Abstract
Context
Second-generation antipsychotics (SGAs) are increasingly used in the treatment of many psychotic and non-psychotic disorders. Unfortunately, SGAs are often associated with substantial weight gain, with no means to predict which patients are at greatest risk.
Objective
To detect alleles of single nucleotide polymorphisms (SNPs) associated with antipsychotic drug-induced weight gain.
Design
Pharmacogenetic association study
Setting
Discovery cohort was collected at a U.S. general psychiatric hospital. Three additional cohorts were collected from psychiatric hospitals in the U.S. and Germany, and from a European antipsychotic drug trial.
Participants
The discovery cohort was comprised of 139 pediatric patients undergoing first exposure to SGA treatment. An additional three cohorts were comprised of 73, 40 and 92 subjects.
Intervention
Patients in the discovery cohort were treated with SGAs for twelve weeks. Additional cohorts were treated for six and twelve weeks.
Main outcome measure
We conducted a genome-wide association study (GWAS) assessing weight gain associated with twelve weeks of SGA treatment in patients undergoing first exposure to antipsychotic treatment. We next genotyped three independent cohorts of subjects assessed for antipsychotic drug-induced weight gain.
Results
GWAS yielded twenty SNPs at a single locus exceeding a statistical threshold of p < 10−5. This locus, near the melanocortin 4 receptor (MC4R) gene, overlaps a region previously identified by large-scale GWAS of obesity in the general population. Effects were recessive, with minor allele homozygotes gaining extreme amounts of weight over the 12-week trial. These results were replicated in three additional cohorts with SNP rs489693 demonstrating consistent recessive effects; meta analysis revealed a genome-wide significant effect (p=5.59×10−12). Moreover, we observed consistent effects on related metabolic indices, including triglycerides, leptin, insulin, and HOMA-IR in our discovery cohort.
Conclusion
These data implicate the MC4R locus in extreme SGA-induced weight gain and related metabolic disturbances. A priori identification of high-risk subjects could lead to alternative treatment strategies in this population.
Introduction
Although second-generation antipsychotic drugs (SGAs) are the cornerstone of treatment for many psychotic and non-psychotic disorders, these medications are associated with substantial weight gain, including the development of obesity and other cardiovascular risk factors.1 These medication effects are important mediating factors in the reduction in life expectancy, estimated to reach 20–30 years, in those with chronic and severe mental illnesses.2 Moreover, SGA-induced weight gain frequently leads to medication non-adherence and decreased quality of life, and adequate treatments to prevent or ameliorate weight gain are lacking.3
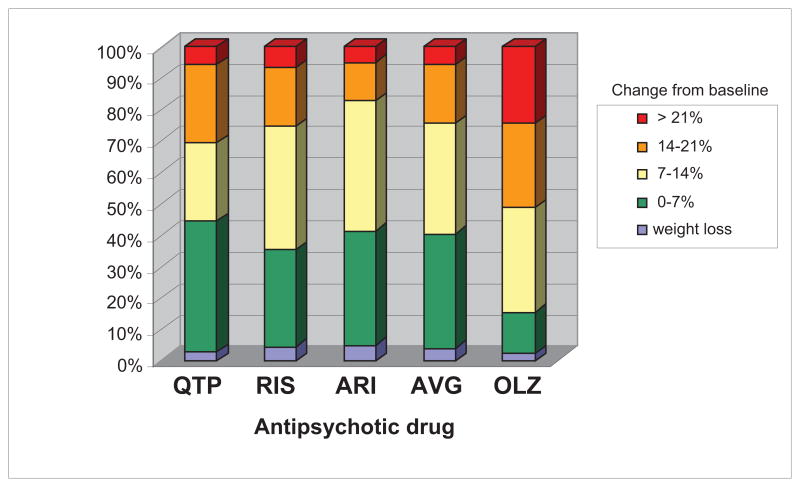
A subgroup of patients experience severe weight gain following exposure to SGAs. In a study4 of the weight and metabolic effects of SGAs in a unique cohort of 272 antipsychotic-naïve pediatric patients beginning initial treatment with one of four SGAs, we found that approximately one-quarter of patients treated with risperidone, quetiapine, or aripiprazole gained more than 14% of their baseline weight, with the top quartile gaining between 15 and 35 pounds, in just 12 weeks of treatment (Figure 1). Olanzapine-treated patients demonstrated a markedly different distribution, with the majority of subjects experiencing extreme weight gain. The amount of weight gain was not related to age, pubertal status, ethnicity, or sex of the subjects, and SGA dosage ranges were relatively restricted. These data are consistent with multiple clinical reports indicating that a significant proportion of patients gain extreme amounts of weight upon exposure to SGAs.5–7
Figure 1.
Distribution of antipsychotic-drug induced weight gain in antipsychotic naïve subjects following 12 weeks of treatment with second-generation antipsychotic drugs. QTP = quetiapine; RIS = risperidone; ARI = aripiprazole; AVG = average of QTP, RIS and ARI; OLZ = olanzapine. The Y-axis represents the percentage of subjects in each of five weight gain categories:e.g.,subjects gaining > 21% of their baseline weight (red); subjects gaining >14% of their baseline weight (orange), etc.
