining and Molecular Engineering
It has been extraordinarily difficult to fundamentally alter ion selectivity or to generate useful action spectrum peak shifts of more than 10–20 nm (for ChRs that remain highly functional) by molecular engineering alone. Such efforts have generally failed, despite motivation to experimentally shift the action spectra of ChRs in order to achieve combinatorial (multichannel) optical control, and despite clear understanding that the distribution of partial negative charges on either end of the all-trans retinal chromophore polyene will contribute to setting the wavelength of photons effectively absorbed.
However, using genomic analysis, in 2008 two new ChRs were reported from the colonial alga Volvox carteri, and one (VChR1) was found to absorb at markedly red-shifted wavelengths (λmax = 540 nm; spiking could be driven even at 589 nm in hippocampal neurons) (Zhang et al., 2008). The expression level of VChR1 was found to be significantly lower than ChR2 in most host cells, although a slower decay of the open state partially compensated for the lower levels of protein expression (Zhang et al., 2008). The next few years witnessed steady improvement in VChR1, including TM domain shuffling, membrane-trafficking enhancement, and point mutations guided by structural models, culminating in the C1V1 family of ChRs (Yizhar et al., 2011b). C1V1 tools contain no ChR2 sequence but express more potent photocurrents than the original ChR2 and allow control of spiking with red light (e.g., 630 nm) and combinatorial excitation even in behaving mammals (Yizhar et al., 2011b).
The difficulty facing molecular engineering efforts highlights the value of crystal-structural information to allow more principled and accurate tuning (and understanding) of opsin properties and also underscores the value of genomic methods. Just as genome prospecting played an instrumental role in the identification of the new ChRs from Volvox carteri, genomic approaches could in principle be used to identify entirely new classes of opsins. Moreover, comparative analysis of opsins from the full range of ecological diversity could shed light on the fundamental mechanisms of ion selectivity, spectral tuning, and photocycle dynamics. To that end, we here report the identification of a panel of new ChRs from the genomes of the algae species Pleodorina starrii, Pyramimonas gelidicola, and Dunaliella salina; report that the D. salina ChR falls into a unique functional class (pure proton channel); and explore the theoretical and practical implications (Figures 4 and 5; Table 1).
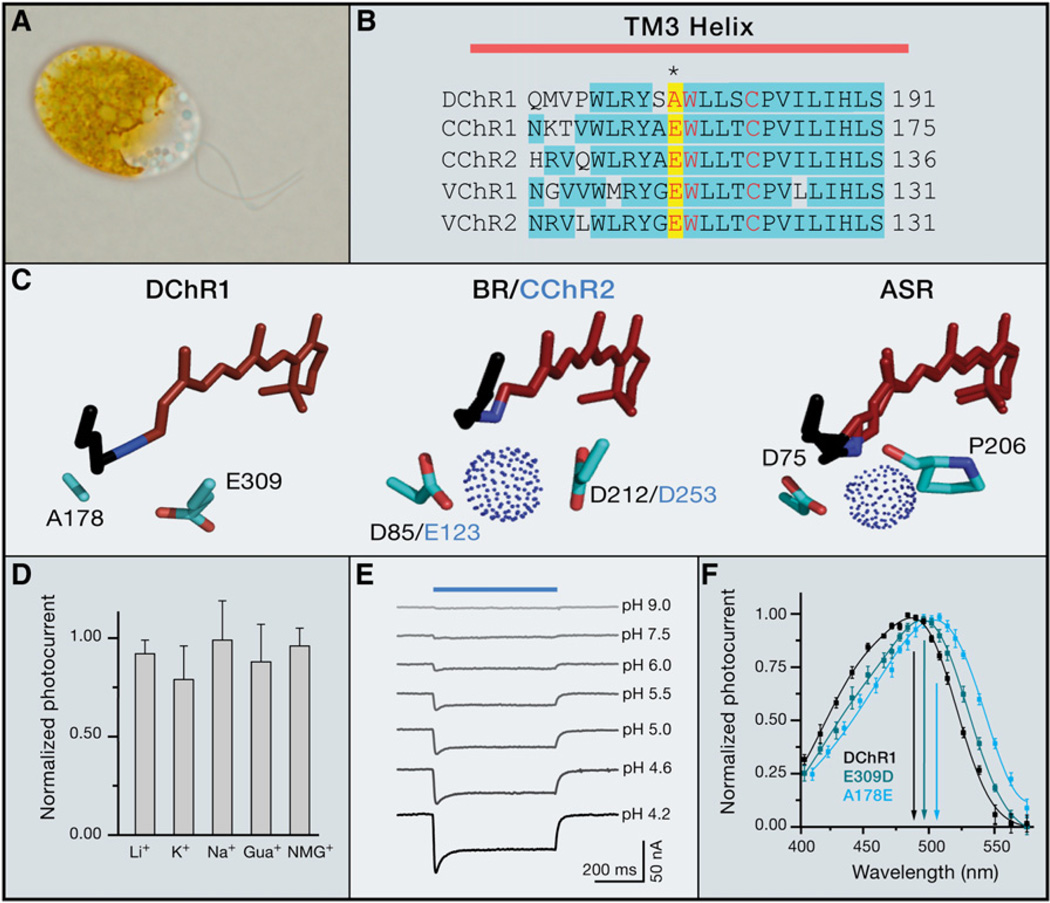
Figure 4. Characterization of a ChR from Dunaliella salina.
