Abstract
Cancers driven by oncogenic Ras proteins encompass some of the most deadly human cancer types, and there is a pressing need to develop therapies for these diseases. While recent studies suggest that mutant Ras proteins may yet be druggable, the most promising and advanced efforts involve inhibitors of Ras effector signaling. Most efforts to target Ras signaling have been aimed at the ERK mitogen-activated protein kinase and the phosphatidylinositol 3-kinase signaling networks. However, to date, no inhibitors of these Ras effector pathways have been effective against RAS mutant cancers. This ineffectiveness is due, in part, to the involvement of additional effectors in Ras-dependent cancer growth, such as the Rac small GTPase and the p21-activated serine-threonine kinases (PAK). PAK proteins are involved in many survival, cell motility, and proliferative pathways in the cell and may present a viable new target in Ras-driven cancers. In this review, we address the role and therapeutic potential of Rac and Group I PAK proteins in driving mutant Ras cancers.
Background
The frequent mutational activation of Ras oncoproteins has stimulated considerable interest in developing therapeutic approaches for blocking aberrant Ras signaling for cancer treatment (1). Disappointingly, despite more than three decades of intensive effort by researchers, no clinically effective anti-Ras drugs have been developed. However, with cancer genome sequencing studies revealing that aberrant Ras signaling remains the most significant driver of cancer growth, there is renewed interest in the search for the elusive anti-Ras therapy (2). Currently, the most tractable approach to inhibit Ras is through ablation of Ras effector signaling. However, given the complex nature of Ras signaling, whether targeting one or multiple Ras effector signaling pathways will be required for effective and long-term therapy is unresolved (3). Currently, most efforts have centered on the two canonical Ras effectors, the Raf serine/threonine protein kinases and the phosphatidylinositol 3-kinases (PI3Ks) (4). Numerous inhibitors of each of these effector pathways are currently under clinical evaluation. In this review, we focus on a lesser-studied Ras effector pathway that leads to activation of the Rac small GTPase. Among the multitude of Rac effectors, there is increasing evidence that the p21-activated serine/threonine kinases (PAKs) may contribute significantly to cancer growth (5, 6). In this review we summarize the evidence for aberrant Rac-PAK signaling in supporting RAS mutant cancer growth, and the status of developing PAK inhibitors for cancer treatment.
Ras effector signaling
Ras proteins (H-Ras, N-Ras, K-Ras4A/B) function as GDP-GTP-regulated on-off switches, cycling between active Ras-GTP-bound and inactive GDP-bound states (1). Guanine nucleotide exchange factors promote the formation of active Ras-GTP, whereas GTPase activating proteins (RasGAPs) stimulate GTP hydrolysis and formation of inactive Ras-GDP (7, 8). Mutant Ras proteins contain single amino acid substitutions at G12, G13 or Q61, making them refractory to RasGAP activity and persistently GTP-bound (1). KRAS mutations comprise 86% of all RAS mutations, followed by NRAS (10%), with HRAS mutation rarely seen in cancer (9).
Ras-GTP binds preferentially to a spectrum of functionally diverse downstream effectors (3). Most attention has been focused on the Raf serine/threonine kinases, which activate the ERK1/2 mitogen-activated protein kinases (Fig. 1). One Raf isoform, B-Raf, is mutated frequently in human cancers (10). PI3Ks comprise the second most studied Ras effector class. PI3K activation causes increased formation of phosphatidylinositol-3,4,5-trisphosphate (PIP3) and activation of the AKT serine/threonine kinase. PI3K is mutated frequently in cancer and PI3K is essential for RAS-driven cancer development (11, 12). There are currently at least 30 inhibitors of the Raf-MEK-ERK pathway and 50 inhibitors of the PI3K-AKT-mTOR pathway under clinical evaluation (clinicaltrials.gov(13)).
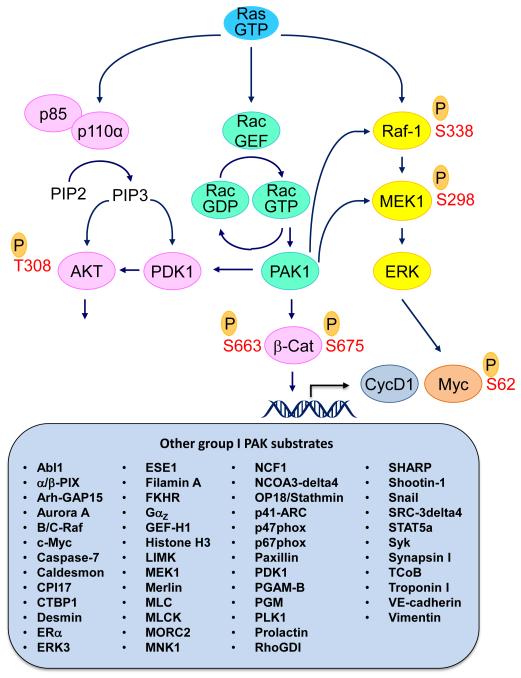
Figure 1.