The use of pharmacogenetic approaches to identify patients at risk for severe SGA-induced weight gain could lead to targeted interventions to ameliorate effects in high-risk individuals, as well as provide data on the molecular substrates of SGA-induced weight gain. To date, however, pharmacogenetic studies of weight gain have been restricted by methodological and technological limitations. In particular, prior studies have typically utilized samples of convenience, including patients with varying and often lengthy prior exposure to antipsychotics, considerably confounding prospectively-observed weight gain. Moreover, non-adherence to treatment, a substantial problem in antipsychotic pharmacotherapy,8 can lead to misclassification of non-adherent subjects as low-risk for weight gain. Finally, only a modest number of genetic loci have been examined, with only one study (in chronically-treated adults) utilizing genome-wide association.9
Therefore, we conducted the first GWAS study of SGA-induced weight gain in patients carefully monitored for medication adherence who were undergoing initial exposure to SGAs. To confirm our results, we next assessed three independent replication cohorts: 1) a cohort of adult subjects undergoing first exposure to a single SGA (clozapine); 2) a cohort of adult subjects treated with the same SGAs as in our discovery sample; 3) a cohort of adult subjects in the first episode of schizophrenia enrolled in a randomized clinical trial of antipsychotic drugs.14
Methods
Subjects: Discovery Cohort
Subjects assessed in the GWAS study were enrolled in an observational cohort study assessing the weight and metabolic effects of SGAs in pediatric psychiatric patients. Participants aged 18 to 19 years, and caregivers of all minor participants aged 4 to 17 years, provided written informed consent; minors aged 9 to 17 years signed informed assent to a protocol approved by the Institutional Review Board of the North Shore-Long Island Jewish Health System. Detailed methods have been previously reported.4
Briefly, subjects undergoing initial treatment with SGA were included if: 1) age was ≤19 years, and 2) prior lifetime exposure to all antipsychotics as a class was ≤1 week. Exclusion criteria included: active or past diagnosis of eating disorder; biochemical evidence of thyroid dysfunction; pregnancy or breastfeeding; any acute non-psychiatric medical disorder. Specific antipsychotic drug choice, drug dosage and titration schedule were based upon clinical indications. To ensure adherence with SGA treatment, antipsychotic plasma levels were measured; individuals with undetectable antipsychotic plasma levels were excluded.
Phenotypic Assessments: Discovery Cohort
Subjects were assessed after 8 or more hours of overnight fasting at baseline and weeks 4, 8 and 12 of treatment. Height was measured using the stadiometer Seca 214. Weight, BMI, and fat mass were assessed by impedantiometry with the Tanita Body Composition Analyzer TBF-310. As shown in Figure 1, weight gain profiles after 12 weeks of treatment for three SGAs (quetiapine, n=36; risperidone, n=135; aripiprazole, n=41) were indistinguishable from each other, but significantly differed from olanzapine (n=45). An omnibus chi-square testing the distributions displayed in Figure 1 (merging the lowest two categories) demonstrated a significant effect of drug (χ2=24.68, df=9, p=0.003). When olanzapine was removed, there was no significant difference across the three remaining drugs (χ2=4.42, df=6, p=0.62), and pairwise comparisons demonstrated each of these medications differed in weight gain distributions compared to olanzapine (all p’s <0.05), but did not differ from each other (all p’s>0.40). Consequently, subjects taking olanzapine were excluded from the planned GWAS analysis to maintain homogeneity of phenotype.10
Fasting blood was drawn between 7 and 11 AM, prior to taking morning medications. Plasma levels were obtained at each post baseline visit (weeks 4, 8 and 12). Glucose and lipid levels were analyzed with the Roche Hitachi 747 chemistry analyzer and insulin level was analyzed via Roche Elecsys 2010 immunochemistry analyzer (Roche Diagnostics). Plasma levels were measured with liquid chromatography at the Cooper Laboratory (NKI, Orangeburg, NY).
DNA Collection, Genotyping, and Quality Control (QC): Discovery Cohort
Of 272 individuals reported in Correll et al. (2009),6 245 (90.1%) provided blood samples for DNA extraction. DNA samples were genotyped on ~1M SNPs using the Illumina Omni1-Quad platform. All QC procedures were performed in SVS 7.3.1 GoldenHelix, Inc., Bozeman, MT), except for cryptic identity/relatedness, which was performed in PLINK 17.11 Of 245 available samples, 16 (6.5%) were eliminated during QC due to low call rates (<97%), sex mismatch, cryptic identity, or cryptic relatedness. Elimination of SNPs with low call rate (<95%), low MAF (<2%), or out of Hardy-Weinberg equilibrium (p<E-6) resulted in 803,582 high-quality autosomal SNPs available for analysis. Mean call rate per sample was 99.7%. Of 229 samples passing QC, 38 were prescribed olanzapine and excluded from GWAS, and 10 were excluded due to demonstrated SGA non-adherence. 139 subjects completed the full study, with 12-week BMI change data and high-quality genotype data available for GWAS analysis (Supplemental Tables 1 and 2).
Statistical analysis: Discovery Cohort
Principal components analysis (PCA) was performed using the EIGENSTRAT method12 implemented in SVS 7.3.1 using default settings, and the 10 top principal components were entered into subsequent GWAS analyses. PCA-corrected correlation/trend tests were performed to test dominant, recessive, and additive models, using 12-week change in BMI as the quantitative dependent measure.
Additional cohorts
To validate the GWAS results from the discovery cohort, we identified an additional cohort of subjects undergoing their first exposure to an SGA with assured adherence with medication. Details on this cohort have been published previously. 13 Briefly, this cohort consists of 73 patients without prior exposure to SGAs, beginning initial treatment with the prototypic SGA, clozapine. Patients were aged 18 to 60 years, diagnosed with schizophrenia according to DSM-IIIR criteria, and were either treatment refractory or intolerant to treatment with typical antipsychotics. Exclusion criteria were pregnancy/breastfeeding, organic brain disorder or severe head injuries, previous medical conditions that required treatment and were not stable, substance dependence, mental retardation, and severe personality disorder. Prior to initiating treatment with clozapine, subjects underwent a medication washout period of 7–14 days. Clozapine dosage was titrated based upon clinical indication and treatment continued for at least 6 weeks. Clozapine plasma levels were monitored to ascertain compliance. Patients underwent weight assessment at baseline and at 6 weeks of treatment.