(A) The halophilic unicellular alga Dunaliella salina.
(B) Sequence homology between the algal ChRs and BR within the third TM helix. The typically conserved E123 position has been replaced with an Ala in DChR1 (and is shown on a yellow background), conserved residues are shown on a blue background, and amino acids likely interacting with the chromophore are shown in red.
(C) Lack of a proton acceptor in DChR1 (A178), compared with BR (D85) and Chlamydomonas ChR2 (CChR2; E123). ASR (Anabaena sensory rhodopsin) has been crystallized with a mixture of all-trans and 13-cis retinal seen as an overlay (Vogeley et al., 2004).
(D) DChR1 photocurrents are unaffected by changes in the extracellular cation composition (sole cation present in each condition shown on category x axis). Cation exchange was performed in 5 mM Mops-NMG, 0.1 mM MgCl2 with 100 mM LiCl, KCl, NaCl, guanidium chloride, or NMG chloride (pH 7.5). We used a human codonadapted DChR sequence (amino acid residues 1–339) as a template for capped RNA synthesis by T7 RNA polymerase (mMessage mMachine, Ambion). Oocyte preparation and injection of capped RNA were carried out as described previously (Berthold et al., 2008), and two-electrode voltage clamp was performed with a Turbo Tec-05 (NPI Electronic) or a GeneClamp 500 (Molecular Devices) amplifier on an oocyte after 3–7 days of the capped RNA injection. Continuous light was provided by a75-W Xenon lamp (Jena Instruments) and delivered to the oocytes via a 3 mm light guide. The light passed through a 500 ± 25 nm broadband filter (Balzers) with an intensity of 46 mW/cm2.
(E) In contrast, DChR1 photocurrent is highly sensitive to changes in the pH environment. Solutions contained 100 mM NMG chloride, 0.1 mM MgCl2, 0.1 mM CaCl2 with 5 mM glycine (pH 9.0), 5 mM Mops-NMG (pH 7.5), 5 mM citrate (pH 6, 5.5, 5.0, 4.6, 4.2).
(F) Introduction or alteration of a proton acceptor (A178E or E309D) in the DChR1 retinal-binding pocket causes a pronounced red-shift in the absorption spectrum. We applied 10 ns laser flashes as described previously (Berthold et al., 2008); solutions for action spectra recording contained 100 mM NaCl, 0.1 mM MgCl2, 0.1 mM CaCl2, and 5 mM citrate (pH 4.2).
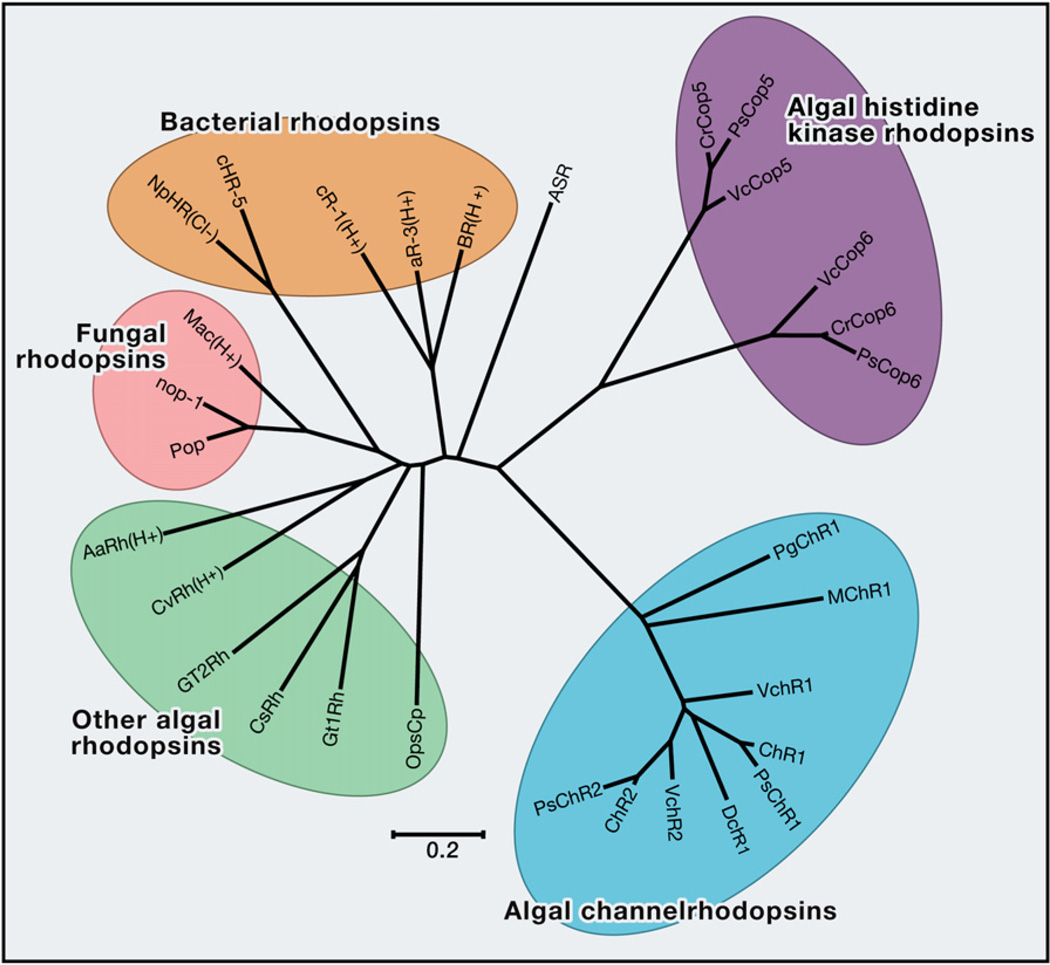
Figure 5. Phylogenetic Analysis of Microbial Opsins.