Ras effector signaling. The importance of the Raf-MEK-ERK and PI3K-AKT-mTOR effector signaling networks are well validated drivers of mutant Ras-dependent cancer growth. Rac is activated by Ras through direct (e.g., Tiam1) or indirect (via PIP3 formation) activation of guanine nucleotide exchange factors for the Rac small GTPase. The group I PAKs comprise one key effector family of Rac. Over 50 substrates of PAK1 have been described (compiled from references 37 and 39). These substrates include components of the ERK MAPK cascade. PAK1 can also function as a scaffold to facilitate AKT activation.
A less-studied Ras effector network results in activation of the Rac small GTPase. This can be mediated through Ras interaction with an RBD-containing RacGEF, Tiam1 (14, 15) (Fig. 1). Another mechanism involves PI3K-mediated formation of PIP3, which then activates Rac-selective GEFs (e.g., P-Rex, Vav) (16, 17).
The three Rac isoforms are members of the Rho branch of the Ras superfamily (18). They are best known for their regulation of actin organization, in particular to regulate lamellipodia induction and promotion of cell migration and pinocytosis. Rac also regulates the formation of reactive oxygen species (19). The recent identification of activated Rac1 mutants in melanoma supports an important driver role for Rac in cancer growth (20), (21).
Like Ras, Rac is a GDP-GTP regulated binary switch, with Rac-GTP engaging multiple effectors (22). While the precise effector(s) that drives Rac-dependent cancer growth remains to be determined, the PAK protein kinases are intriguing candidates. Below, we summarize the evidence for the importance of the Rac-PAK effector signaling pathway in Ras-driven cancer development and growth.
Rac and Ras in cancer
Early studies identified upregulated Rac activation in H-Ras-transformed rodent fibroblasts (15, 23, 24). These were followed by studies where dominant negative Rac1 mutants, which sequester and inactivate RacGEFs, impaired the growth of H-Ras-transformed rodent fibroblasts (25-27). Subsequent genetically-engineered mouse model studies found that tissue-restricted genetic loss of Rac1 impaired mutant Kras-driven lung (28) and pancreatic (29) cancer development. Furthermore, in a mutant Kras-driven model of papilloma development, tumor tissue exhibited increased levels of Rac-GTP, and loss of one Rac1 copy alone was sufficient to reduce tumor growth and increase survival (30).
The key effectors that drive Rac-dependent cancer growth remain to be elucidated. In an early study utilizing effector-binding mutants of Rac1 to study the effectors important for transformation of NIH3T3 mouse fibroblasts, PAK1 was found to be dispensable (31). These analyses suggested that Rac1 regulates at least four distinct effector-mediated functions and that multiple pathways may contribute to Rac1-induced cellular transformation. However, since subsequent studies identified cell type and species differences in the effectors involved in Ras-mediated transformation (32, 33), a reevaluation of the role of PAK1 in Rac-dependent cancer growth in human cancers is clearly merited. Another Rac1 activity, upregulation of reactive oxygen species, in which PAK1 is also involved (34), has been suggested to contribute to Rac1-mediated growth regulation
PAK activation in RAS mutant cancer
PAKs comprise a family of six proteins divided into two sub-groups: group I comprises PAK1-3 and group II contains PAK4-6 (35). Since group I PAKs are Rac and Cdc42 effectors, whereas group II PAKs are Cdc42 only, we focus on the group I PAKs. Although the group I PAKs share strong sequence identity in their kinase domains (92-95%), PAK1 is thus far the most studied family member, so we focus primarily on PAK1 in this review.
Though PAK1 activity can be deregulated by a diversity of mechanisms in cancer that include gene amplification and increased gene transcription (5, 36), here we focus on activation of PAK downstream of Ras, RacGEFs, and Rac. While in the inactive conformation within the cytosol, PAK1-3 form head-to-tail homodimers with the N-terminal autoinhibitory domain (AID) of one monomer inserted within the C-terminal kinase domain of another. Upon binding of Rac1-GTP to the GTPase binding domain (GBD) of group I PAKs, a conformational change releases the AID from the kinase domain leading to autophosphorylation at multiple serine/threonines and activation of PAK catalytic activity, allowing phosphorylation of substrates (35). Additionally, plasma membrane-associated Rac binding facilitates PAK plasma membrane recruitment, where PAKs can interact with effectors.