Additionally, a second replication cohort of 40 subjects was collected from the Charite University of Medicine, Berlin, Germany.13 Subjects 18–62 years old were diagnosed with schizophrenia or schizoaffective disorder according to DSM-IV criteria. Hospital admission was either due to first manifestation or relapse of psychosis with significant deterioration. Exclusion criteria were the same as described above. Patients were not excluded if they had undergone previous antipsychotic treatment. Patients underwent 6 weeks of treatment with risperidone, quetiapine or aripiprazole. Antipsychotic drug choice, drug dosage and titration schedule were based upon clinical indications (Table 1). Subjects’ weight was assessed at baseline and following 6 weeks of treatment.
Table 1a.
ASSOCIATION OF TOP CHROMOSOME 18 SNPS WITH ANTIPSYCHOTIC-INDUCED WEIGHT GAIN IN DISCOVERY COHORT GWAS
| rsNumber | Position | Minor allele frequency | p value | ΔBMI
|
||
|---|---|---|---|---|---|---|
| Minor allele homozygotes | Heterozygotes | Major allele homozygotes | ||||
| rs8092668 | 55934091 | 22.66% | 1.30E-06 | 3.939 | 1.856 | 1.568 |
| rs1942879 | 55939970 | 33.81% | 4.63E-06 | 3.201 | 1.696 | 1.519 |
| rs952044 | 55949090 | 33.33% | 5.57E-06 | 3.201 | 1.687 | 1.548 |
| rs66723169 | 55959958 | 19.15% | 5.45E-06 | 3.827 | 1.846 | 1.613 |
| rs129678781 | 55977550 | 19.15% | 3.60E-07 | 4.153 | 1.787 | 1.613 |
| rs65671601 | 55980115 | 22.34% | 8.16E-07 | 4.277 | 1.952 | 1.512 |
| rs4768282 | 56003567 | 28.01% | 3.29E-03 | 2.674 | 1.865 | 1.574 |
| rs619825 | 56010046 | 45.74% | 4.62E-06 | 2.625 | 1.663 | 1.430 |
| rs1942876 | 56022246 | 42.20% | 1.20E-07 | 2.886 | 1.662 | 1.454 |
| rs996022 | 56023341 | 41.84% | 1.32E-07 | 2.925 | 1.667 | 1.454 |
| rs12955983 | 56023969 | 21.58% | 4.09E-06 | 3.760 | 1.900 | 1.560 |
| rs11663816 | 56027207 | 21.99% | 3.17E-06 | 3.760 | 1.864 | 1.567 |
| rs17175602 | 56033697 | 21.63% | 3.17E-06 | 3.760 | 1.898 | 1.554 |
| rs4896931 | 56033767 | 34.40% | 2.80E-07 | 3.333 | 1.797 | 1.382 |
| rs6467491 | 56034105 | 44.33% | 3.09E-07 | 2.884 | 1.624 | 1.417 |
| rs694780 | 56034525 | 44.33% | 3.09E-07 | 2.884 | 1.624 | 1.417 |
| rs12957325 | 56035596 | 21.28% | 3.17E-06 | 3.760 | 1.938 | 1.538 |
| rs129701341 | 56035730 | 21.43% | 3.26E-06 | 3.760 | 1.927 | 1.554 |
| rs11660069 | 56036393 | 21.63% | 3.17E-06 | 3.760 | 1.898 | 1.554 |
| rs603940 | 56036763 | 44.33% | 3.28E-06 | 2.806 | 1.657 | 1.428 |
| rs581401 | 56036944 | 44.33% | 3.28E-06 | 2.806 | 1.657 | 1.428 |
Finally, a third replication cohort of patients treated in their first episode of schizophrenia was collected as part of the European Union First Episode Schizophrenia Trial (EUFEST). (Note that only a subset of patients enrolled in the larger trial provided DNA samples).14 Because there was an insufficient number of non-Caucasian subjects available to conduct meaningful covariates analyses, only Caucasian subjects were included for this report. Patients were excluded if more than 2 years had passed since the onset of positive symptoms or if any antipsychotic drug had been used for more than 2 weeks in the previous year, or for 6 weeks at any time. Patients were randomly assigned to one of 4 antipsychotics: haloperidol, amisulpride, quetiapine, or ziprasidone (as in the discovery cohort, subjects assigned to olanzapine were excluded from the present study). Patients were excluded from pharmacogenetic analyses if non-adherence to medication was reported. Weight was assessed at baseline and after 12 weeks of treatment as part of a longer trial; results are reported for a total n=92 subjects meeting above criteria (Table 1).
Genotyping of Additional Cohorts
Genotyping of the second two cohorts was completed subsequent to analysis of the GWAS results from the discovery cohort and was comprised of 5 SNPs highlighted in Table 1a. As shown in Table 1 and Supplementary Figure 1, the SNPs identified by GWAS of the discovery cohort were in strong linkage disequilibrium, with D′=1 in most instances. Thus there was considerable redundancy which obviated the need to test more SNPs in the replication cohorts. However, there was some difference in minor allele frequencies across the SNPs in Table 1, with three modes (~21%, ~34%, ~44%) as depicted in Table 1b. The 5 SNPs chosen for replication in the first two replication cohorts were selected based upon providing a comprehensive assessment across this frequency distribution. SNPs rs1942786 and rs996022, as well as other SNPs in the region, were not selected because they were either in near complete LD with other selected SNPs, or at very low allele frequencies, or failed in assay development. No other SNPs from Table 1 were successfully genotyped in the replication cohorts.