Phylogenetic tree of the microbial opsins from algae, bacteria, and fungi. The tree was constructed by the neighbor-joining method based on amino acid sequences using MEGA5 (Tamura et l., 2011). The scale bar indicates the number of substitutions per site. H+ and Cl− indicate proton and chloride pumping capability, respectively. Detailed opsin information is listed in Table 1. Sequences are provided in the Supplemental Information (“Sequences”).
Table 1.
Phylogenetics of Microbial Opsins
| Abbreviation | Name or Type | Species | Accession Number | Reference |
|---|---|---|---|---|
| Channelrhodopsin | ||||
| ChR1 | channelrhodopsin-1 | Chlamydomonas reinhardtii | AF385748 | (Nagel et al., 2002) |
| ChR2 | channelrhodopsin-2 | Chlamydomonas reinhardtii | EF474017; AAM15777 | (Nagel et al., 2003) |
| VChR1 | channelrhodopsin-1 | Volvox carteri | EU622855 | (Zhang et al., 2008) |
| VChR2 | channelrhodopsin-2 | Volvox carteri | ABZ90903 | (Kianianmomeni et al., 2009) |
| MChR1 | channelrhodopsin-1 | Mesostigma viride | JF922293 | (Govorunova et al., 2011) |
| DChR1 | channelrhodopsin-1 | Dunaliella salina | JQ241364 | present report |
| PsChR1 | channelrhodopsin-1 | Pleodorina starrii | JQ249903 | present report |
| PsChR2 | channelrhodopsin-2 | Pleodorina starrii | JQ249904 | present report |
| PgChR1 | channelrhodopsin-1 | Pyramimonas gelidicola | JQ241366 | present report |
| Proton Pump | ||||
| AaRh | rhodopsin |
Acetabularia acetabulum |
AAY82897 | (Tsunoda et al., 2006) |
| CvRh | rhodopsin | Chlorella vulgaris | JQ255360 | present report |
| Gt1Rh | rhodopsin | Guillardia theta | ABA08437 | (Sineshchekov et al., 2005) |
| GtR3 | rhodopsin | Guillardia theta | JQ241365 | (Gradinaru et al., 2010) |
| Pop | bacteriorhodopsin-like |
Podospora Anserine S mat+ |
XP_001904282 | (Espagne et al., 2008) |
| Mac | L. Maculans rhodopsin | Leptosphaeria maculans | AAG01180 | (Idnurm and Howlett, 2001) |
| aR-3/Arch | archaerhodopsin-3 | Halorubrum sodomense | BAA09452 | (Ihara et al., 1999) |
| cR-1 | cruxrhodopsin-1 | Haloarcula argentinensis | BAA06678 | (Tateno et al., 1994) |
| BR | bacteriorhodopsin | Halobacterium salinarum | CAA23744 | (Dunn et al., 1981) |
| Cl− Pump | ||||
| NpHR | halorhodopsin |
Natronomonas pharaonis |
EF474018 | (Lanyi et al., 1990) |
| cHR-5 | halorhodopsin |
Haloarcula marismortui ATCC 43049 |
AAV46572 | (Baliga et al., 2004) |
| Histidine Kinase Rhodopsin | ||||
| ASR | Anabaena sensory rhodopsin | Anabaena sp. | 1XIO_A | (Vogeley et al., 2004) |
| CrCop5 | chlamyopsin-5 | Chlamydomonas reinhardtii | AAQ16277 | (Kateriya et al., 2004) |
| CrCop6 | chlamyopsin-6 | Chlamydomonas reinhardtii | XP_001698789 | (Kateriya et al., 2004) |
| VcCop5 | volvoxopsin-5 | Volvox carteri | XP_002954798 | (Prochnik et al., 2010) |
| VcCop6 | volvoxopsin-6 | Volvox carteri | XP_002957065 | (Prochnik et al., 2010) |
| PsCop5 | pleopsin-5 | Pleodorina starri | JQ249905 | present report |
| PsCop6 | pleopsin-6 | Pleodorina starri | JQ249906 | present report |
| Gt2Rh | rhodopsin-2 | Guillardia theta | ABA08438 | (Sineshchekov et al., 2005) |
| Other | ||||
| CsRh | rhodopsin | Cryptomonas sp. S2 | ABA08439 | (Sineshchekov et al., 2005) |
| OpsCp | rhodopsin | Cyanophora paradoxa | ACV05065 | (Frassanito et al., 2010) |
| nop-1 | rhodopsin-1 | Neurospora crassa OR74A | XP_959421 | (Bieszke et al., 1999) |
Typically found in hyper-saline environments such as evaporation salt fields, the unicellular (oval with two flagella) green alga Dunaliella salina is salt tolerant. Despite belonging to the same order as the green algae Chlamydomonas reinhardtii and Volvox carteri, Dunaliella can appear reddish due to the accumulation of high levels of carotenoid molecules (Figure 4A). We hypothesized that a Dunaliella ChR might have unusual properties and engaged in efforts to clone ChRs from this flagellated algal species. Although we were only able to identify one opsin gene (DChR1) with homology to ChRs and VChRs from Dunaliella expressed sequence tags (ESTs), future work based on full genomic information may reveal additional DChRs.