PAK effector signaling in human cancer
Group I PAKs regulate a spectrum of catalytically diverse substrates (5, 6). The precise substrates critical for PAK-dependent cancer growth remain to be fully understood and the interplay of multiple substrates is likely involved. In particular, PAK1 facilitates cross-talk with both the Raf and PI3K effector signaling networks. PAK1 can enhance ERK signaling by phosphorylation of Raf-1 (S338) and MEK1 (S298) (37-40). PAK1 also regulates the PI3K-AKT-mTOR pathway, where PAK1 exhibits a kinase-independent scaffolding function to facilitate PDK1-mediated recruitment of AKT to the plasma membrane to facilitate AKT activation (41). The physiologic relevance of PAK1 cross-talk with ERK and AKT signaling is supported by the observation that genetic or pharmacologic ablation of PAK1 impaired both ERK and AKT activation in Kras-driven skin tumors (42). Pharmacologic inhibitors of the Raf and PI3K pathways have been ineffective in RAS mutant cancer cells, in part, due to kinome reprogramming mechanisms that stimulate signaling activities that overcome inhibitor action (43-46). Consequently, combination targeting of PAKs and members of these pathways, such as MEK, ERK, PI3K or AKT, may help overcome these resistance mechanisms. However, PAK1 cross-talk with these Ras effector pathways can be context-dependent as PAK1 suppression in KRAS mutant colon carcinoma cells impaired anchorage-dependent and –independent proliferation, but not ERK or AKT activation (47).
PAKs are also capable of influencing transcription of genes that promote cell cycle progression and cell survival. In breast cancer and colon cancer cell lines, PAK1 can phosphorylate β-catenin on S663 and S675, stabilizing β-catenin and promoting nuclear translocation and transcriptional stimulation of TCF-responsive genes, including CCND1 and MYC (48, 49).
PAKs enhance cell survival by phosphorylating proteins associated with apoptosis. PAK1 phosphorylates Bad on S111 to prevent Bcl2 binding and induction of apoptosis (50). Additionally, PAK1 can phosphorylate induce relocalization of Raf-1 to the mitochondria where it also inhibits BAD by phosphorylating BAD on S112 (50).
PAKs are also critical mediators of the cytoskeleton and cell motility. PAK1 and PAK2 phosphorylate LIM-kinase on T508, which in turn phosphorylates cofilin to prevent actin depolymerization (51, 52). Additionally, PAK1 can phosphorylate the p41-ARC subunit of the Arp2/3 complex to promote actin nucleation and cell motility (53, 54). PAKs are also involved in microtubule reorganization through both tubulin cofactor B (TCoB), a protein responsible for assembling tubulin heterodimers (55), and through the inactivation of stathmin, which is normally responsible for destabilizing microtubules at the leading edge of cells (56-58).
Metabolism is a critically important factor to the survival of cancer cells because of their high energy demands, and PAKs play a role in driving several metabolic processes that aid tumor cell growth and survival. Increased macropinocytosis to facilitate increased extracellular protein and lipid uptake is one consequence of the high metabolic requirements of cancer cells (59). PAK1 was found to be necessary and sufficient for growth factor- and Rac-induced macropinocytosis in NIH3T3 fibroblasts (60). Rac and PAK1 were found to be both necessary for Bladder cancer cell macropinocytotic uptake of Bacille Calmette-Guerin, a strain of bacteria used in the treatment of bladder carcinoma (61). Additionally, bacterial uptake was also stimulated by activated K-Ras or H-Ras and this activity was blocked by pharmacologic inhibition of group I PAKs (IPA-3). This study suggests that the activity state of PAKs in cancer cells could be a determinate for efficient uptake of cancer therapeutics. Similarly, in pancreatic cancer cells, K-Ras-dependent stimulation of macropinocytosis and uptake of albumen (62) may provide a basis for the efficacy of albumen-bound (nab) paclitaxel for the treatment of this cancer. It will also be important to assess a role for Rac-PAK signaling in K-Ras-dependent macropinocytosis to determine whether pharmacologic inhibition of PAK1 may be an approach in blocking cancer cell metabolism.
While PAKs are canonically thought of as functioning in the cytosol or at the plasma membrane, they do contain several nuclear localization signals (NLS) and play several roles within the nucleus. In zebrafish, PAK1 nuclear import is essential for development (63). In cancer cells, increased nuclear accumulation of PAK1 has been associated with advanced tumor stage in colorectal and breast tumors (64, 65). In breast tumors, increased nuclear PAK1 is capable of phosphorylating ERα at S305 and causing it to become active in a ligand-independent manner, leading to tamoxifen resistance (66). Finally, PAK1 can translocate to the nucleus to drive transcription of fibronectin, which is crucial for supporting pancreatic cancer cell growth and migration (67).
