Abstract
WWOX is a candidate tumour suppressor gene that exhibits LOH or homozygous deletion in several tumour types. As well as the predominant full-length transcript (variant 1) there also exist alternatively spliced transcripts found previously only in malignant tissue. It has been suggested that proteins encoded by these variants may interfere with normal WWOX function in a dominant negative fashion. The most prevalent alternate transcript demonstrated in ovarian cancer is variant 4, which lacks exons 6-8. Here, we report the first comparison of the mRNA expression of WWOX variants 1 and 4 in human ovarian tumours and normal ovaries and correlate expression with clinical data. We demonstrate significantly lower WWOX variant 1 expression in tumours than in normal ovaries. This reduction was not associated with any specific clinical subgroup. Variant 4 was expressed at low levels, and significantly associated with high grade and advanced stage ovarian cancer. Furthermore, tumours co-expressing variant 4 and relatively high levels of variant 1 showed significantly worse survival than tumours expressing variant 1 alone. However, variant 4 was also frequently identified in non-malignant ovarian tissue. These results support the role of WWOX variant 1 as a suppressor of ovarian tumouri-genesis, but the role of variant 4 remains speculative.
Keywords: WWOX, tumour suppressor gene, FRA16D, ovaria carcinoma
Introduction
Loss of heterozygosity (LOH) of chromosome 16q has been described in ovarian, breast, prostate and other cancers (1-6). Later studies identified 16q23-24 as a region of particularly high allelic loss (7-10), suggesting that there was a tumour suppressor gene in this region. Recently the WWOX gene was mapped to 16q23.3-24.1 (11). The 2.2 kb WWOX transcript is encoded by 9 exons and spans a region of >1 Mb. This region encompasses FRA16D (12,13), the second most frequently expressed common fragile site in the human genome.
Functional evidence exists for the role of WWOX as a tumour suppressor. Overexpression of the gene in breast cancer cell lines with low baseline WWOX expression resulted in strong inhibition of anchorage-independent growth in soft agar and dramatic suppression of tumourigenicity in vivo (14). Studies with the murine homologue, WOX1, suggest that it interacts with p53 and plays a role in apoptosis (15). However, examples of mutations in the human WWOX gene are sparse (11,16). One somatic leucine to proline missense mutation, two residues from the putative active site, was identified in an esophageal cancer that also showed LOH, suggesting possible inactivation of both alleles (17). Homozygous deletions disrupting both alleles of the WWOX gene have been reported in a variety of cancer cell lines and in human primary tumour material (16), and high frequency of allelic loss involving WWOX has been shown in ovarian, esophageal (17) and non-small cell lung cancers (18).
In light of the paucity of mutations, we chose to investigate other mechanisms of WWOX dysregulation, such as reduced expression or the production of aberrant isoforms, that may play a role in cancer. Alternative mRNA variants of WWOX with unique 3’-terminal exons have been described (19), and aberrant variants of WWOX have been identified in cancer cell lines and tumour tissue (14,16-18,20,21). In ovarian tumours the predominant aberrant transcript is the Δ6-8 (variant 4) transcript that skips exons 6-8 (16). These variants lack the sequence encoding the enzymatic oxidoreductase domain, but retain the sequence encoding the protein binding WW-domains, leading some investigators to postulate that they may function in a dominant negative fashion by sequestering WWOX-binding proteins. During the preparation of this manuscript, two studies have reported the use of antibodies that recognise WWOX short-form proteins (21,22). Watanabe et al (22) could only detect alternate transcript expression if the proteasome was blocked. In contrast, Ishii et al (21) did identify the presence of WWOX short form proteins in haematopoietic malignancies, although no indication of the specificity of the antibody was given. In this report, we demonstrate that the sensitivity of a WWOX-specific antibody is orders of magnitude below that required to detect the WWOX short-form proteins in solid tumours. We therefore used quantitative RT-PCR to investigate the mRNA expression levels of full length WWOX (variant 1) and the predominant alternate transcript Δ6-8 (variant 4) in malignant and normal ovarian tissues. We further determined whether the expression of these variants correlated with clinico-pathological factors or patient survival. We report that the expression of WWOX variant 1 in ovarian tumours is significantly reduced as compared with normal ovary tissue, supporting a tumour suppressor role for WWOX. However, the clinical significance of the alternate variant 4 transcript remains uncertain.
Materials and methods
PEOl transfections
The WWOX variant 1 coding region was inserted into the pEF6/V5-His TOPO®-cloning vector (Invitrogen, Paisley, UK) and this construct was stably transfected into a hygromycin-tagged derivative of the PEOl ovarian cancer cell line (PEOlhygl.6) using Effectene Reagent (Qiagen, Crawley, UK).
