Abstract
Malignant peripheral nerve sheath tumours (MPNSTs), which develop sporadically or from neurofibromatosis, recur frequently with high metastatic potential and poor outcome. The polycomb group protein enhancer of zeste homologue 2 (EZH2) is an important regulator for various human malignancies. However, the function of EZH2 in MPNSTs is unknown. Here we report that the EZH2–miR-30d–KPNB1 signalling pathway is critical for MPNST tumour cell survival in vitro and tumourigenicity in vivo. Up-regulated EZH2 in MPNST inhibits miR-30d transcription via promoter binding activity, leading to enhanced expression of the nuclear transport receptor KPNB1 that is inhibited by miR-30d targeting of KPNB1 3′ UTR region. Furthermore, inhibition of EZH2 or KPNB1, or miR-30d over-expression, induces MPNST cell apoptosis in vitro and suppresses tumourigenesis in vivo. More importantly, forced over-expression of KPNB1 rescues MPNST cell apoptosis induced by EZH2 knockdown. Immunohistochemical analyses show that EZH2 and KPNB1 over-expression is observed in human MPNST specimens and is negatively associated with miR-30d expression. Our findings identify a novel signalling pathway involved in MPNST tumourigenesis, and also suggest that EZH2–miR-30d–KPNB1 signalling represents multiple potential therapeutic targetable nodes for MPNST.
Keywords: EZH2, miR-30d, KPNB1, MPNST, tumourigenesis
Introduction
Malignant peripheral nerve sheath tumours (MPNSTs) are a rare spindle cell malignancy that develops from large peripheral nerves and is the most common cause of mortality in neurofibromatosis type 1 (NF1), a common autosomal dominant disorder with an incidence of 1 in 3500 people [1,2]. Approximately half of MPNSTs arise in patients with NF1, whereas the remaining MPNSTs are sporadic and develop de novo [3]. For NF1 patients, the lifetime risk of developing MPNST is 8–13% [1]. MPNSTs have high local recurrence rates and a significant potential for metastasis, which dictates their unfavourable prognosis. Surgical resection is the mainstay of MPNST therapy, and the benefit of radiotherapy and systemic chemotherapy is limited. Due to the lack of therapeutic options, the 5 year survival rate for MPNST patients is in the range 35–50% and the 10 year disease-specific survival rate is only 7.5% [4]. This therefore highlights an urgent need for novel MPNST targeted chemotherapeutics.
Molecular and genetic studies of NF1 have determined that mutations of the NF1 tumour suppressor gene and inactivation of the NF1 protein Neurofibromin, a negative regulator of oncogenic RAS signalling, contributes to NF1, benign neurofibromas and MPNST pathogenesis [1]. Bi-allelic inactivation of the NF1 gene is required for the progression of NF1 to plexiform neurofibroma, which occurs in 34% of NF1 cases and is the precursor lesion of NF1-related MPNST [5]. In most cases of sporadic MPNSTs, mutations of the NF1 gene have also been found [3]. RAS inhibition of plexiform neurofibromas has been examined in clinical trials. Tipifarnib, a farnesyl transferase inhibitor that blocks RAS's ability to bind to the membrane where it is activated, was unsuccessful in a phase II clinical trial as a promising therapy for plexi-form neurofibroma, probably because of the alternative RAS prenylation by geranylgeranyl transferase [1,6]. Recently, mTOR, AKT, MET and HDAC were identified as potential drug targets for MPNST, and targeting these proteins with small-molecular inhibitors has demonstrated anti-tumour effects in MPNST [7–10]. Despite our understanding of Neurofibromin and RAS signalling in NF1, the molecular events involved in the tumourigenesis of MPNST are still poorly understood. A genetic mouse model with NF1 loss in stem/progenitor cells of peripheral nerves develops plexiform neurofibromas but not MPNSTs [11,12], suggesting that signalling pathways other than NF1/RAS may be involved in MPNST development.
Enhancer of zeste homolog 2 (EZH2) is a his-tone methyltransferase that catalyses the trimethylation of histone H3 lysine 27 (H3K27me3) [13]. EZH2, together with two other core proteins, SUZ12 and EED, forms polycomb-repressor complex 2 (PRC2), which functions as a transcription repressor and plays an important role in coordinating gene expression and repression during many physiological and developmental processes [14]. These processes include stem cell maintenance, cell senescence, cell differentiation and cell fate determination [15]. Not surprisingly, deregulation of EZH2 has been found to be involved in human diseases, including diabetes and cancers [15,16]. EZH2 has been identified as an onco-gene, most notably in breast and prostate cancers, by epigenetically inhibiting various tumour suppressor genes [15,17,18]. Recently, the epigenetic regulation of tumour suppressive microRNAs has emerged as critical signalling pathways involved in tumourigenesis. It has been shown that EZH2 directly inhibits miR-29, miR-181 and miR-200 families, which in turn targets EZH2 and other PRC2 proteins in B cell lymphomas and prostate cancer [19,20].
The function of EZH2 is undefined in mesenchyme-originating MPNST. Here we investigated the function and molecular mechanisms of EZH2 and microRNAs directly regulated by EZH2 in MPNST pathogenesis. Our findings enhance the biological functional knowledge of EZH2 and microRNAs, and have the potential to provide novel therapeutic approaches for MPNST patients.