Role of PAK in RAS-driven cancer development and growth
The first evidence for a role for PAK1 in Ras-dependent growth transformation comes from studies in model cell systems. Ectopic expression of a kinase-dead PAK1 dominant negative mutant impaired H-Ras and Rac1 growth transformation of rat 3Y1 fibroblasts (68) or H-Ras transformation of Rat-1 but not NIH3T3 mouse fibroblasts (69, 70). Similarly, dominant negative Rac1 and kinase-dead PAK1 inhibited K-Ras transformation of MT4H1 rat Schwann cells (71). Recently, using a mouse model of Kras-driven skin squamous cell carcinoma formation, genetic ablation of Pak1 strongly impaired tumor initiation and progression (42). Together, with the validated role of Rac1 in Ras-driven oncogenesis, these observations implicate the Rac-PAK effector pathway as a target for the development of anti-Ras therapeutic strategies. Like Ras, Rac is not considered a highly tractable drug target. Therefore, below we focus on the development of PAK inhibitors for cancer treatment.
Clinical-Translational Advances
PAK inhibitor development is still largely at the preclinical stage, with only one PAK inhibitor evaluated in clinical trials (72). Due to the high sequence identity of the kinase domains, most attempts thus far have yielded molecules with a high affinity for all group I PAK members, and in some cases, inhibitory activity for both group I and II PAKs. Early stage ATP-competitive PAK inhibitors (e.g., staurosporine, A-FL172) lacked selectivity for PAK. The first PAK inhibitor to reach clinical trials was a pan-PAK inhibitor, PF-3758309 (73). This compound was identified originally as a hit in a screen for inhibitors of PAK4, but it proved to effectively inhibit all PAK family members, in addition to other protein kinases. Preclinical evaluation showed anti-tumor activity against multiple human tumor cell lines, leading to Phase I evaluation in patients with solid tumors. Unfortunately, this trial was stopped in phase I due to pharmacokinetic issues. More recently, derivatives of PF-3758309 have been described with much improved pharmacologic properties, raising hope that this class of compound may yet have clinical utility (74).
More recently, Licciulli et al. describe the discovery of a small-molecule pyridopyrimidinone, FRAX597, that potently inhibits group I PAKs by preventing ATP-binding and hydrolysis (75). FRAX597 exhibited high specificity and potency for Group I PAKs, although potent inhibition of other kinases was also seen. When evaluated in vivo, FRAX597 inhibited the tumorigenic growth of NF2-null Schwann cells. NF2 loss causes Rac1 and PAK1 activation indicating that this compound could be viable therapeutic strategy for treating PAK-dependent tumors. FRAX597 treatment also phenocopied genetic loss of Pak1 and impaired Kras-driven skin tumorigenesis (42). Interestingly, in this mouse model, both genetic and pharmacologic inhibition of PAK1 resulted in reduction of ERK and AKT activity, supporting the importance of PAK1 signaling cross-talk with these two Ras effector pathways.
Peterson and colleagues performed a screen to identify small molecule allosteric inhibitors of Cdc42 activation of group I PAKs. The results of this screen led to the development of IPA-3 (2,2′-dihydroxy-1,1′dinaphthyldsulfide), which interacts with the PBD/AID region of group I PAKs and prevents their activation by GTPase binding (76, 77). IPA-3 showed strong selectivity for group I PAKs, with no inhibitory activity for group II PAKs or more than 200 other protein kinases evaluated. However, inability of IPA-3 to inhibit already activated PAK1, the μM IC50 of this compound and it’s rapid metabolism to a toxic compound, due to the reduction of the disulfide bond it contains, limit the ability to transition IPA-3 as a clinically useful chemical platform.
Conclusions
With increasing experimental evidence validating a driver role PAKs in tumor growth and invasion, a key issue for the clinical advancement of PAK inhibitors will be defining genetic and/or biochemical markers that identify those cancers that will respond to anti-PAK therapy. The position of PAK downstream of mutant K-Ras and Rac, in addition to PAK signaling cross-talk with the key Ras effector pathways, support PAK inhibitors as a therapeutic strategy for RAS mutant cancers. Given the involvement of multiple effectors in driving RAS-dependent cancer growth, PAK inhibition in combination with inhibitors of Raf or PI3K effector signaling will likely be required. Currently, pharmacologic inhibitors of PAK1 also inhibit other Group I PAKs; whether PAK1-selective inhibitors are more desirable and possible to develop are issues that remain to be resolved. Of the spectrum of PAK substrates, which substrate(s) will provide a reliable biomarker for PAK inhibitor anti-tumor activity also remains unclear. A survey of the patent literature indicates that more PAK inhibitors are in the pipeline (78). As more potent and selective inhibitors become available, the answers to many of these unresolved questions will likely be addressed.