Despite high homology with other known ChRs, the DChR1 sequence contained several notable features (Figure 4B). First, one of the residues that is thought to contribute to the complex counterion of the RSB, E123 in ChR2 as discussed above, is replaced by Ala in the DChR1 TM3 (Figures 4B and 4C); from structural modeling (Figure 4C), we expect that the counterion function is assumed by E309 in DChR1, a position that plays only a minor role in BR (D212) or Anabaena sensory rhodopsin (ASR) (Vogeley et al., 2004). Even more remarkably, DChR1 photocurrents are exclusively carried by protons, unlike any other known ChR, and were completely unaffected by changes in the extracellular cation composition (Figure 4D). Consequently, the photocurrent was highly sensitive to changes in the pH environment and completely vanished at high pH (Figure 4E).
Full understanding of structure-function relationships will require high-resolution crystal structures in multiple photocycle states. However, directed mutagenesis studies here demonstrate that DChR1 has a different counterion arrangement and ion selectivity compared to other known ChRs. The strict H+ selectivity of DChR1 was not mediated by the unusual RSBH counterion, as substitution of A178 with the more typical putative counterion Glu as found in ChR2 only red-shifted the activation spectrum (Figure 4F, from 475 to 510 nm) with minimal effect on current amplitude or kinetics. Similarly, replacing E309 with Asp caused a slight spectral shift and a slight current increase, whereas replacing the charged E309 by Ala rendered the protein almost totally inactive (Figure 4F).
Given typical electrochemical proton gradients, the DChR1 H+ current is opposite in direction to the H+ current generated by BR pump activity; therefore, DChR1 and BR could enable interventions such as bidirectional control of cellular pH, for example in manipulating the pH of intracellular compartments (mitochondria and synaptic vesicles). DChR1 therefore defines a new class of microbial opsin—a light-activated proton channel—unlike any other microbial opsin including ChR1 and ChR2. These findings illustrate the diversity of function likely to be present within the vast array of microbial opsin genomes.
To represent this diversity, in Figure 5 and Table 1 we have collected known microbial opsin sequences along with key properties and phylogenetic relationships and constructed a phylogenetic tree to display the primary sequence relationships among these distinct opsin genes. Although the development and application of microbial opsins as optogenetic tools has already profoundly improved our ability to perturb and understand biological systems, much remains to be learned regarding mechanisms by which these proteins respond to different light stimuli and select distinct ions for transport.
Indeed, as shown here, the microbial genomes continue to surprise and enrich us with fundamentally new optogenetic tools and capabilities. Continued investigation of this ecological diversity (Figure 5) and the functional phylogenetics of microbial opsins will enable the discovery and engineering of new variants with further wavelength-shifted absorption spectra and ion selectivity. Further rapid expansion of the optogenetic toolkit will in turn spur increased understanding of structure-function relationships and mechanisms of these ancient and powerful molecular machines.
Supplementary Material
ACKNOWLEDGMENTS
Full funding information for K.D. is listed at http://www.optogenetics.org/funding and includes the Gatsby Charitable Foundation and the Keck, Snyder, Woo, and Yu Foundations, as well as CIRM, NIMH, NINDS, NIDA, and the DARPA REPAIR program. F.Z. is supported by the NIH TR01, Robert Metcalfe, Michael Boylan, and the McKnight, Gates, Simons, and Damon Runyon Foundations. L.E.F. is supported by the NIH MSTP program. P.H. is supported by the DFG. We thank the entire Deisseroth laboratory for their support and Dr. Duc Tran for assistance. Genome and transcriptome sequencing and assembly were performed by the Department of Energy (DOE) Joint Genome Institute and supported by the DOE’s Office of Science under Contract No. DE-AC02-05CH11231.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information contains sequences and can be found with this article online at doi:10.1016/j.cell.2011.12.004.