Western blot analysis
Cells grown in monolayer culture were harvested and lysed with lysis buffer supplemented with IX Complete protease inhibitors (Roche, UK) and 0.5 mM phenylmethylsulfonyl fluoride. Following incubation on ice for 15 min, the lysate was centrifuged at 13,000 rpm for 5 min, and the postnuclear supernatant was harvested and sampled for quantitation of protein concentration, using the BioRad protein reagent. Forty micrograms of the lysate were then mixed with SDS-PAGE sample buffer, boiled for 5 min, and subjected to electrophoresis in 10% SDS-PAGE gels under reducing conditions. The separated proteins were electrophoretically transferred to Trans-Blot transfer membrane (BioRad, UK). Blots were incubated with primary anti-WWOX (a polyclonal antibody, raised in rabbit using as antigen a fusion protein of GST bound to the WW-domains of WWOX) or anti-GAPDH (Abeam, Cambridge UK) antibodies overnight at 4°C. Immuno-complexes were visualized with the BM chemiluminescence detection kit (Roche) using horseradish peroxidase-conjugated secondary antibodies. Quantitative values for WWOX and GAPDH proteins were obtained by densitometric analysis using a gel scanner (UVP Life Sciences, Cambridge, UK) and analyzed by Labworks gel analysis software (UVP Life Sciences, Cambridge, UK). This provided integrated absorbance values.
Preparing RNA from transfected PEOl cell lines
DNase I-treated total RNA from the transfected PEOl cell lines was prepared using the Absolutely RNA RT-PCR Miniprep Kit (Stratagene, CA). First strand cDNA was prepared from 2 μg of DNase-treated RNA using a first strand cDNA synthesis kit (Roche).
Preparing RNA from human tissues
Primary ovarian tumour material and non-malignant tissues were obtained from patients undergoing gynaecological surgery in Lothian University Hospitals NHS Trust, Scotland, UK. Institutional ethical approval was granted for this work by the Lothian University National Health Service Trust Medicine/Clinical Oncology Research Ethics Subcommittee. Tissue samples were excised, transferred on ice for section then transferred into liquid nitrogen. Eighty-three consecutive tumour samples were taken from this bank for analysis. Twelve were excluded before analysis: 3 on histological grounds (primary peritoneal, cystadenoma, pseudomyxoma peritonei); 4 samples were not obtained at the time of primary surgery; extraction of RNA was unsatisfactory in 2 cases; 2 patients had concurrent malignancies at the time of diagnosis and 1 patient had no available clinical information. Tumour characteristics of the remaining 71 tumours are described in Table I.
Table I.
Grade, stage and histology of 71 epithelial ovarian tumours.
| Tumour characteristic | No. of patients |
|---|---|
| Grade | |
| 1 | 3 |
| 2 | 16 |
| 3 | 49 |
| Unknown | 3 |
| Stage | |
| I | 11 |
| II | 6 |
| III | 41 |
| IV | 10 |
| Unknown | 3 |
| Histology | |
| Serous papillary | 36 |
| Endometrioid | 13 |
| Clear cell | 10 |
| Mucinous | 6 |
| Mixed serous papillary/Endometrioid | 4 |
| Unknown | 2 |
Thirteen non-malignant tissue samples were taken from our tissue bank for analysis. These patients underwent bilateral oophorectomies for suspected malignancy but were found to have various benign histologies such as serous cystadenoma, fibrothecoma, ovarian fibroma, endometriosis and salpingitis. On each occasion, the apparently normal contralateral ovary was used for our analysis. DNase I-treated total RNA and first strand cDNA were prepared as for the transfected PEO1 cell lines.
Non-quantitative RT-PCR
Non-quantitative RT-PCR was performed to analyse WWOX expression in normal ovary and ovarian tumour samples. Reactions (25 (μl) contained 2 μl first strand cDNA, 200 nM dNTPs, 1X PCR buffer, 1 U Taq Gold (Applied Biosystems, UK) or Pic Taq (Cancer Research UK), MgCl2 and 0.2 μM (Taq Gold) or 0.8 μM (Pic Taq) of each primer (magnesium concentrations, primer sequences and cycling conditions in Table II). Initiation steps of 95°C for 12 min (Taq Gold) and 94°C for 3 min (Pic Taq) were used pre-cycling.
Table II.