Acknowledgments
Grant Support
This work was supported by NIH grants (CA042978, CA179193, and CA175747; to C.J. Der); the Lustgarten Pancreatic Cancer Foundation (to C.J. Der); the 2012 Pancreatic Cancer Action Network-AACR Innovative Grant, supported by Tempur-Pedic Retailers, Grant Number 12-60-25-DER (to C.J. Der); and the Ruth L. Kirschstein National Research Service Award Individual Predoctoral Fellowship (1F31CA180628; to N.M. Baker).
Footnotes
Disclosure of Potential Conflicts of Interest
C.J. Der reports receiving a commercial research grant from Onconova. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Cox AD, Der CJ. Ras history: The saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging Ras Back in the ring. Cancer Cell. 2014;25:272–81. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–57. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines AT, Xu D, Der CJ. Inhibition of Ras for cancer treatment: the search continues. Future Med Chem. 2011;3:1787–808. doi: 10.4155/fmc.11.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radu M, Semenova G, Kosoff R, Chernoff J. PAK signalling during the development and progression of cancer. Nat Rev Cancer. 2014;14:13–25. doi: 10.1038/nrc3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King H, Nicholas NS, Wells CM. Role of p-21-activated kinases in cancer progression. Int Rev Cell Mol Biol. 2014;309:347–87. doi: 10.1016/B978-0-12-800255-1.00007-7. [DOI] [PubMed] [Google Scholar]
- 7.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–77. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 8.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–80. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 9.COSMIC: Catalogue Of Somatic Mutations In Cancer. 2014 [cited; Available from: http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/
- 10.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 11.Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–68. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 12.Castellano E, Sheridan C, Thin MZ, Nye E, Spencer-Dene B, Diefenbacher ME, et al. Requirement for interaction of PI3-kinase p110alpha with RAS in lung tumor maintenance. Cancer Cell. 2013;24:617–30. doi: 10.1016/j.ccr.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.2014 ClinicalTrials.gov: A service of the U.S. National Institutes of Health. [cited; Available from: http://clinicaltrials.gov.
- 14.Malliri A, van der Kammen RA, Clark K, van der Valk M, Michiels F, Collard JG. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature. 2002;417:867–71. doi: 10.1038/nature00848. [DOI] [PubMed] [Google Scholar]
- 15.Lambert JM, Lambert QT, Reuther GW, Malliri A, Siderovski DP, Sondek J, et al. Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat Cell Biol. 2002;4:621–5. doi: 10.1038/ncb833. [DOI] [PubMed] [Google Scholar]
- 16.Welch HC, Coadwell WJ, Ellson CD, Ferguson GJ, Andrews SR, Erdjument-Bromage H, et al. P-Rex1, a PtdIns(3,4,5)P3- and Gbetagamma-regulated guanine-nucleotide exchange factor for Rac. Cell. 2002;108:809–21. doi: 10.1016/s0092-8674(02)00663-3. [DOI] [PubMed] [Google Scholar]
- 17.Han J, Luby-Phelps K, Das B, Shu X, Xia Y, Mosteller RD, et al. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science. 1998;279:558–60. doi: 10.1126/science.279.5350.558. [DOI] [PubMed] [Google Scholar]
- 18.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–6. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 19.Archer H, Bar-Sagi D. Ras and Rac as activators of reactive oxygen species (ROS) Methods Mol Biol. 2002;189:67–73. doi: 10.1385/1-59259-281-3:067. [DOI] [PubMed] [Google Scholar]
- 20.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bustelo XR, Sauzeau V, Berenjeno IM. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays. 2007;29:356–70. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bar-Sagi D, Feramisco JR. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science. 1986;233:1061–8. doi: 10.1126/science.3090687. [DOI] [PubMed] [Google Scholar]
- 24.Walsh AB, Bar-Sagi D. Differential activation of the Rac pathway by Ha-Ras and K-Ras. J Biol Chem. 2001;276:15609–15. doi: 10.1074/jbc.M010573200. [DOI] [PubMed] [Google Scholar]