Materials and methods
Patient tissue specimens, cell lines and immunohistochemical and western blot analyses
All normal, neurofibroma and MPNST patient samples were obtained from MD Anderson Cancer Center Neuro-oncology Department with patient consent after approval by the Institutional Review Board of the University of Texas MD Anderson Cancer Center. Neurofibroma and MPNST tissue microarray (TMA) information has been published previously [3]. Immunohistochemical analyses of EZH2 and KPNB1 was performed as previously described [3]. For TMA analyses, each biomarker was scored by two independent observers (PZ and AL or GA) after excluding arrayed specimens with insufficient tumour tissue or those that detached from the slide. Intensity was graded as none (= 0), weak/low (=1), or moderate-to-strong/high (=2–3), and staining distribution (the percentage of positive tumour cells) was estimated. Human NF1-related MPNST cell lines ST88-14, T265, MPNST642 and S462 and non-NF1-related sporadic human MPNST cell lines STS26T and MPNST724 were maintained as previously described [7]; Primary cultured normal human Schwann cells served as controls and were purchased from ScienCell Research Laboratories. Authentication of cell lines was performed by short tandem repeat DNA fingerprinting. Western blot analyses were done according to standard protocols, with minor modifications due to antibody optimization [21]. Commercially available antibodies were used for all immunoblot and immunohistochemical detection of EZH2 (D2C9, Cell Signalling), KPNB1 (NB100-81650, Novus Biologicals), cleaved PARP (ab32064, Abcam), GAPDH–HRP (ab9483, Abcam) and actin–HRP (I-19, Santa Cruz).
Cell culture, EZH2 knockdown
MPNST cell lines were cultured in 1 Dulbecco's modified Eagle's medium (DMEM) ×supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin. For siRNA knockdown and shRNA-mediated gene knockdown, a negative scrambled control siRNA or siRNA pool targeting EZH2 (Dharmacon) and a scrambled negative control or two different EZH2 shRNA targeting constructs (GI318845 and GI318846, Origene) were transfected into MPNST cells with lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Cells transfected with siRNAs were harvested after 48 h and subjected to qRT–PCR, western blot, cell cycle or apoptosis analyses. After 48 h, cells transfected with shRNA constructs (GFP-positive) were isolated using flowcytometry sorting. Sorted cells were cultured for western blot, cell cycle, apoptosis analyses and in vivo animal experiments.
Gene and miRNA microarray and qRT–PCR analyses
Total RNA, including miRNA, was isolated from cells using Trizol (Invitrogen). After quality assurance with a 2100 Bioanalyser (Agilent), procedures at the genomic core facility at MD Anderson were conducted to result in eventual hybridization and reporting of raw data for the cDNA microarray and the miRNA microarray. Human HT 12 v. 4 arrays (Illumina) were used for the cDNA microarray, and the MDACC v. 5.0 platform [22] was used for the miRNA array. Array data were quantile normalized; negative values were replaced with the minimum non-negative value across samples. Within each cell line, miRNA expression values were centred on the average of the corresponding control, and mRNA expression values were centred to standard deviations from the median. Two-sided t-tests defined significance of differential expression, and fold change was computed as the average fold change for each cell line. The GEO Database Accession Nos for the microRNA and mRNA microarray data are GSE47477 and GSE46904, respectively.
For qRT–PCR analyses of miRNAs, total RNA was isolated using Trizol, and reverse-transcribed to cDNA using the RT2 miRNA First Strand kit (Qiagen). Then, quantitative PCR (q-PCR) was performed with miR-30d specific primers (Qiagen). SNORD47 was used as a normalizing control for miRNA qRT–PCR analyses. For qRT–PCR analyses of mRNAs, first-strand cDNA was synthesized using a reverse transcription kit (Invitrogen) with oligo-dT primers. EZH2, KPNB1 and Actin primers were purchased from RealtimePrimers.
Cell viability assay and cell cycle and apoptosis analyses
In vitro cell viability was measured using a CellTiter96 Aqueous Non-Radioactive Cell Proliferation Assay kit (Promega), according to their standard protocol. Cell cycle analyses were performed as described previously [21]. For the cell apoptosis assays, cells were stained with annexin V and PI and the protocol of an Apoptosis Detection kit (BD Pharmingen), according to the manufacturer's recommendations.
Promoter activity analyses and ChIP assays
Promoter regions of miR-30d were amplified by genomic PCR with use of specific primers and cloned into the pGL vector directionally at NheI and BglII sites (see supplementary material, Table S3). For the promoter activity assay, empty pGL vector, pGL-miR-30d promoter were co-transfected with a scrambled siRNA control or an EZH2 siRNA into MPNST cells. The pRL vector was used as an internal control. After 48 h, cells were lysed and subjected to luciferase assays, using a Dual Luciferase Assay kit (Promega) according to the manufacturer's instructions. ChIP was conducted with the use of a ChromaFlash One-step ChIP kit (Epigenek) according to the manufacturer's instructions. A total of 2 × 106 cells and 3 μg anti-EZH2 or IgG antibody were used per immunoprecipitation. Primers for NUP214 and miR-30d promoters are listed in Table S3 (see supplementary material).
miR-30d and KPNB1 over-expression and reporter activity assays
To over-express miR-30d in MPNST cells, a miRNASelect pEP-hsa-miR-30d expression vector and an empty control vector were purchased (Cell Biolabs). KPNB1 constitutive expression construct was purchased from Origene. MPNST cells were transfected with either construct, using Lipofectamine 2000. After 48 h, cells were collected and subjected to qRT–PCR, western blot and cell cycle analyses. The mutant KPNB1 3′ UTR reporter was generated using a site-directed mutagenesis kit (Clontech) from the wild-type KPNB1 reporter construct (pLightSwitch 3′ UTR system, SwitchGear Genomics). For miR-30d reporter analyses, a negative control reporter, a wild-type KPNB1 3′ UTR reporter or a mutant KPNB1 3′ UTR reporter was transfected into MPNST cells with Lipofectamine 2000. After 48 h, reporter activity was assessed using LightSwitch luciferase assay reagents (SwitchGear Genomics).