Table 1b.
ASSOCIATION OF TOP SNPS WITH ANTIPSYCHOTIC-INDUCED WEIGHT GAIN IN THREE COHORTS
| rsNumber | Position | Minor allele frequency | Discovery p value | Replication 1 p value | Replication 2 p value | Replication 3 p value |
|---|---|---|---|---|---|---|
| rs489693 | 56033767 | 34.40% | 2.80E-07 | 0.00014 | 0.007 | .042 |
| rs646749 | 56034105 | 44.33% | 3.09E-07 | 0.00026 | 0.092 | |
| rs12970134 | 56035730 | 21.43% | 3.26E-06 | 0.143 | 0.007 |
SNPs genotyped in additional cohorts. Rs 12967878 and rs6567160 produced an insufficient number of minor allele homozygotes to test recessive effects in the additional cohorts.
Proxy SNP for rs17782313
All genotyping was performed using TaqMan SNP genotyping assays (Applied Biosystems, Foster City, CA). Two independent researchers confirmed calling of genotypes and 10% of the sample was re-genotyped to assure concordance. The concordance rate was 99.5% and discordant genotypes were treated as missing data in the statistical analysis. Samples with >2 missing genotype calls were excluded and are not included in Table 1b.
Finally, genotyping of the EUFEST cohort was performed after all prior analyses had been completed as part of an ongoing study of antipsychotic drug efficacy. Genotyping was performed on the Illumina Omni-1Quad platform with QC parameters as described above. For purposes of the present study, only rs469893 was examined. Rs469893 was selected after analysis of the above replication cohorts revealed that it was significant in both replication cohorts (p values of 0.00014 and 0.007, respectively). No other SNP yielded a p value less than p = 0.05 in all three prior cohorts and were therefore not selected for follow-up.
Statistical Analysis of Additional Cohorts
Based upon the results from the GWAS of the discovery cohort, we tested for recessive effects for each SNP by comparing minor allele homozygotes with all others using t-tests, with change in weight across the 6- or 12-week trial as the dependent measure. Effects of potential confounds, including sex, race, and baseline weight were tested using ANCOVA. Meta-analysis of p-values derived from t-tests was conducted using Stouffer’s z trend test, an extension of Fisher’s method which permits weighting for sample sizes and effect directions, as implemented in MetaP (http://compute1.lsrc.duke.edu/softwares/MetaP/metap.php).
Results
Discovery Cohort
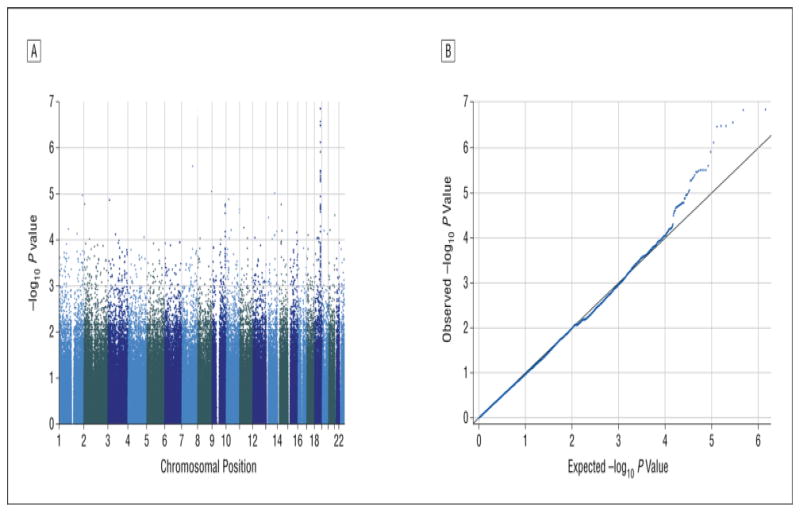
GWAS of the BMI-change phenotype revealed a striking genotypic effect under the recessive model (Figure 2a). Twenty SNPs at a single locus exceeded a statistical threshold of p< E-5 (Table 1a), with no evidence of inflation of test statistics due to population effects (λGenomic Control (GC) = 1; Q–Q plot; Figure 2b). This locus, extending from coordinates 55934091–56036944 on Chromosome 18, is approximately 190kb downstream from the MC4R gene and overlaps the region previously identified by large-scale GWAS studies of obesity and BMI in the general population.15,16 For each of these SNPs, the minor allele homozygotes gained significantly more weight over the 12-week trial than either heterozygotes or common allele homozygotes, which did not differ from each other. Results did not substantially change when drug assignment, sex, or race were entered into a regression model. Importantly, distribution of drug assignment across the minor allele homozygotes did not differ from the proportions in the group as a whole (χ2=0.86, df=2, p=0.65). There was also no significant correlation between baseline BMI and delta BMI (r=−.032; p=.71). Moreover, results did not substantially change when baseline BMI was added to a regression, and there was no significant association between genotype at any of the top GWAS SNPs and baseline BMI (for example, p=.32 for rs489693).