REFERENCES
- Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- Baliga NS, Bonneau R, Facciotti MT, Pan M, Glusman G, Deutsch EW, Shannon P, Chiu Y, Weng RS, Gan RR, et al. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 2004;14:2221–2234. doi: 10.1101/gr.2700304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamann C, Kirsch T, Nagel G, Bamberg E. Spectral characteristics of the photocycle of channelrhodopsin-2 and its implication for channel function. J. Mol. Biol. 2008;375:686–694. doi: 10.1016/j.jmb.2007.10.072. [DOI] [PubMed] [Google Scholar]
- Bamann C, Gueta R, Kleinlogel S, Nagel G, Bamberg E. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry. 2010;49:267–278. doi: 10.1021/bi901634p. [DOI] [PubMed] [Google Scholar]
- Bamberg E, Hegemann P, Oesterhelt D. Reconstitution of halorhodopsin in black lipid membranes. Prog. Clin. Biol. Res. 1984;164:73–79. [PubMed] [Google Scholar]
- Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat. Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- Berthold P, Tsunoda SP, Ernst OP, Mages W, Gradmann D, Hegemann P. Channelrhodopsin-1 initiates phototaxis and photo-phobic responses in chlamydomonas by immediate light-induced depolarization. Plant Cell. 2008;20:1665–1677. doi: 10.1105/tpc.108.057919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieszke JA, Braun EL, Bean LE, Kang S, Natvig DO, Borkovich KA. The nop-1 gene of Neurospora crassa encodes a seven trans-membrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc. Natl. Acad. Sci. USA. 1999;96:8034–8039. doi: 10.1073/pnas.96.14.8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogomolni RA, Spudich JL. Identification of a third rhodopsin-like pigment in phototactic Halobacterium halobium. Proc. Natl. Acad. Sci. USA. 1982;79:6250–6254. doi: 10.1073/pnas.79.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Büldt G, Heberle J, Dencher NA, Sass HJ. Structure, dynamics, and function of bacteriorhodopsin. J. Protein Chem. 1998;17:536–538. [PubMed] [Google Scholar]
- Chen X, Spudich JL. Demonstration of 2:2 stoichiometry in the functional SRI-HtrI signaling complex in Halobacterium membranes by gene fusion analysis. Biochemistry. 2002;41:3891–3896. doi: 10.1021/bi015966h. [DOI] [PubMed] [Google Scholar]
- Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, Henninger MA, Belfort GM, Lin Y, Monahan PE, Boyden ES. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K. Optogenetics. Nat. Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Feng G, Majewska AK, Miesenböck G, Ting A, Schnitzer MJ. Next-generation optical technologies for illuminating genetically targeted brain circuits. J. Neurosci. 2006;26:10380–10386. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn R, McCoy J, Simsek M, Majumdar A, Chang SH, Rajbhandary UL, Khorana HG. The bacteriorhodopsin gene. Proc. Natl. Acad. Sci. USA. 1981;78:6744–6748. doi: 10.1073/pnas.78.11.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlenbeck S, Gradmann D, Braun FJ, Hegemann P. Evidence for a light-induced H(+) conductance in the eye of the green alga Chlamy-domonas reinhardtii. Biophys. J. 2002;82:740–751. doi: 10.1016/S0006-3495(02)75436-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espagne E, Lespinet O, Malagnac F, Da Silva C, Jaillon O, Porcel BM, Couloux A, Aury JM, Ségurens B, Poulain J, et al. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 2008;9:R77. doi: 10.1186/gb-2008-9-5-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essen LO. Halorhodopsin: light-driven ion pumping made simple? Curr. Opin. Struct. Biol. 2002;12:516–522. doi: 10.1016/s0959-440x(02)00356-1. [DOI] [PubMed] [Google Scholar]
- Facciotti MT, Rouhani S, Burkard FT, Betancourt FM, Downing KH, Rose RB, McDermott G, Glaeser RM. Structure of an early intermediate in the M-state phase of the bacteriorhodopsin photocycle. Biophys. J. 2001;81:3442–3455. doi: 10.1016/S0006-3495(01)75976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldbauer K, Zimmermann D, Pintschovius V, Spitz J, Bamann C, Bamberg E. Channelrhodopsin-2 is a leaky proton pump. Proc. Natl. Acad. Sci. USA. 2009;106:12317–12322. doi: 10.1073/pnas.0905852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frassanito AM, Barsanti L, Passarelli V, Evangelista V, Gualtieri P. A rhodopsin-like protein in Cyanophora paradoxa: gene sequence and protein immunolocalization. Cell. Mol. Life Sci. 2010;67:965–971. doi: 10.1007/s00018-009-0225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geibel S, Friedrich T, Ormos P, Wood PG, Nagel G, Bamberg E. The voltage-dependent proton pumping in bacteriorhodopsin is characterized by optoelectric behavior. Biophys. J. 2001;81:2059–2068. doi: 10.1016/S0006-3495(01)75855-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova EG, Jung KH, Sineshchekov OA, Spudich JL. Chlamydomonas sensory rhodopsins A and B: cellular content and role in photophobic responses. Biophys. J. 2004;86:2342–2349. doi: 10.1016/S0006-3495(04)74291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorunova EG, Spudich EN, Lane CE, Sineshchekov OA, Spudich JL. New channelrhodopsin with a red-shifted spectrum and rapid kinetics from Mesostigma viride. MBio. 2011;2:e00115–e11. doi: 10.1128/mBio.00115-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Thompson KR, Zhang F, Mogri M, Kay K, Schneider MB, Deisseroth K. Targeting and readout strategies for fast optical neural control in vitro and in vivo. J. Neurosci. 