Mice xenograft experiment
All animal procedures and care was approved by MD Anderson's Institutional Animal Care and Usage Committee. The MPNST xenograft mouse model using MPNST724 cells has been described previously [7]. For each mouse, 2 × 106 MPNST724 cells that stably expressed either a negative control vector or shRNA targeting EZH2 (GFP-positive) were used. Cell suspensions in PBS were injected subcutaneously into the flanks of 6 week-old female hairless SCID mice (n = 6/arm) and growth was measured twice weekly using a caliper.
Statistics
Data were analysed by means of a two-sided unpaired t-test, using GraphPad software (Prism 6.0). Data are shown as mean ± SD of multiple independent experiments. p < 0.05 was considered statistically significant.
Results
EZH2 expression is significantly up-regulated in MPNST
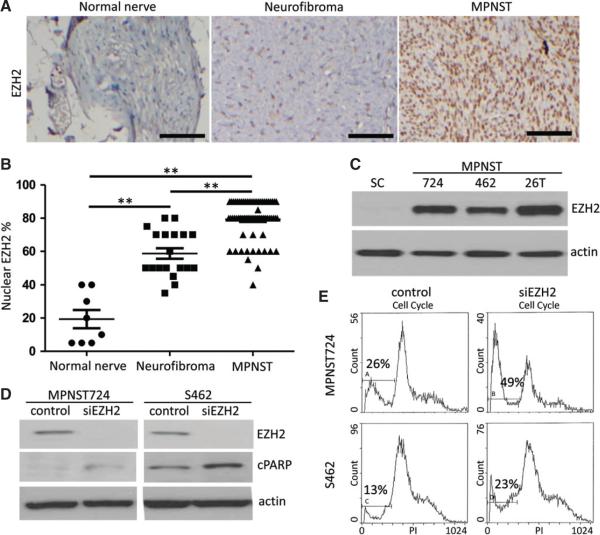
To investigate the role of EZH2 in MPNST tumours, we first examined EZH2 protein expression in human normal nerve tissues, neurofibromas and MPNST patient specimens. As shown in Figure 1A, immunohistochemical staining of EZH2 on a tissue microarray (TMA) showed that the protein expression of EZH2 was noted in nuclei of all normal nerves, neurofibromas and MPNSTs. However, the level of nuclear EZH2 expression was significantly higher in MPNSTs than in neurofibromas and normal nerves (p < 0.01) (Figure 1B). In addition, nuclear EZH2 expression in neurofibromas was also significantly higher than that of normal nerves (p < 0.01) (Figure 1B). Immunohistochemical analyses also showed that the nuclear EZH2 expression intensity in normal nerves and neurofibromas were significantly lower when compared with MPNSTs (see supplementary material, Figure S1). These results suggested that up-regulation of EZH2 expression may play a pivotal role in the development of MPNSTs. Next, we compared the protein expression of EZH2 in the MPNST cell lines MPNST724, S462 and STS26T versus normal human Schwann cells. Western blot analyses revealed that EZH2 expression was substantially enhanced in MPNST cell lines when compared with Schwann cells (Figure 1C).
Figure 1.
EZH2 is over-expressed in neurofibroma and MPNST specimens compared with normal nerves, and transient EZH2 knockdown induces MPNST cell apoptosis. (A) Representative immunohistochemical staining image of EZH2 in a normal nerve, a neurofibroma, and a MPNST specimen on a TMA. Scale bars = 100 μm. (B) Scatter plots depicting nuclear EZH2 expression percentages in normal nerve (n = 8), neurofibroma (n = 19) and MPNST (n = 68) specimens, as determined by immunohistochemical staining (**p < 0.01, Student's t-test). (C) Western blot analyses of EZH2 expression in human normal Schwann cells (SC) and in the MPNST cell lines MPNST724 (724), S462 (462) and STS26T (26T). Actin served as a loading control. (D) Western blot analyses of EZH2, cleaved PARP (cPARP), and actin in MPNST724 and S462 cells transfected with negative control siRNA or EZH2 siRNA (siEZH2). (E) Cell cycle analyses of MPNST724 and S462 cells transfected with negative control siRNA or EZH2 siRNA (siEZH2).
EZH2 knockdown induces apoptosis in MPNST cells in vitro and their tumourigenicity in vivo
To investigate the functional role of EZH2 in MPNST cells, we transiently knocked down EZH2 in the non-NF1 sporadic MPNST cell line MPNST724 and the NF1-related S462 cell line (Figure 1D). Cell cycle analyses showed that transient EZH2 inhibition induced the increased sub-G1 population in both cell lines (Figure 1E). Western blot analyses also showed an increased cleaved PARP as a marker of apoptosis after transient knockdown of EZH2 (Figure 1D). This suggested that transient knockdown of EZH2 may increase the apoptotic rates of MPNST724 and S462 cells in vitro.
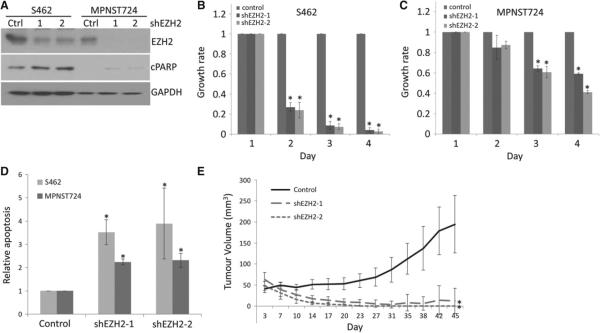
To further examine the function of silencing EZH2 in MPNST cells in vitro and in vivo, we stably knocked down EZH2 in MPNST cell lines. Figure 2A shows that for MPNST724 and S462 cell lines EZH2 protein expression significantly decreased, using two EZH2-targeting shRNAs compared with the non-targeting shRNA. Additionally, cleaved PARP increased in EZH2 shRNA-transduced MPNST cells, suggesting increased apoptosis. Consistently, MTT assays indicated that stable EZH2 knockdown inhibited growth and viability of MPNST724 and S462 cells in vitro (Figure 2B, C). Annexin V/propidium iodide staining assays showed that both EZH2 shRNAs significantly increased the annexin V-positive population in both MPNST cell lines (Figure 2D; see also supplementary material, Figure S2), suggesting that the stable knockdown of EZH2 also induces MPNST cell apoptosis. Next, we investigated the tumour-inhibitory effects of EZH2 knockdown in vivo using an MPNST xenograft mouse model [7]. MPNST724 cells that stably expressed a non-targeting shRNA control, or either of the two EZH2 shRNAs, were injected subcutaneously into the flanks of NOD/SCID mice. Tumour growth results indicated that EZH2-targeting shRNAs significantly inhibited MPNST tumourigenicity in vivo (Figure 2E).