Figure 2. Genome-wide Association Study Results.
a. Manhattan plot displaying statistical significance levels (−log10P-values) of correlation/trend tests for change in BMI in the discovery cohort, plotted by chromosomal position for all autosomal SNPs. Peak values are observed on chromosome 18, between positions 55.934MB and 56.037MB, as detailed in Table 1A.
b. Q–Q plot displaying statistical significance levels (−log10P-values) of correlation/trend tests for change in BMI in the discovery cohort, plotted against expected values under the null hypothesis. With the exception of the most strongly associated SNPs on chromosome 18, there is no deviation from the diagonal (λGC=1.00).
Additional cohorts
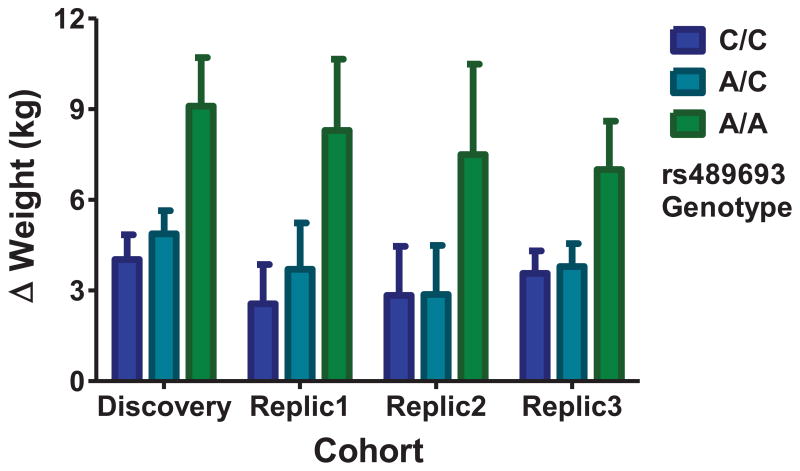
SNPs emerging from the GWAS are in strong linkage disequilibrium (D′=1 for most SNPs, Supplemental Figure 1, upper panel), but vary in minor allele frequency (Table 1a), resulting in variable degrees of r2 (Supplemental Figure 1, lower panel). We genotyped 5 SNPs, representing the various levels of minor allele frequency evident at this locus (Table 1a, highlighted SNPs) in the first two replication cohorts. However, two SNPs (rs6567160 and rs12967878) produced an insufficient number of minor allele homozygotes (n<5) in either cohort to test recessive effects. Results for the remaining 3 SNPs are displayed in Table 1b; r2 values for these three SNPs in the discovery cohort were of moderate strength (rs489693/rs646749, r2=0.66; rs489693/rs12970134, r2=0.52; rs646749/rs12770134, r2=0.34). Of these three SNPs, rs489693 yielded consistent and statistically significant recessive effects across each cohort. Results did not change substantially when race, sex, or baseline weight were added to an ANCOVA model. We next examined rs489693 in a third replication cohort, and again obtained statistically significant recessive effects which remained significant when sex, baseline weight, and study drug were added using ANCOVA. Meta-analysis across all 4 cohorts yielded a strong, genome-wide significant effect (Stouffer’s z trend, p=5.59×10−12). To graphically demonstrate the effects, Figure 3 plots the mean change in weight in each cohort as a function of genotype at rs489693 (percent weight change displayed in supplemental figure 2). Baseline weight and BMI did not significantly differ between rs489693 genotype groups in any of the cohorts.
Figure 3.
rs489693 genotype and antipsychotic-induced weight gain in four cohorts of subjects.
Metabolic indices
Finally, we examined the relationship of rs489693 genotype to SGA-induced changes in metabolic indices in our discovery cohort (Table 2). Minor allele homozygotes had significantly greater increases in triglycerides, leptin, insulin, HOMA-IR and total fat mass compared to the group of heterozygotes and common allele homozygotes. Other measures approached significance (p> 0.05 and <0.10), including changes in total cholesterol and HDL cholesterol.
Table 2.
RS489693 GENOTYPE AND METABOLIC CHANGES IN ANTIPSYCHOTIC NAÏVE SUBJECTS FOLLOWING 12 WEEKS OF TREATMENT WITH SECOND GENERATION ANTIPSYCHOTIC DRUGS.
| Metabolic Index | rs489693 genotype | Mean(SEM) | p value (2-tailed) |
|---|---|---|---|
| Fat mass (kg) | AC/CC AA |
4.87(0.46) 10.03(1.63) |
<0.001 |
| Triglycerides (mg/dl) | AC/CC AA |
7.29 (5.60) 51.67(17.5) |
0.011 |
| Total cholesterol (mg/dl) | AC/CC AA |
3.18(2.25) 15.73(5.73) |
0.066 |
| HDL cholesterol (mg/dl) | AC/CC AA |
0.03(0.81) −2.87(1.18) |
0.052 |
| LDL cholesterol (mg/dl) | AC/CC AA |
1.78(1.75) 7.50(4.70) |
0.292 |
| Glucose (mg/dl) | AC/CC AA |
1.56(0.86) 3.07(2.74) |
0.566 |
| Insulin (ulU/ml) | AC/CC AA |
0.35(0.75) 4.91(1.88) |
0.043 |
| HOMA-IR | AC/CC AA |
0.12(0.17) 1.23(0.49) |
0.033 |
| Leptin (ng/ml) | AC/CC AA |
3.40(0.68) 8.27(3.10) |
0.028 |
Discussion
We conducted the first genome-wide association study of SGA-induced weight gain in an antipsychotic-naïve cohort of pediatric subjects, and identified evidence of recessive effects at multiple SNPs located at chromosome 18q21.32. This peak directly overlaps a region that has been repeatedly identified as a predictor of weight and BMI in healthy individuals (Supplemental Figure 1), and has been implicated in obesity, Type-II diabetes, and related phenotypes.15,16 SNP rs489693 demonstrated statistically significant recessive effects in three additional independent cohorts, with minor allele homozygotes at risk for extreme weight gain following a short duration of treatment in all cohorts, as well as consistent effects on related metabolic indices in our discovery cohort. This locus is approximately 190kb downstream from MC4R, the melanocortin 4 receptor gene, which has previously been identified as a candidate for weight-related phenotypes, as mutations in this gene have been linked to extreme obesity in children and adolescents and MC4R knockout mice develop obesity.17
A major strength of our approach was the assessment of subjects undergoing their first exposure to antipsychotic drug treatment, unlike prior GWAS studies of weight change induced by antipsychotics9. Baseline weight variability due to prior treatment with agents known to induce substantial weight gain was therefore minimized, and provided us with substantially enhanced power to detect the effects of genetic variation on a complex weight regulation phenotype. Moreover, the use of antipsychotic plasma levels to ensure medication compliance reduced phenotypic variation due to the nuisance (non-genetic) effects of medication non-adherence, thereby enhancing the strength of the genotype-phenotype relationships. This effect may be particularly important in psychotic disorders, in which non-compliance with treatment is estimated to occur in 40% or more of patients.