2007;27:14231–14238. doi: 10.1523/JNEUROSCI.3578-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 2008;36:129–139. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Zhang F, Ramakrishnan C, Mattis J, Prakash R, Diester I, Goshen I, Thompson KR, Deisseroth K. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Yizhar O, Berndt A, Sohal VS, Deisseroth K, Hegemann P. Ultrafast optogenetic control. Nat. Neurosci. 2010;13:387–392. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- Harz H, Nonnengasser C, Hegemann P. The photoreceptor current of the green alga chlamydomonas. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1992;338:39–52. [Google Scholar]
- Haupts U, Tittor J, Bamberg E, Oesterhelt D. General concept for ion translocation by halobacterial retinal proteins: the isomerization/switch/ transfer (IST) model. Biochemistry. 1997;36:2–7. doi: 10.1021/bi962014g. [DOI] [PubMed] [Google Scholar]
- Hegemann P, Fuhrmann M, Kateriya S. Algal sensory photoreceptors. J. Phycol. 2001;37:668–676. [Google Scholar]
- Hildebrand E, Dencher N. Two photosystems controlling behavioural responses of Halobacterium halobium. Nature. 1975;257:46–48. doi: 10.1038/257046a0. [DOI] [PubMed] [Google Scholar]
- Idnurm A, Howlett BJ. Characterization of an opsin gene from the ascomycete Leptosphaeria maculans. Genome. 2001;44:167–171. doi: 10.1139/g00-113. [DOI] [PubMed] [Google Scholar]
- Ihara K, Umemura T, Katagiri I, Kitajima-Ihara T, Sugiyama Y, Kimura Y, Mukohata Y. Evolution of the archaeal rhodopsins: evolution rate changes by gene duplication and functional differentiation. J. Mol. Biol. 1999;285:163–174. doi: 10.1006/jmbi.1998.2286. [DOI] [PubMed] [Google Scholar]
- Ishizuka T, Kakuda M, Araki R, Yawo H. Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci. Res. 2006;54:85–94. doi: 10.1016/j.neures.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Kateriya S, Nagel G, Bamberg E, Hegemann P. “Vision” in single-celled algae. News Physiol. Sci. 2004;19:133–137. doi: 10.1152/nips.01517.2004. [DOI] [PubMed] [Google Scholar]
- Kianianmomeni A, Stehfest K, Nematollahi G, Hegemann P, Hallmann A. Channelrhodopsins of Volvox carteri are photochromic proteins that are specifically expressed in somatic cells under control of light, temperature, and the sex inducer. Plant Physiol. 2009;151:347–366. doi: 10.1104/pp.109.143297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Hwa J, Garriga P, Reeves PJ, RajBhandary UL, Khorana HG. Light-driven activation of beta 2-adrenergic receptor signaling by a chimeric rhodopsin containing the beta 2-adrenergic receptor cytoplasmic loops. Biochemistry. 2005;44:2284–2292. doi: 10.1021/bi048328i. [DOI] [PubMed] [Google Scholar]
- Kleinlogel S, Feldbauer K, Dempski RE, Fotis H, Wood PG, Bamann C, Bamberg E. Ultra light-sensitive and fast neuronal activation with the Ca2+-permeable channelrhodopsin CatCh. Nat. Neurosci. 2011;14:513–518. doi: 10.1038/nn.2776. [DOI] [PubMed] [Google Scholar]
- Kolbe M, Besir H, Essen LO, Oesterhelt D. Structure of the light-driven chloride pump halorhodopsin at 1.8 A resolution. Science. 2000;288:1390–1396. doi: 10.1126/science.288.5470.1390. [DOI] [PubMed] [Google Scholar]
- Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. [Published online November 27, 2011];Nat. Methods. 2011 doi: 10.1038/nmeth.1782. 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyi JK. Bacteriorhodopsin. Annu. Rev. Physiol. 2004;66:665–688. doi: 10.1146/annurev.physiol.66.032102.150049. [DOI] [PubMed] [Google Scholar]
- Lanyi JK, Duschl A, Hatfield GW, May K, Oesterhelt D. The primary structure of a halorhodopsin from Natronobacterium pharaonis. Structural, functional and evolutionary implications for bacterial rhodopsins and halorhodopsins. J. Biol. Chem. 1990;265:1253–1260. [PubMed] [Google Scholar]
- Li X, Gutierrez DV, Hanson MG, Han J, Mark MD, Chiel H, Hegemann P, Landmesser LT, Herlitze S. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. USA. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JY, Lin MZ, Steinbach P, Tsien RY. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys. J. 2009;96:1803–1814. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lörinczi E, Verhoefen MK, Wachtveitl J, Woerner AC, Glaubitz C, Engelhard M, Bamberg E, Friedrich T. Voltage- and pH-depen-dent changes in vectoriality of photocurrents mediated by wild-type and mutant proteorhodopsins upon expression in Xenopus oocytes. J. Mol. Biol. 2009;393:320–341. doi: 10.1016/j.jmb.2009.07.055. [DOI] [PubMed] [Google Scholar]
- Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 A resolution. J. Mol. Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- Man D, Wang W, Sabehi G, Aravind L, Post AF, Massana R, Spudich EN, Spudich JL, Béjà O. Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 2003;22:1725–1731. doi: 10.1093/emboj/cdg183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marwan W, Oesterhelt D. Signal formation in the halobacterial photophobic response mediated by a fourth retinal protein (P480) J. Mol. Biol. 1987;195:333–342. doi: 10.1016/0022-2836(87)90654-1. [DOI] [PubMed] [Google Scholar]
- Matsuno-Yagi A, Mukohata Y. Two possible rolesofbacteriorhodopsin; a comparative study of strains of Halobacterium halobium differing in pigmentation. Biochem. Biophys. Res. Commun. 1977;78:237–243. doi: 10.1016/0006-291x(77)91245-1. [DOI] [PubMed] [Google Scholar]
- Michel H, Oesterhelt D. Light-induced changes of the pH gradient and the membrane potential in H. halobium. FEBS Lett. 1976;65:175–178. doi: 10.1016/0014-5793(76)80473-5. [DOI] [PubMed] [Google Scholar]
- Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, Hegemann P. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G, Brauner M, Liewald JF, Adeishvili N, Bamberg E, Gott-schalk A. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr. Biol. 2005;15:2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D, Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat. New Biol. 1971;233:149–152. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D, Hegemann P, Tittor J. The photocycle of the chloride pump halorhodopsin. II: Quantum yields and a kinetic model. EMBO J. 1985;4:2351–2356. doi: 10.1002/j.1460-2075.1985.tb03938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Maejima T, Liu C, Deneris E, Herlitze S. Substitution of 5-HT1A receptor signaling by a light-activated G protein-coupled receptor. J. Biol. Chem. 2010;285:30825–30836. doi: 10.1074/jbc.M110.147298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329:223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racker E, Stoeckenius W. Reconstitution of purple membrane vesicles catalyzing light-driven proton uptake and adenosine triphosphate formation. J. Biol. Chem. 1974;249:662–663. [PubMed] [Google Scholar]
- Radu I, Bamann C, Nack M, Nagel G, Bamberg E, Heberle J. Conformational changes of channelrhodopsin-2. J. Am. Chem. Soc. 2009;131:7313–7319. doi: 10.1021/ja8084274. [DOI] [PubMed] [Google Scholar]
- Ritter E, Stehfest K, Berndt A, Hegemann P, Bartl FJ. Monitoring light-induced structural changes of Channelrhodopsin-2 by UV-visible and Fourier transform infrared spectroscopy. J. Biol. Chem. 2008;283:35033–35041. doi: 10.1074/jbc.M806353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakmar TP. Structure of rhodopsin and the superfamily of seven-helical receptors: the same and not the same. Curr. Opin. Cell Biol. 2002;14:189–195. doi: 10.1016/s0955-0674(02)00306-x. [DOI] [PubMed] [Google Scholar]
- Scharf B, Engelhard M. Blue halorhodopsin from Natronobacterium pharaonis: wavelength regulation by anions. Biochemistry. 1994;33:6387–6393. doi: 10.1021/bi00187a002. [DOI] [PubMed] [Google Scholar]
- Scharf BE. Summary of useful methods for two-component system research. Curr. Opin. Microbiol. 2010;13:246–252. doi: 10.1016/j.mib.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Schobert B, Lanyi JK. Halorhodopsin is a light-driven chloride pump. J. Biol. Chem. 1982;257:10306–10313. [PubMed] [Google Scholar]
- Schoenenberger P, Gerosa D, Oertner TG. Temporal control of immediate early gene induction by light. PLoS ONE. 2009;4:e8185. doi: 10.1371/journal.pone.0008185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder-Lang S, Schwärzel M, Seifert R, Strünker T, Kateriya S, Looser J, Watanabe M, Kaupp UB, Hegemann P, Nagel G. Fast manipulation of cellular cAMP level by light in vivo. Nat. Methods. 2007;4:39–42. doi: 10.1038/nmeth975. [DOI] [PubMed] [Google Scholar]
- Shichida Y, Yamashita T. Diversity of visual pigments from the viewpoint of G protein activation—comparison with other G protein-coupled receptors. Photochem. Photobiol. Sci. 2003;2:1237–1246. doi: 10.1039/b300434a. [DOI] [PubMed] [Google Scholar]
- Sineshchekov OA, Govorunova EG. Rhodopsin-mediated photosensing in green flagellated algae. Trends Plant Sci. 1999;4:58–63. doi: 10.1016/s1360-1385(98)01370-3. [DOI] [PubMed] [Google Scholar]
- Sineshchekov OA, Jung KH, Spudich JL. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 2002;99:8689–8694. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sineshchekov OA, Govorunova EG, Jung KH, Zauner S, Maier UG, Spudich JL. Rhodopsin-mediated photoreception in cryptophyte flagellates. Biophys. J. 2005;89:4310–4319. doi: 10.1529/biophysj.105.070920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JL. Variations on a molecular switch: transport and sensory signalling by archaeal rhodopsins. Mol. Microbiol. 1998;28:1051–1058. doi: 10.1046/j.1365-2958.1998.00859.x. [DOI] [PubMed] [Google Scholar]