Figure 2.
Stable EZH2 knockdown results in MPNST cell growth inhibition and apoptosis in vitro and tumourigenicity inhibition in vivo. (A) Western blot analyses of EZH2, cPARP and GAPDH in S462 and MPNST724 cells stably transduced with a non-targeting control (Ctrl) or two shRNA constructs targeting EZH2 (shEZH2). (B, C) Cell growth analyses of S462 and MPNST724 cells expressing a shRNA control, shEZH2-1 or shEZH2-2. Data are shown as mean ± SD; n = 3; *p < 0.05, Student's t-test. (D) Cell apoptosis analyses of MPNST724 and S462 cells with shRNA control, shEZH2-1 or shEZH2-2. Data are shown as mean ± SD; n = 3; *p < 0.05, Student's t-test. (E) Tumour growth of an MPNST724 xenograft mouse model injected with cells either bearing a shRNA control or shEZH2 constructs; 2 × 107 cellswere transduced to obtain 1.2 × 107 transduced cells (GFP+) for animal implantation. Data are shown as mean ± SD; n = 6; *p < 0.05, Student's ttest.
EZH2 regulates microRNA expression in MPNST cells
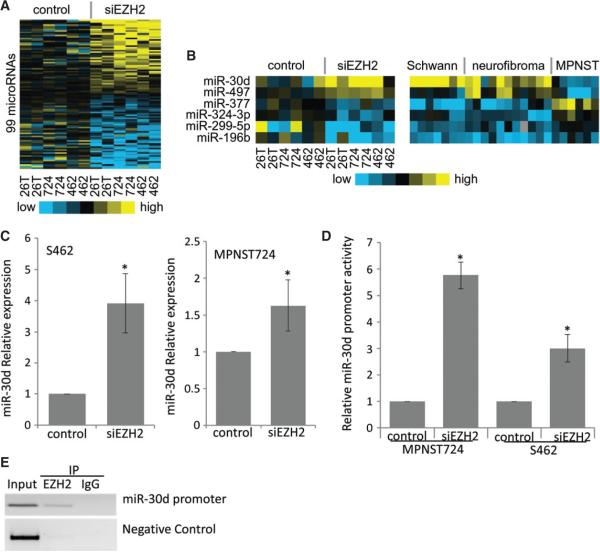
To illustrate the unique molecular mechanisms by which EZH2 regulates MPNST cells, we performed high-throughput microRNA (miRNA) microarray analyses of MPNST724, S462 and STS26T cells transfected with a non-targeting control siRNA or an EZH2 siRNA. After qRT–PCR confirmation of EZH2 knockdown, we performed miRNA microarray analyses. Data analyses showed that 99 miRNAs were significantly up- or down-regulated (46 and 53 miRNAs, respectively) at least ± 1.5-fold with EZH2 knockdown in all three MPNST cell lines (p < 0.05) (Figure 3A; see also supplementary material, Table S1). To further screen for miRNAs that may be particularly relevant to the malignant behaviours of MPNST, we compared the EZH2-regulated miRNAs that we found in our study to miRNA expression data of schwannoma, neurofibroma and MPNST specimens from another study [23]. We found that miR-30d and miR-497, which were up-regulated after EZH2 silencing, were decreased in MPNSTs compared with schwannomas or neurofibromas (Figure 3B). In contrast, four miRNAs (miR-377, miR-324-3p, miR-299-5p and miR-196b) were down-regulated with EZH2 silencing in MPNST cells, but were increased in MPNSTs compared with schwannomas or neurofibromas. Since EZH2 is a transcription repressor [13], we focused on miRNAs that were up-regulated following EZH2 suppression, leading to the investigation of miR-30d's contribution to MPNST pathobiology.
Figure 3.
Expression of miRNAs that are modulated with EZH2 knockdown in MPNST cells. (A) Heat map of miRNA expression data showing EZH2-regulated miRNA expression following EZH2 knockdown (two repeats) in MPNST724 (724), S462 (462) and STS26T (26T) cells. (B) Heat map of miRNA expression data changed with EZH2 knockdown in MPNST724 (724), S462 (462) and STS26T (26T) cells that were also differentially expressed in schwannoma, neurofibroma and MPNST patient samples. (C) qRT–PCR analyses of miR-30d in S462 (left) and MPNST724 (right) cells transfected with a negative control or EZH2 siRNA. miR-30d expression was normalized to SNORD47. Data are shown as mean ± SD; n = 3; *p < 0.05, Student's t-test. (D) Luciferase activities of miR-30d promoter constructs in MPNST724 and S462 cells transfected with a negative control or EZH2 siRNA (siEZH2). Data are shown as mean ± SD; n = 3; *p < 0.05, Student's t-test. (E) Chromatin immunoprecipitation analyses showing the specific EZH2 interaction with miR-30d promoter regions. The NUP214 promoter served as a negative control.