While sample size in the discovery cohort was small in comparison to GWAS studies of complex disease entities and quantitative traits in the general population, GWAS of pharmacogenetic phenotypes have, in some instances, demonstrated extremely robust effects in small samples.18,19 While our initial GWAS result did not meet conventional thresholds for genome-wide significance, the possibility that our result is a false positive is substantially reduced by three factors: 1) the convergence of results across four independent cohorts, resulting in a meta-analytic p-value several orders of magnitude beyond genome-wide thresholds; 2) the inherent biological plausibility of MC4R for weight gain, and 3) the high prior probability for association to this genomic region based on numerous previous GWAS of obesity and related phenotypes in the general population.15,16,20–26 Similarly, while our discovery sample included subjects from multiple ethnic groups, the likelihood that results are artifacts of population stratification is greatly reduced by several factors: 1) PCA-correction of the GWAS analysis resulted in no evidence of population stratification (λGC=1.00); 2) results were replicated in an ethnically homogeneous German sample; 3) the overlapping obesity locus from general population GWAS studies has been replicated in African-ancestry populations.27,28
It should be noted, however, that the GWAS signal for antipsychotic-induced weight gain is not precisely the same as that identified in general population studies. First, our genotypic effects followed a recessive pattern (Figure 3); heterozygotes did not substantially differ from common allele homozygotes. Moreover, no SNPs on any chromosome exceeded a statistical threshold of p<10−06 for analyses testing the dominant or additive models in our cohort. By contrast, GWAS effects reported in the general population literature are additive, with heterozygotes intermediate to the two homozygous groups. Second, while this genomic region is marked by considerable linkage disequilibrium with multiple SNPs achieving nominal associations in both our GWAS and general population studies of obesity, specific SNP effects differ. For example, a proxy for the strongest additive correlate of general population obesity was not amongst the top 20 recessive SNPs in our cohort, although it was nominally significant (see Table 1). Similarly, the SNP in our study (rs489693) has not emerged as the most strongly associated SNP in general population studies, except for a single study of waist circumference.20 Further research with larger samples will be needed to test for multiple, independent allelic effects at this locus, as has been reported in a recent study of obesity.29
Our results may inform the design of GWAS studies seeking to identify risk alleles for complex phenotypes, such as obesity, that are mediated by a plethora of genetic and environmental factors. In GWAS studies of weight, samples sizes in the thousands were necessary to achieve statistically significant results, presumably because of the vast numbers of unmeasured (and uncontrolled) environmental factors working over variable amounts of time to influence the ultimate weight phenotype. In the current study, the critical environmental factor predisposing individuals to weight gain was antipsychotic drug administration over a short period of time. The experimental control of this one factor provided us with sufficient environmental homogeneity to detect genome-wide significant results in a study of slightly more than one hundred, rather than thousands, of subjects. Future studies of complex phenotypes may benefit from consideration of pharmacological or other environmental “challenge” paradigms for the detection of susceptibility alleles.
These data have potential clinical implications. For example, a priori identification of those subjects at increased risk of severe weight gain could lead to alternative treatments other than SGAs – particularly in patients not suffering from an Axis I psychotic disorder. Of note, recent data from the 2007 National Ambulatory Medical Care survey30 indicate that antipsychotic drugs (most commonly quetiapine and risperidone) were prescribed in 21.3% of patient visits for anxiety disorders, with the largest increase in new patient visits, despite the fact that there is little evidence for these drugs’s efficacy in anxiety. Therefore, it might be plausible to consider pharmacotherapeutic strategies that would not include antipsychotic drugs in those non-psychotic individuals who carry the high risk genotype for weight gain, as well as increased behavioral and psychosocial interventions focused on dietary and exercise habits. Finally, research on the co-administration of MC4R agonists, of which several are being developed,31 in this subset of patients could be informative for the development of ameliorative strategies.
Supplementary Material
Acknowledgments
Each of the authors contributed to the design and execution of the study. A.K.M., C.C., N.C., T.L. and J.K. were responsible for the primary drafting of the manuscript. D.J.M., P.G., A.L., A.T., J.L., H.M., J.K., W.F., R. K., and R. O. were responsible for critical revisions and approval of the final manuscript. T.L. performed the statistical analyses. A.K.M. had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
A.K.M. is a consultant to Vanda, Eli Lilly, Wyeth, and PGx Health. He has grant support from Eli Lilly and serves on the speaker’s bureau for Merck, Sunovion, Inc., and Bristol-Myers Squibb.