- 25.Khosravi-Far R, Solski PA, Clark GJ, Kinch MS, Der CJ. Activation of Rac1, RhoA, and mitogen-activated protein kinases is required for Ras transformation. Molecular Cell Biol. 1995;15:6443–53. doi: 10.1128/mcb.15.11.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu RG, Chen J, Kirn D, McCormick F, Symons M. An essential role for Rac in Ras transformation. Nature. 1995;374:457–9. doi: 10.1038/374457a0. [DOI] [PubMed] [Google Scholar]
- 27.Joneson T, White MA, Wigler MH, Bar-Sagi D. Stimulation of membrane ruffling and MAP kinase activation by distinct effectors of RAS. Science. 1996;271:810–2. doi: 10.1126/science.271.5250.810. [DOI] [PubMed] [Google Scholar]
- 28.Kissil JL, Walmsley MJ, Hanlon L, Haigis KM, Bender Kim CF, Sweet-Cordero A, et al. Requirement for Rac1 in a K-ras induced lung cancer in the mouse. Cancer Res. 2007;67:8089–94. doi: 10.1158/0008-5472.CAN-07-2300. [DOI] [PubMed] [Google Scholar]
- 29.Heid I, Lubeseder-Martellato C, Sipos B, Mazur PK, Lesina M, Schmid RM, et al. Early requirement of Rac1 in a mouse model of pancreatic cancer. Gastroenterology. 2011;141:719–30. 30 e1–7. doi: 10.1053/j.gastro.2011.04.043. [DOI] [PubMed] [Google Scholar]
- 30.Samuel MS, Lourenco FC, Olson MF. K-Ras mediated murine epidermal tumorigenesis is dependent upon and associated with elevated Rac1 activity. PloS One. 2011;6:e17143. doi: 10.1371/journal.pone.0017143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westwick JK, Lambert QT, Clark GJ, Symons M, Van Aelst L, Pestell RG, et al. Rac regulation of transformation, gene expression, and actin organization by multiple, PAK-independent pathways. Mol Cell Biol. 1997;17:1324–35. doi: 10.1128/mcb.17.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamad NM, Elconin JH, Karnoub AE, Bai W, Rich JN, Abraham RT, et al. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 2002;16:2045–57. doi: 10.1101/gad.993902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–83. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Ahmed S, Prigmore E, Govind S, Veryard C, Kozma R, Wientjes FB, et al. Cryptic Rac-binding and p21(Cdc42Hs/Rac)-activated kinase phosphorylation sites of NADPH oxidase component p67(phox) J Biol Chem. 1998;273:15693–701. doi: 10.1074/jbc.273.25.15693. [DOI] [PubMed] [Google Scholar]
- 35.Arias-Romero LE, Chernoff J. A tale of two Paks. Biology of the cell / under the auspices of the European. J Cell Biol. 2008;100:97–108. doi: 10.1042/BC20070109. [DOI] [PubMed] [Google Scholar]
- 36.Ye DZ, Field J. PAK signaling in cancer. Cell Logist. 2012;2:105–16. doi: 10.4161/cl.21882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaudhary A, King WG, Mattaliano MD, Frost JA, Diaz B, Morrison DK, et al. Phosphatidylinositol 3-kinase regulates Raf1 through Pak phosphorylation of serine 338. Curr Biol. 2000;10:551–4. doi: 10.1016/s0960-9822(00)00475-9. [DOI] [PubMed] [Google Scholar]
- 38.Coles LC, Shaw PE. PAK1 primes MEK1 for phosphorylation by Raf-1 kinase during cross-cascade activation of the ERK pathway. Oncogene. 2002;21:2236–44. doi: 10.1038/sj.onc.1205302. [DOI] [PubMed] [Google Scholar]
- 39.Sun H, King AJ, Diaz HB, Marshall MS. Regulation of the protein kinase Raf-1 by oncogenic Ras through phosphatidylinositol 3-kinase, Cdc42/Rac and Pak. Curr Biol. 2000;10:281–4. doi: 10.1016/s0960-9822(00)00359-6. [DOI] [PubMed] [Google Scholar]
- 40.Eblen ST, Slack JK, Weber MJ, Catling AD. Rac-PAK signaling stimulates extracellular signal-regulated kinase (ERK) activation by regulating formation of MEK1-ERK complexes. Mol Cell Biol. 2002;22:6023–33. doi: 10.1128/MCB.22.17.6023-6033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higuchi M, Onishi K, Kikuchi C, Gotoh Y. Scaffolding function of PAK in the PDK1-Akt pathway. Nat Cell Biol. 2008;10:1356–64. doi: 10.1038/ncb1795. [DOI] [PubMed] [Google Scholar]
- 42.Chow HY, Jubb AM, Koch JN, Jaffer ZM, Stepanova D, Campbell DA, et al. p21-Activated kinase 1 is required for efficient tumor formation and progression in a Ras-mediated skin cancer model. Cancer Res. 2012;72:5966–75. doi: 10.1158/0008-5472.CAN-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Little AS, Smith PD, Cook SJ. Mechanisms of acquired resistance to ERK1/2 pathway inhibitors. Oncogene. 2013;32:1207–15. doi: 10.1038/onc.2012.160. [DOI] [PubMed] [Google Scholar]