EZH2 inhibits miR-30d expression by binding to their promoters in MPNST cells
First, we confirmed the miRNA microarray results by qRT–PCR analyses. In an independent transient EZH2 knockdown experiment, miR-30d expression increased significantly when compared with the non-targeting siRNA controls in both the MPNST724 and S462 cell lines (Figure 3C; see also supplementary material, Figure S3). To investigate the mechanism of EZH2 regulation of miR-30d, we cloned the promoter region of miR-30d (ch8q23.3-8q24.3) from genomic DNA of MPNST cells by PCR. We cloned > 3 kb upstream of the miR-30d transcription start site into a pGL vector as miR-30d's promoter. To determine whether EZH2 can inhibit miR-30d expression at the promoter level, we co-transfected a non-targeting siRNA or siEZH2 along with either miR-30d promoter construct into MPNST724 and S462 cells. The pRL construct was used as an internal control. Promoter activity results showed that EZH2 knockdown significantly increased promoter activity of miR-30d in both cell lines, suggesting that EZH2 suppresses the expression of miR-30d at the transcription level (Figure 3D).
To determine whether EZH2 binds directly to miR-30d promoters, we performed chromatin immunoprecipitation analyses using S462 cells. the results showed that immunoprecipitation of EZH2 specifically brought down miR-30d promoter DNA (Figure 3E). The promoter of NUP214 served as a negative control for EZH2 binding. These data demonstrated that EZH2 inhibited miR-30d expression directly through binding to their promoters as a part of the PRC2–EED–EZH2 complex.
miR-30d targets KPNB1 and induces MPNST cell apoptosis
To investigate the functional role of miR-30d in MPNST cells, we transfected MPNST724 and S462 cells with either a miR-30d over-expression vector or an empty vector control. After qRT–PCR confirmation of miR-30d over-expression (see supplementary material, Figure S4A), we analysed the cell cycle profile of cells. We found that miR-30d over-expression induced a significant increase in the sub-G1 population (Figure 4A; see also supplementary material, Figure S4B). These data suggested that miR-30d over-expression may increase apoptosis in MPNST cells.
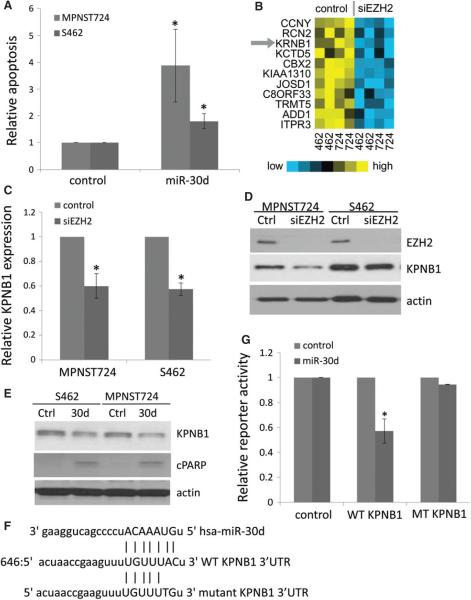
Figure 4.
Over-expression of miR-30d induced cell death and suppressed KPNB1 through 3′ UTR targeting. (A) Apoptosis analyses of MPNST724 and S462 cells transfected with negative control or miR-30d over-expression vector. Data are shown as mean ± SD; n = 3; *p < 0.05, Student's t-test. (B) Heat map of mRNAs down-regulated with EZH2 knockdown that are also targeted by miR-30d in MPNST724 (724) and S462 (462) cells. (C) qRT–PCR analyses of KPNB1 expression in MPNST724 and S462 cells transfected with a control or EZH2 siRNA. KPNB1 expression is normalized to actin. Data are shown as mean ± SD; n = 3; *p < 0.05, Student's t-test. (D) Western blot analyses of EZH2, KPNB1 and actin in MPNST724 and S462 cells transfected with a control or EZH2 siRNA. (E) Western blot analyses of KPNB1 and actin in MPNST724 and S462 cells transfected with a control (Ctrl) or miR-30d mimic (30d). (F) Putative miR-30d target site in the wild-type KPNB1 3′ UTR region (WT) and a mutant KPNB1 3′ UTR construct sequence. (G) Effects of the miR-30d mimic on luciferase reporter activities of the vector control, wild-type KPNB1 3′ UTR (WT KPNB1) and the mutant KPNB1 3′ UTR (MT KPNB1). Data are shown as mean ± SD; n = 3; *p < 0.03, Student's t-test.
We next investigated miR-30d's functionally relevant targets for MPNST tumour cell survival. Given that EZH2 knockdown up-regulates miR-30d expression, we predicted that miR-30d target genes may be down-regulated with siRNA-mediated knockdown of EZH2 in MPNST cells relative to control cells. Therefore, we performed cDNA microarray analyses with RNA samples isolated from control and EZH2 knockdown MPNST724 and S462 cells. Microarray analyses with EZH2 knockdown showed that 31 genes, including EZH2 itself, were significantly down-regulated by EZH2 silencing (p < 0.01, fold > 1.2) (see supplementary material, Table S2). We then crossed these 31 genes with miR-30d's potential target genes, which were discovered using miRanda and TargetScan databases, and found that 11 of those genes were miR-30d potential targets (Figure 4B). Amongst these genes, KPNB1 was of interest as a regulator of cell survival [24]. KPNB1, also known as Importin β1 or Karyopherin β1, is an important component of the nucleocytoplasmic transporter complex that mediates shuttling of cytoplasmic proteins into the nucleus [25]. Therefore, we focused on elucidating the possible interconnections between KPNB1, miR-30d and EZH2 in MPNST cells.
First, we confirmed that KPNB1 mRNA and protein expression levels decreased with the transient knockdown of EZH2 in MPNST724 and S462 cells by qRT–PCR and western blot analyses (Figure 4C, D). Consistently, we also found that KPNB1 was down-regulated with stable EZH2 knockdown in MPNST724 and S462 cells (see supplementary material, Figure S4C). Next, we tested the hypothesis that miR-30d might target KPNB1 in MPNST cells, using a negative control vector and a miR-30d over-expression construct. Immunoblotting procedures revealed that MPNST724 and S462 cells transfected with the miR-30d over-expression suppressed KPNB1 protein expression and increased apoptosis marker cleaved PARP (Figure 4E).