C.C. has received grants from BMS, Janssen and Janssen, Otsuka, NARSAD, AACAP, Feinstein Institute for Medical Research, and NIMH. He has received consultant fee or honorarium from Actelion, Alexza, AstraZeneca, Boehringer Ingelheim, Biotis, BMS, Cephalon, Desitin, Eli Lilly, IntraCellular Therapies, MedAvante, Ortho-McNeil/Janssen/J&J, GlaxoSmithKline, Hoffmann-La Roche, Lunbeck, Medicure, Merck, Novartis, Otsuka, Pfizer, Schering-Plough, Sunovion Inc, Takeda, and Vanda. He has been a consultant to AstraZeneca, BMS, Cephalon, Lundbeck, Medicure, Otsuka, Supernus, Medscape, Asante, Physicians Postgraduate Press, Network for Continuing Medical Education, OptumHealth, PeerView, Veritasime, Albert Einstein College of medicine Center for Continuous Medical Education, UCLA Center for Continuous Medical Education and CME LLC. He has been an advisory board member for Actelion, Alexza, AstraZeneca, Bristol-Myers Squibb, IntraCellular Therapies, Lundbeck, MedAvante, Merck, Novartis, Otsuka, Pfizer, Schering-Plough, Sunovion Inc,
Takeda, and Vanda. He also has served on the speaker’s bureau for AstraZeneca, Bristol- Myers Squibb, Eli Lilly, Merck, Otsuka, and Pfizer. He also received payment for manuscript preparation and development of educational presentations from PeerView, Physicians Postgraduate Press, UCLA Center for Continuous Medical Education, Veritasime, Albert Einstein College of Medicine Center for Continuous Medical Education, Asante, CME LLC, Medscape, Network for Continuing Medical Education, OptumHealth, PeerView, Physicians Postgraduate Press, UCLA Center for Continuous Medical Education, and Veritasime.
N.C. has nothing to disclose.
D.J.M. has grants from CIHR and NARSAD.
P.G. has nothing to disclose.
A.L. has nothing to disclose.
A.T. has nothing to disclose.
J.K. receives honorarium or consulting fees from AstraZeneca, Boehringer-Ingelheim, Bristol-Myers Squibb, Sunovion Inc, Eli Lilly, Lundbeck, Intracellular Therapeutics, Janssen, Johnson & Johnson, Merck, Novartis, Otsuka, Pfizer, Takeda, Wyeth, Vanda, and Roche. He has received travel support from Bristol-Myers Squibb, Eli Lilly, Lundbeck, Janssen, Merck, Otsuka, Pfizer, and Roche. He also receives fees for participation in review activities from Otsuka.
W.F. receives research support from Otsuka, Pfizer, Janssen, Alkermes, and Eli Lilly. He receives honoraria for consulting with Lundbeck, Roche, BMS, Otsuka, Janssen, Pfizer, Unitedbiosource, MedAvante, Sunovion, Merck, Neurosearch, Amgen, and Endo Pharmaceuticals. He receives speaker honoraria from Lundbeck, Sunovion, Janssen, Eli Lilly, Otsuka, Astra Zeneca, Roche, and Sunovion. He also holds stock in MedAvante.
R.K is a consultant and, DSMB, and receives grants or honoraria from Astra-Zeneca, BMS, Envivo, Lilly, Janssen-Cilag, Otsuka, Gedeon Richter, Roche, Sunovion.
R. O. has nothing to disclose.
J.L. is a consultant to AstraZeneca, Bioline, Cephalon, GlaxoSmithKline, Intracellular Therapies, Eli Lilly, Forest Laboratories, Janssen, Otsuka, Pfizer, Pierre Fabre, Psychogenics, and Wyeth. He has active or pending grants from Allon, AstraZeneca, Bristol-Myers Squibb, GlaxoSmithKline, Forest Laboratories, Janssen, Merck, Novartis, Pfizer, Sunovion Inc, Targacept, Wyeth, Eli Lilly, and Roche. He also has patents from Repligen and Royalties from Oxford Publishing, Wiley Publishing, and American Psychiatric Publishing, Inc.
T.L. is a consultant to Eli Lilly.
J.K. has grant support from Cdn Inst. Health Res, CIHR, NIMH, and Eli Lilly corp. He is a consultant to Sanofi-Aventis and Dainippon Sumitomo. He is also on the speaker’s bureau for Eli Lilly and has a patent from Theragenetics.
Funding This work was supported by NIH grants P50MH080173 (A.K.M.), P30MH090590 (J.K.), NARSAD Independent Investigator Award (A.K.M.), and a NARSAD Young Investigator Award (D.J.M.). Funding sources did not have any role in the design of study and approval of manuscript.