- 44.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–21. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graves LM, Duncan JS, Whittle MC, Johnson GL. The dynamic nature of the kinome. Biochem J. 2013;450:1–8. doi: 10.1042/BJ20121456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burris HA., 3rd Overcoming acquired resistance to anticancer therapy: focus on the PI3K/AKT/mTOR pathway. Cancer Chemother Pharmacol. 2013;71:829–42. doi: 10.1007/s00280-012-2043-3. [DOI] [PubMed] [Google Scholar]
- 47.Tabusa H, Brooks T, Massey AJ. Knockdown of PAK4 or PAK1 inhibits the proliferation of mutant KRAS colon cancer cells independently of RAF/MEK/ERK and PI3K/AKT signaling. Mol Cancer Res. 2013;11:109–21. doi: 10.1158/1541-7786.MCR-12-0466. [DOI] [PubMed] [Google Scholar]
- 48.Arias-Romero LE, Villamar-Cruz O, Huang M, Hoeflich KP, Chernoff J. Pak1 kinase links ErbB2 to beta-catenin in transformation of breast epithelial cells. Cancer Res. 2013;73:3671–82. doi: 10.1158/0008-5472.CAN-12-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu G, Wang Y, Huang B, Liang J, Ding Y, Xu A, et al. A Rac1/PAK1 cascade controls beta-catenin activation in colon cancer cells. Oncogene. 2012;31:1001–12. doi: 10.1038/onc.2011.294. [DOI] [PubMed] [Google Scholar]
- 50.Ye DZ, Jin S, Zhuo Y, Field J. p21-Activated kinase 1 (Pak1) phosphorylates BAD directly at serine 111 in vitro and indirectly through Raf-1 at serine 112. PLoS One. 2011;6:e27637. doi: 10.1371/journal.pone.0027637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Misra UK, Deedwania R, Pizzo SV. Binding of activated alpha2-macroglobulin to its cell surface receptor GRP78 in 1-LN prostate cancer cells regulates PAK-2-dependent activation of LIMK. J Biol Chem. 2005;280:26278–86. doi: 10.1074/jbc.M414467200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bagheri-Yarmand R, Mazumdar A, Sahin AA, Kumar R. LIM kinase 1 increases tumor metastasis of human breast cancer cells via regulation of the urokinase-type plasminogen activator system. Int J Cancer. 2006;118:2703–10. doi: 10.1002/ijc.21650. [DOI] [PubMed] [Google Scholar]
- 53.Vadlamudi RK, Li F, Barnes CJ, Bagheri-Yarmand R, Kumar R. p41-Arc subunit of human Arp2/3 complex is a p21-activated kinase-1-interacting substrate. EMBO Rep. 2004;5:154–60. doi: 10.1038/sj.embor.7400079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Webb BA, Eves R, Crawley SW, Zhou S, Cote GP, Mak AS. PAK1 induces podosome formation in A7r5 vascular smooth muscle cells in a PAK-interacting exchange factor-dependent manner. Am J Physiol Cell Physiol. 2005;289:C898–907. doi: 10.1152/ajpcell.00095.2005. [DOI] [PubMed] [Google Scholar]
- 55.Vadlamudi RK, Barnes CJ, Rayala S, Li F, Balasenthil S, Marcus S, et al. p21-activated kinase 1 regulates microtubule dynamics by phosphorylating tubulin cofactor B. Mol Cell Biol. 2005;25:3726–36. doi: 10.1128/MCB.25.9.3726-3736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Banerjee M, Worth D, Prowse DM, Nikolic M. Pak1 phosphorylation on t212 affects microtubules in cells undergoing mitosis. Curr Biol. 2002;12:1233–9. doi: 10.1016/s0960-9822(02)00956-9. [DOI] [PubMed] [Google Scholar]
- 57.Takahashi K, Suzuki K. Membrane transport of WAVE2 and lamellipodia formation require Pak1 that mediates phosphorylation and recruitment of stathmin/Op18 to Pak1-WAVE2-kinesin complex. Cell Signal. 2009;21:695–703. doi: 10.1016/j.cellsig.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 58.Morimura S, Takahashi K. Rac1 and Stathmin but Not EB1 Are Required for Invasion of Breast Cancer Cells in Response to IGF-I. Int J Cell Biol. 2011;2011:615912. doi: 10.1155/2011/615912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39:91–100. doi: 10.1016/j.tibs.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dharmawardhane S, Schurmann A, Sells MA, Chernoff J, Schmid SL, Bokoch GM. Regulation of macropinocytosis by p21-activated kinase-1. Mol Biol Cell. 2000;11:3341–52. doi: 10.1091/mbc.11.10.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Redelman-Sidi G, Iyer G, Solit DB, Glickman MS. Oncogenic activation of Pak1-dependent pathway of macropinocytosis determines BCG entry into bladder cancer cells. Cancer Res. 2013;73:1156–67. doi: 10.1158/0008-5472.CAN-12-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–7. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lightcap CM, Kari G, Arias-Romero LE, Chernoff J, Rodeck U, Williams JC. Interaction with LC8 is required for Pak1 nuclear import and is indispensable for zebrafish development. PloS One. 2009;4:e6025. doi: 10.1371/journal.pone.0006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qing H, Gong W, Che Y, Wang X, Peng L, Liang Y, et al. PAK1-dependent MAPK pathway activation is required for colorectal cancer cell proliferation. Tumour Biol. 2012;33:985–94. doi: 10.1007/s13277-012-0327-1. [DOI] [PubMed] [Google Scholar]