To further investigate the mechanism by which miR-30d targets KPNB1, we searched for a potential binding site of miR-30d in KPNB1's 3′ UTR, using TargetScan. One potential miR-30d binding site was identified within the 3′ UTR region of KPNB1 mRNA (Figure 4F). To confirm that miR-30d targets KPNB1's 3′ UTR, we used a reporter construct, pLightSwitch, containing KPNB1's 3′ UTR. In order to unequivocally show that miR-30d specifically targets KPNB1 at the afore-mentioned region (Figure 4F), we mutated the first two nucleotides of the potential miR-30d 'seed region’ in the wild-type KPNB1 3′ UTR reporter construct, such that miR-30d would not be able to bind and target KPNB1 (Figure 4F). Then, we co-transfected different combinations of the empty vector mimic control, the miR-30d mimic, the negative control reporter, the wild-type KPNB1 3′ UTR reporter and the mutated KPNB1 3′ UTR reporter constructs into S462 cells. The results indicated that the miR-30d mimic significantly reduced the activity of the wild-type KPNB1 3′ UTR reporter while having little effect on mutant KPNB1's 3′ UTR reporter activity (Figure 4G). These results indicated that miR-30d binds and targets KPNB1 within its 3′ UTR at 646 bp from the translation stop site, thereby inhibiting its expression.
Knockdown of KPNB1 induces apoptosis in MPNST cells, and KPNB1 over-expression rescues MPNST cell apoptosis induced by EZH2 knockdown
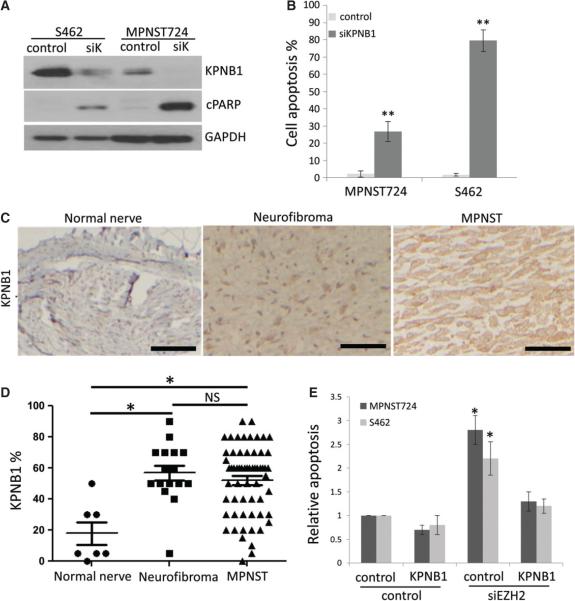
To determine whether KPNB1 may be an important contributor to MPNST pathobiology, we transiently knocked down KPNB1 using siRNA-mediated interference and evaluated the effects on markers of apoptosis and cell viability of MPNST724 and S462 cells in vitro. Immunoblotting revealed that cleaved PARP increased following the depletion of KPNB1 in MPNST cells (Figure 5A). Next, we performed AnnexinV/PI analyses. The results showed that KPNB1 knockdown in MPNST cells significantly increased MPNST724 and S462 cell apoptosis (Annexin V+) (Figure 5B; see also supplementary material, Figure 5A), suggesting that KPNB1 is a crucial modulator of MPNST cell survival.
Figure 5.
KPNB1 knockdown induces cell apoptosis in MPNST cells and KPNB1 force expression rescued MPNST cell apoptosis induced from EZH2 knockdown. (A) Western blot analyses of KPNB1, cPARP and GAPDH in MPNST724 and S462 cells transfected with control or KPNB1 siRNA (siK). (B) Cell apoptosis assay of MPNST724 and S462 cells transfected with control siRNA or KPNB1 siRNA (siKPNB1). Data are shown as mean ± SD; n = 3; **p < 0.005, Student's t-test. (C) Representative immunohistochemical staining image of KPNB1 in normal nerve, neurofibroma and MPNST specimens on a TMA. Scale bars = 100 μm. (D) Scatter plots depicting the nuclear and cytoplasmic KPNB1 expression percentage in normal nerve (n = 8), neurofibroma (n = 19) and MPNST (n = 68) specimens, as determined by IHC (*p < 0.05; NS, not significant; Student's t-test). (E) Apoptosis analyses of MPNST724 and S462 cells transfected with negative control or siEZH2 combined with negative control or KPNB1 over-expression vector. Data are shown as mean + SD; n = 3; *p < 0.05, Student's t-test.
To assess whether KPNB1 was over-expressed in MPNST clinical samples, we performed immunohistochemical analyses for KPNB1 expression on the same TMA used for EZH2 expression evaluation (Figure 1A). The results showed that KPNB1 was expressed in normal nerves, neurofibromas and MPNST tissues (Figure 5C), but the percentage of KPNB1 expression was significantly higher in neurofibroma and MPNST than in normal nerve specimens (p < 0.005) (Figure 5C, D). The percentage of KPNB1 expression between neurofibroma and MPNST tissues did not show significant differences (Figure 5D). KPNB1 staining intensity was significantly higher in MPNSTs than normal nerve and neurofibroma specimens (see supplementary material, Figure S5B). These results suggest a positive correlation between the varying degrees of nuclear EZH2 and KPNB1 protein expression in MPNSTs, neurofibromas and normal nerve patient specimens.