References
- 1.De Hert M, Dekker JM, Wood D, Kahl KG, Holt RI, Möller HJ. Cardiovascular disease and diabetes in people with severe mental illness position statement from the European Psychiatric Association (EPA), supported by the European Association for the Study of Diabetes (EASD) and the European Society of Cardiology (ESC) Eur Psychiatry. 2009;24:412–24. doi: 10.1016/j.eurpsy.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Tiihonen J, Lonnquist J, Wahlbeck K, et al. 11-year follow-up of motality in patients with schizophrenia: a population-based cohort study (FIN11 study) Lancet. 2009;374:620–7. doi: 10.1016/S0140-6736(09)60742-X. [DOI] [PubMed] [Google Scholar]
- 3.Maayan L, Correll CU. Management of antipsychotic-related weight gain. Expert Rev Neurother. 2010;10:1175–200. doi: 10.1586/ern.10.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Correll CU, Manu P, Olshanskiy V, Napolitano B, Kane JM, Malhotra AK. Cardiometabolic risk of second-generation antipsychotic medications during first-time use in children and adolescents. JAMA. 2009;302:1765–73. doi: 10.1001/jama.2009.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sikich L, Frazier JA, McClellan J, et al. Double-blind comparison of first- and second-generation antipsychotics in early-onset schizophrenia and schizo-affective disorder: findings from the treatment of early-onset schizophrenia spectrum disorders (TEOSS) study. Am J Psychiatry. 2008;165:1420–31. doi: 10.1176/appi.ajp.2008.08050756. [DOI] [PubMed] [Google Scholar]
- 6.Martin A, Scahill L, Anderson GM, et al. Weight and leptin changes among risperidone-treated youths with autism: 6-month prospective data. Am J Psychiatry. 2004;16:1125–7. doi: 10.1176/appi.ajp.161.6.1125. [DOI] [PubMed] [Google Scholar]
- 7.Robinson DG, Woerner MG, Napolitano B, et al. Randomized comparison of olanzapine versus risperidone for the treatment of first-episode schizophrenia: 4-month outcomes. Am J Psychiatry. 2006;163:2096–102. doi: 10.1176/ajp.2006.163.12.2096. [DOI] [PubMed] [Google Scholar]
- 8.Kane JM. Treatment adherence and long-term outcomes. CNS Spectr. 2007;12:21–6. doi: 10.1017/s1092852900026304. [DOI] [PubMed] [Google Scholar]
- 9.Adkins DE, Aberg K, McClay JL, et al. Genomewide pharmacogenomic study of metabolic side effects to antipsychotic drugs. Mol Psychiatry. 2011;16:321–32. doi: 10.1038/mp.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Contopoulos-Ioannidis DG, Kouri I, Ioannidis JP. Pharmacogenetics of the response to beta 2 agonist drugs: a systematic overview of the field. Pharmacogenomics. 2007;8:933–58. doi: 10.2217/14622416.8.8.933. [DOI] [PubMed] [Google Scholar]
- 11.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 13.Tiwari AK, Zai CC, Likhodi O, et al. A common polymorphism in the cannabinoid receptor 1 (CNR1) gene is associated with antipsychotic-induced weight gain in Schizophrenia. Neuropsychopharmacol. 2010;35:1315–24. doi: 10.1038/npp.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahn RS, Fleischhacker WW, Boter H, et al. Effectiveness of antipsychotic drugs in first-episode schizophrenia and schizophreniform disorder: an open randomised clinical trial. Lancet. 2008;37:1085–97. doi: 10.1016/S0140-6736(08)60486-9. [DOI] [PubMed] [Google Scholar]
- 15.Loos RJ, Lindgren CM, Li S, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–75. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chambers JC, Elliot P, Zabaneh D, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40:716–8. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 17.Huszar D, Lynch CA, Fairchild-Huntress V, et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–41. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 18.Daly AK, Donaldson PT, Bhatnagar P, et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat Genet. 2009;41:816–9. doi: 10.1038/ng.379. [DOI] [PubMed] [Google Scholar]
- 19.Shuldiner AR, O’Connell JR, Bliden KP, Gandhi A, Ryan K. Association of cytochrome P450–2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–57. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heard-Costa NL, Zillikens MC, Monda KL, et al. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE consortium. PLoS Genet. 2009;5:e1000539. doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 22.Zabaneh D, Balding DJ. A genome-wide association study of the metabolic syndrome in Indian Asian men. PLoS One. 2010;5:e11961. doi: 10.1371/journal.pone.0011961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyre D, Delplanque J, Chèvre JC, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–9. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 24.Willer CJ, Speliotes EK, Loos RJ, et al. Genetic Investigation of Anthropometric Traits Consortium. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scherag A, Dina C, Hinney A, et al. Two new Loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and German study groups. PLoS Genet. 2010;6:e1000916. doi: 10.1371/journal.pgen.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang SJ, Chiang CW, Palmer CD, et al. Genome-wide association of anthropometric traits in African- and African-derived populations. Hum Mol Genet. 2010;19:2725–38. doi: 10.1093/hmg/ddq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu G, Zhu H, Lagou V, et al. Common variants near melanocortin 4 receptor are associated with general and visceral adiposity in European- and African-American youth. J Pediatr. 2010;156:598–605.e1. doi: 10.1016/j.jpeds.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scherag A, Jarick I, Grothe J, et al. A Investigation of a genome wide association signal for obesity: synthetic association and haplotype analyses at the melanocortin 4 receptor gene locus. PLoS One. 2010;5:e13967. doi: 10.1371/journal.pone.0013967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Comer JS, Mojtabai R, Olfson M. National trends in the antipsychotic treatment of psychiatric outpatients with anxiety disorders. Am J Psychiatry. 2011;168:1057–1065. doi: 10.1176/appi.ajp.2011.11010087. [DOI] [PubMed] [Google Scholar]
- 31.Nargund RP, Strack AM, Fong TM. Melanocortin-4 receptor (MC4R) agonists for the treatment of obesity. J Med Chem. 2006;49:4035–43. doi: 10.1021/jm058241a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.