- 65.Rayala SK, Molli PR, Kumar R. Nuclear p21-activated kinase 1 in breast cancer packs off tamoxifen sensitivity. Cancer Res. 2006;66:5985–8. doi: 10.1158/0008-5472.CAN-06-0978. [DOI] [PubMed] [Google Scholar]
- 66.Holm C, Rayala S, Jirstrom K, Stal O, Kumar R, Landberg G. Association between Pak1 expression and subcellular localization and tamoxifen resistance in breast cancer patients. J Natl Cancer Inst. 2006;98:671–80. doi: 10.1093/jnci/djj185. [DOI] [PubMed] [Google Scholar]
- 67.Jagadeeshan S, Krishnamoorthy YR, Singhal M, Subramanian A, Mavuluri J, Lakshmi A, et al. Transcriptional regulation of fibronectin by p21-activated kinase-1 modulates pancreatic tumorigenesis. Oncogene. 2014 Feb 24; doi: 10.1038/onc.2013.576. doi: 10.1038/onc.2013.576. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 68.Osada S, Izawa M, Koyama T, Hirai S, Ohno S. A domain containing the Cdc42/Rac interactive binding (CRIB) region of p65PAK inhibits transcriptional activation and cell transformation mediated by the Ras-Rac pathway. FEBS Lett. 1997;404:227–33. doi: 10.1016/s0014-5793(97)00139-7. [DOI] [PubMed] [Google Scholar]
- 69.Tang Y, Yu J, Field J. Signals from the Ras, Rac, and Rho GTPases converge on the Pak protein kinase in Rat-1 fibroblasts. Mol Cell Biol. 1999;19:1881–91. doi: 10.1128/mcb.19.3.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang Y, Chen Z, Ambrose D, Liu J, Gibbs JB, Chernoff J, et al. Kinase-deficient Pak1 mutants inhibit Ras transformation of Rat-1 fibroblasts. Molecular Cell Biol. 1997;17:4454–64. doi: 10.1128/mcb.17.8.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tang Y, Marwaha S, Rutkowski JL, Tennekoon GI, Phillips PC, Field J. A role for Pak protein kinases in Schwann cell transformation. PNAS. 1998;95:5139–44. doi: 10.1073/pnas.95.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rudolph J, Crawford JJ, Hoeflich KP, Chernoff J. Inhibitors of the Ras Superfamily G-proteins, Part B. p21-activated kinase inhibitors. The Enzymes. 2013:157–80. doi: 10.1016/B978-0-12-420146-0.00007-X. [DOI] [PubMed] [Google Scholar]
- 73.Murray BW, Guo C, Piraino J, Westwick JK, Zhang C, Lamerdin J, et al. Small-molecule p21-activated kinase inhibitor PF-3758309 is a potent inhibitor of oncogenic signaling and tumor growth. PNAS. 2010;107:9446–51. doi: 10.1073/pnas.0911863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo C, McAlpine I, Zhang J, Knighton DD, Kephart S, Johnson MC, et al. Discovery of pyrroloaminopyrazoles as novel PAK inhibitors. J Med Chem. 2012;55:4728–39. doi: 10.1021/jm300204j. [DOI] [PubMed] [Google Scholar]
- 75.Licciulli S, Maksimoska J, Zhou C, Troutman S, Kota S, Liu Q, et al. FRAX597, a small molecule inhibitor of the p21-activated kinases, inhibits tumorigenesis of neurofibromatosis type 2 (NF2)-associated Schwannomas. J Biol Chem. 2013;288:29105–14. doi: 10.1074/jbc.M113.510933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deacon SW, Beeser A, Fukui JA, Rennefahrt UE, Myers C, Chernoff J, et al. An isoform-selective, small-molecule inhibitor targets the autoregulatory mechanism of p21-activated kinase. Chem Biol. 2008;15:322–31. doi: 10.1016/j.chembiol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Viaud J, Peterson JR. An allosteric kinase inhibitor binds the p21-activated kinase autoregulatory domain covalently. Mol Cancer Ther. 2009;8:2559–65. doi: 10.1158/1535-7163.MCT-09-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crawford JJ, Hoeflich KP, Rudolph J. p21-Activated kinase inhibitors: a patent review. Expert Opin Ther Pat. 2012;22:293–310. doi: 10.1517/13543776.2012.668758. [DOI] [PubMed] [Google Scholar]