To investigate whether KPNB1 is a critical regulator for MPNST cell apoptosis induced by EZH2 knockdown, we performed a rescue experiment to test whether KPNB1 over-expression decreased cell apoptosis induced by EZH2 in MPNST724 and S462 cells. The results showed that KPNB1 forced expression significantly reduced cell apoptosis in EZH2 knockdown MPNST cells (Figure 5E), suggesting that KPNB1 is an important mediator of EZH2 effects on MPNST cells.
Correlation between EZH2, miR-30d, and KPNB1 in MPNST cells
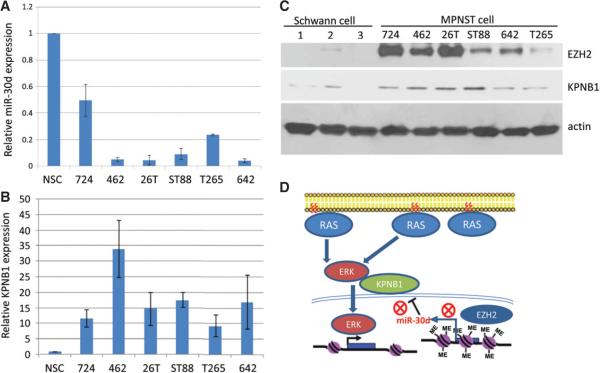
Finally, we evaluated a larger panel of MPNST cell lines for miR-30d and KPNB1 expression by qRT–PCR, and immunoblot analyses for EZH2 and KPNB1 in the same cell line panel. Results showed that miR-30d and KPNB1 had an inverse correlation at the mRNA levels (Figure 6A, B) and that EZH2 and KPNB1 were positively correlated at the protein level in multiple human normal Schwann cells and in the panel of MPNST cell lines (Figure 6C). Positive correlation between EZH2 and KPNB1 expression was also found in TMA MPNST patient samples (see supplementary material, Figure S5C).
Figure 6.
EZH2, KPNB1, and miR-30d expression correlates in human normal Schwann cells and MPNST cell lines. (A, B) qRT–PCR analyses of miR-30d and KPNB1 expression in human normal Schwann cells (NSCs) and MPNST cell lines MPNST724 (724), S462 (462), STS26T (26T), ST88-14 (ST88), T265 and MPNST642 (642). miR-30d and KPNB1 expression are normalized to SNORD47 and actin, respectively. Data are shown as mean ± SD; n = 3. (C) Western blot analyses for EZH2, KPNB1 and actin in human normal Schwann cells and in the MPNST cell lines MPNST724 (724), S462 (462), STS26T (26T), ST88-14 (ST88), MPNST642 (642) and T265. (D) Schematic model of EZH2-regulated miR-30d expression that targets KPNB1 in MPNST cells. EZH2 over-expression inhibits miR-30d transcription by binding to its promoter. Lower miR-30d expression results in KPNB1 up-regulation, which may increase the nuclear translocation of ERK proteins thereby promoting cell growth and survival.
Discussion
As a crucial epigenetic regulator of gene expression, EZH2 has been found to be functionally involved in many aspects of multiple carcinomas, such as tumour initiation, tumour cell survival, chemoresistance, invasiveness, metastasis and angiogenesis [17]. Until only recently, EZH2 function has been implicated in mesenchyme-originating sarcomas [26]. However, the function of EZH2 in MPNST is entirely unknown. Our studies demonstrate that EZH2 plays an oncogenic role in both NF1-related and non-NF1-related MPNST tumourigenesis. We found that EZH2 modulates the expression of miR-30d that directly targets oncogenic signalling of the nuclear transporter protein KPNB1 in MPNST (Figure 6D). Not only do the results of this study provide possible additional insights of EZH2 that may be relevant to other human malignancies, but they also highlight the importance of performing additional preclinical studies in MPNST that target EZH2, since this may represent a novel therapeutic approach for MPNST treatment in the clinic. Further studies will be conducted to elucidate more detailed mechanisms of EZH2 regulation on miR-30d transcription and KPNB1 expression in MPNST cells.
Another novel finding from our study was that EZH2 positively regulates nuclear transporter receptor protein KPNB1 through miR-30d. KPNB1 is a member of the karyopherin/importin-β family of nuclear transport factors that mediates the nuclear import of proteins through binding nuclear localization signals [27]. Previous studies have shown that KPNB1 is over-expressed in colon, breast, lung, ovarian and cervical cancer specimens compared with normal tissues [24,25,28]. Smith et al (2010) reported that lower KPNB1 expression in normal cells restricts the nuclear translocation of phospho-ERK after pathway stimulation, thereby limiting cell proliferation and maintaining a differentiated phenotype of ovarian and mammary epithelial cells, suggesting that KPNB1 levels are a rate-limiting step of RAS–MAPK pathway activation. Given that both NF1-related and non-NF1-related MPNSTs have up-regulated RAS activity mediating activation of downstream genes inside the nucleus, it is not surprising that EZH2-up-regulated KPNB1 may contribute to MPNST pathogenesis. It also suggests that targeting EZH2 may serve as an alternative approach to suppressing RAS signalling in MPNST and other cancers that are dependent on RAS activation for tumour progression.
Supplementary Material
Acknowledgements
We thank Drs Xiaoyan Ma, Keila Torres, Yiqun Zhang, John M Slopis and Ian E McCutchmeon for technical assistance and providing clinical specimens. This study was supported by grants from the Mike Hogg Foundation (to PZ), the National Cancer Institute (NCI; SARC Grant No. Sarcoma SPORE U54CA168512, to REP) and the National Institutes of Health (Grant No. P30 CA125123, to CJC). The MD Anderson Cancer Center cell-line characterization core facility and flow cytometry core facility are supported by the NCI (Cancer Center Support Grant No. CA16672).
Footnotes
No conflicts of interest were declared.
Author contributions
PZ and REP contributed to design of the research; and PZ, JG, CJC, GAAS, DRI, AL, XL and CL performed the experiments.