- Spudich JL. The multitalented microbial sensory rhodopsins. Trends Microbiol. 2006;14:480–487. doi: 10.1016/j.tim.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Spudich JL, Bogomolni RA. Mechanism of colour discrimination by a bacterial sensory rhodopsin. Nature. 1984;312:509–513. doi: 10.1038/312509a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JL, Yang CS, Jung KH, Spudich EN. Retinylidene proteins: structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 2000;16:365–392. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- Stehfest K, Hegemann P. Evolution of the channelrhodopsin photocycle model. ChemPhysChem. 2010;11:1120–1126. doi: 10.1002/cphc.200900980. [DOI] [PubMed] [Google Scholar]
- Stierl M, Stumpf P, Udwari D, Gueta R, Hagedorn R, Losi A, Gärtner W, Petereit L, Efetova M, Schwarzel M, et al. Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J. Biol. Chem. 2011;286:1181–1188. doi: 10.1074/jbc.M110.185496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Yamasaki K, Fujita S, Oda K, Iseki M, Yoshida K, Watanabe M, Daiyasu H, Toh H, Asamizu E, et al. Archaeal-type rhodopsins in Chlamydomonas: model structure and intracellular localization. Biochem. Biophys. Res. Commun. 2003;301:711–717. doi: 10.1016/s0006-291x(02)03079-6. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Mochizuki Y, Kamo N, Kobatake Y. Evidence that the long-lifetime photointermediate of s-rhodopsin is a receptor for negative phototaxis in Halobacterium halobium. Biochem. Biophys. Res. Commun. 1985;127:99–105. doi: 10.1016/s0006-291x(85)80131-5. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno M, Ihara K, Mukohata Y. The novel ion pump rhodopsins from Haloarcula form a family independent from both the bacteriorhodopsin and archaerhodopsin families/tribes. Arch. Biochem. Biophys. 1994;315:127–132. doi: 10.1006/abbi.1994.1480. [DOI] [PubMed] [Google Scholar]
- Tomioka H, Takahashi T, Kamo N, Kobatake Y. Flash spectro-photometric identification of a fourth rhodopsin-like pigment in Halobacterium halobium. Biochem. Biophys. Res. Commun. 1986;139:389–395. doi: 10.1016/s0006-291x(86)80003-1. [DOI] [PubMed] [Google Scholar]
- Tsunoda SP, Hegemann P. Glu 87 of channelrhodopsin-1 causes pH-dependent color tuning and fast photocurrent inactivation. Photochem. Photobiol. 2009;85:564–569. doi: 10.1111/j.1751-1097.2008.00519.x. [DOI] [PubMed] [Google Scholar]
- Tsunoda SP, Ewers D, Gazzarrini S, Moroni A, Gradmann D, Hegemann P. H+ -pumping rhodopsin from the marine alga Acetabularia. Biophys. J. 2006;91:1471–1479. doi: 10.1529/biophysj.106.086421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Váró G, Brown LS, Lakatos M, Lanyi JK. Characterization of the photochemical reaction cycle of proteorhodopsin. Biophys. J. 2003;84:1202–1207. doi: 10.1016/S0006-3495(03)74934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogeley L, Sineshchekov OA, Trivedi VD, Sasaki J, Spudich JL, Luecke H. Anabaena sensory rhodopsin: a photochromic color sensor at 2.0 A. Science. 2004;306:1390–1393. doi: 10.1126/science.1103943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Sugiyama Y, Hikima T, Sugano E, Tomita H, Takahashi T, Ishizuka T, Yawo H. Molecular determinants differentiating photocurrent properties of two channelrhodopsins from chlamydomonas. J. Biol. Chem. 2009;284:5685–5696. doi: 10.1074/jbc.M807632200. [DOI] [PubMed] [Google Scholar]
- Wang WW, Sineshchekov OA, Spudich EN, Spudich JL. Spectroscopic and photochemical characterization of a deep ocean proteorhodopsin. J. Biol. Chem. 2003;278:33985–33991. doi: 10.1074/jbc.M305716200. [DOI] [PubMed] [Google Scholar]
- Witten IB, Lin SC, Brodsky M, Prakash R, Diester I, Anikeeva P, Gradinaru V, Ramakrishnan C, Deisseroth K. Cholinergic interneurons control local circuit activity and cocaine conditioning. Science. 2010;330:1677–1681. doi: 10.1126/science.1193771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011a;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O’Shea DJ, Sohal VS, Goshen I, Finkelstein J, Paz JT, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011b;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitsugu M, Yamada J, Kandori H. Color-changing mutation in the E-F loop of proteorhodopsin. Biochemistry. 2009;48:4324–4330. doi: 10.1021/bi900228a. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang LP, Boyden ES, Deisseroth K. Channelrho-dopsin-2 and optical control of excitable cells. Nat. Methods. 2006;3:785–792. doi: 10.1038/nmeth936. [DOI] [PubMed] [Google Scholar]
- Zhang F, Aravanis AM, Adamantidis A, de Lecea L, Deisseroth K. Circuit-breakers: optical technologies for probing neural signals and systems. Nat. Rev. Neurosci. 2007a;8:577–581. doi: 10.1038/nrn2192. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, Deisseroth K. Multimodal fast optical interrogation of neural circuitry. Nature. 2007b;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- Zhang F, Prigge M, Beyrière F, Tsunoda SP, Mattis J, Yizhar O, Hegemann P, Deisseroth K. Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nat. Neurosci. 2008;11:631–633. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gradinaru V, Adamantidis AR, Durand R, Airan RD, de Lecea L, Deisseroth K. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nat. Protoc. 2010;5:439–456. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.