References
- 1.Katz D, Lazar A, Lev D. Malignant peripheral nerve sheath tumour (MPNST): the clinical implications of cellular signalling pathways. Expert Rev Mol Med. 2009;11:e30. doi: 10.1017/S1462399409001227. [DOI] [PubMed] [Google Scholar]
- 2.Widemann BC. Current status of sporadic and neurofi bromatosis type 1-associated malignant peripheral nerve sheath tumors. Curr Oncol Rep. 2009;11:322–328. doi: 10.1007/s11912-009-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zou C, Smith KD, Liu J, et al. Clinical, pathological, and molecular variables predictive of malignant peripheral nerve sheath tumor outcome. Ann Surg. 2009;249(6):1014–1022. doi: 10.1097/SLA.0b013e3181a77e9a. [DOI] [PubMed] [Google Scholar]
- 4.Anghileri M, Miceli R, Fiore M, et al. Malignant peripheral nerve sheath tumors: prognostic factors and survival in a series of patients treated at a single institution. Cancer. 2006;107(5):1065–1074. doi: 10.1002/cncr.22098. [DOI] [PubMed] [Google Scholar]
- 5.Tucker T, Wolkenstein P, Revuz J, et al. Association between benign and malignant peripheral nerve sheath tumors in NF1. Neurology. 2005;65(2):205–211. doi: 10.1212/01.wnl.0000168830.79997.13. [DOI] [PubMed] [Google Scholar]
- 6.Kissil JL, Blakeley JO, Ferner RE, et al. What's new in neurofibromatosis? Proceedings from the 2009 NF conference: new frontiers. Am J Med Genet A 2010. 152A(2):269–283. doi: 10.1002/ajmg.a.33189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou CY, Smith KD, Zhu Q-S, et al. Dual targeting of AKT and mammalian target of rapamycin: a potential therapeutic approach for malignant peripheral nerve sheath tumor. Mol Cancer Therap. 2009;8(5):1157–1168. doi: 10.1158/1535-7163.MCT-08-1008. [DOI] [PubMed] [Google Scholar]
- 8.Lopez G, Torres K, Liu J, et al. Autophagic survival in resistance to histone deacetylase inhibitors: novel strategies to treat malignant peripheral nerve sheath tumors. Cancer Res. 2011;71(1):185–196. doi: 10.1158/0008-5472.CAN-10-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansson G, Mahller YY, Collins MH, et al. Effective in vivo targeting of the mammalian target of rapamycin pathway in malignant peripheral nerve sheath tumors. Mol Cancer Ther. 2008;7(5):1237–1245. doi: 10.1158/1535-7163.MCT-07-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torres KE, Zhu Q-S, Bill K, et al. Activated MET is a molecular prognosticator and potential therapeutic target for malignant peripheral nerve sheath tumors. Clin Cancer Res. 2011;17(12):3943–3955. doi: 10.1158/1078-0432.CCR-11-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng H, Chang L, Patel N, et al. Induction of abnormal proliferation by nonmyelinating Schwann cells triggers neurofibroma formation. Cancer Cell. 2008;13(2):117–128. doi: 10.1016/j.ccr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Wu J, Williams JP, Rizvi TA, et al. Plexiform and dermal neurofibromas and pigmentation are caused by Nf1 loss in desert hedgehog-expressing cells. Cancer Cell. 2008;13(2):105–116. doi: 10.1016/j.ccr.2007.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14(2):155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Simon J, Lange C. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647(1–2):21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Chen H, Gu X, Liu Y, et al. PDGF signalling controls age-dependent proliferation in pancreatic (bgr)-cells. Nature. 2011;478(7369):349–355. doi: 10.1038/nature10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crea F, Fornaro L, Bocci G, et al. EZH2 inhibition: targeting the crossroad of tumor invasion and angiogenesis. Cancer Metast Rev. 2012;31(3–4):753–761. doi: 10.1007/s10555-012-9387-3. [DOI] [PubMed] [Google Scholar]
- 18.Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br J Cancer. 2012;106(2):243–247. doi: 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Zhao X, Fiskus W, et al. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B cell lymphomas. Cancer Cell. 2012;22(4):506–523. doi: 10.1016/j.ccr.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Cao Q, Mani R-S, Ateeq B, et al. Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer Cell. 2011;20(2):187–199. doi: 10.1016/j.ccr.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang P, Bill K, Liu J, et al. MiR-155 is a liposarcoma oncogene that targets casein kinase-1α and enhances β-catenin signaling. Cancer Res. 2012;72(7):1751–1762. doi: 10.1158/0008-5472.CAN-11-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C-G, Calin GA, Volinia S, et al. MicroRNA expression profiling using microarrays. Nat Protoc. 2008;3(4):563–578. doi: 10.1038/nprot.2008.14. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian S, Thayanithy V, West RB, et al. Genome-wide transcriptome analyses reveal p53 inactivation mediated loss of miR-34a expression in malignant peripheral nerve sheath tumours. J Pathol. 2010;220(1):58–70. doi: 10.1002/path.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Watt PJ, Maske CP, Hendricks DT, et al. The Karyopherin proteins, Crm1 and Karyopherin-β1, are overexpressed in cervical cancer and are critical for cancer cell survival and proliferation. Int J Cancer. 2009;124(8):1829–1840. doi: 10.1002/ijc.24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318(5855):1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 26.Ciarapica R, Miele L, Giordano A, et al. Enhancer of zeste homolog 2 (EZH2 ) in pediatric soft tissue sarcomas: first implications. BMC Med. 2011;9(1):63. doi: 10.1186/1741-7015-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strom AC, Weis K. Importin-β-like nuclear transport receptors. Genome Biol. 2001;2(6) doi: 10.1186/gb-2001-2-6-reviews3008. reviews 3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon IK, Jans DA. Regulation of nuclear transport: central role in development and transformation? Traffic. 2005;6(3):173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.