PCR primers and conditions.
| PCR | Primers | Sequences (5′-3′) For/Rev | [Mg2+]/Taqa | Cycling conditions |
|---|---|---|---|---|
| Non-quantitative RT-PCR | 8F2+Z2 (exons 8 and 9) | ACTATTGGGCGATGCTGGCT CGTTCTTGGATCAGCCTCTC |
2.5 mM Gold | 95°C 30 see; 65°C 30 sec; 72°C 45 sec; 40 cycles TDb |
| 7F2+8R2 (exons 7 and 8) | CACCAAAGATGGCCTGGA TGGACCTGTTATAAGCCAGCATCG |
2.0 mM Pic | 94°C 30 sec; 65°C 30 sec; 72°C 30 sec; 30 cycles TD | |
| Ex4/4+Z2 (exons 4-9) | TTTCACTGGCAAAGTGGTTG CGTTCTTGGATCAGCCTCTC |
2.5 mM Gold | 95°C 30 see; 57°C 30 sec; 72°C 75 sec; 35 cycles | |
| Ex1/1+CodR (5′UTR-3′UTR) | GAGTTCCTGAGCGAGTGGAC ACTTTCAAACAGGCCACCAC |
2.5 mM Gold | 95°C 30 sec; 57°C 30 sec; 72°C 75 sec; 35 cycles | |
| Exon-specific PCR on genomic DNA | Exon 1F+R (exon 1) | GGAGACTGGATTTCAGCTTC CCCTGGACCCTTTCCCT |
1.6 mM Pic | 94°C 30 sec; 65°C 30 sec; 72°C 30 sec; 30 cycles TD |
| Exon 2F+R (exon 2) | GTCCTCTTTCTCCTTCTTCC CAATAACCTGTCACCTCTCT |
1.6 mM Pic | 94°C 30 sec; 55°C 30 sec; 72°C 30 sec; 35 cycles | |
| Exon 3F+R (exon 3) | GTCTTTACTTCTCCCTGGCACC GCGGGGAAAATAGAAGAATA |
1.6 mM Pic | 94°C 30 sec; 56°C 30 sec; 72°C 30 sec; 35 cycles | |
| Exon 4F+R (exon 4) | CTTTCTCTTTTGGGCAGC GCAGTCCCAAAGATAAATAAC |
1.6 mM Pic | 94°C 30 sec; 58°C 30 sec; 72°C 30 sec; 35 cycles | |
| Exon 5F+R (exon 5) | AGGACTCTACCCCACAAC ACACACTCCACTGAAATC |
2.0 mM Pic | 94°C 30 sec; 68°C 30 sec; 72°C 30 sec; 40 cycles TD | |
| Exon 6F+R (exon 6) | ATTAAACAGGGGAATTCCGAC TCTCCCAATTGTGTTCATCTG |
1.6 mM Pic | 94°C 30 sec; 63°C 30 sec; 72°C 30 sec; 30 cycles TD | |
| Exon 6aF+R (exon 6a) | TAGGAGGTGTTGGAAGAAGG CACCTGAAGAGTCGTAAAGC |
1.6 mM Pic | 94°C 30 sec; 56vC 30 sec; 72°C 30 sec; 35 cycles | |
| Exon 7F+R2 (exon 7) | ACATCCATGGATCCCGAAG TGATTCACTTGAAAGGTGGTCT |
1.6 mM Pic | 94°C 30 sec; 55°C 30 sec; 72°C 30 sec; 40 cycles | |
| Exon 7F2+R (exon 7) | CACCAAAGATGGCCTGGA TGGTATGAGAAAGGGGATAAGTG |
1.6 mM Pic | 94°C 30 sec; 65°C 30 sec; 72°C 30 sec; 30 cycles TD | |
| Exon 8F+R (exon 8) | TGCACCCAGCATTCCTTAGATTTCC ACCAGACTCATGCCCGCAAG |
1.6 mM Pic | 94°C 30 sec; 65°C 30 sec; 72°C 30 sec; 30 cycles TD | |
| Exon 9F+R (exon 9) | GACGCCATCTCATCACTCC TTTACTTTCAAACGGCCACC |
1.6 mM Pic | 94°C 30 sec; 65°C 30 sec; 72°C 30 sec; 40 cycles TD | |
| Quantitative real-time RT-PCR | β-actin F+R | CTACGTCGCCCTGGACTTCGAGC GATGGAGCCGCCGATCCACACGG |
SYBR | 95°C 15 sec; 57°C 60 sec; 85°C 15 sec (aquire); 40 cycles |
| 8F2+Z2 (exons 8 and 9) | ACTATTGGGCGATGCTGGCT CGTTCTTGGATCAGCCTCTC |
SYBR | 95°C 30 sec; 67°C 30 sec; 72°C 45 sec; 85°C 15 sec (aquire); 45 cycles TD | |
| Δ6-8 F+R (exons 4 and 5/9 junct) | GGTTGTGGTCACTGGAGCTAA CAGCTCCCTGTTGCCATTC |
SYBR | 95°C 15 sec; 67vC 60 sec; 78°C 15 sec (aquire); 45 cycles TD |
Primer sequences, magnesium concentrations, type of Taq DNA polymerase and cycling conditions used for non-quantitative RT-PCRs, exon-specific PCRs on genomic DNA and quantitative real time RT-PCR.
Gold, Taq Gold; Pic, Pic Taq; SYBR, SYBR Green PCR Mastermix (containing MgCl2).
TD, annealing temperature decreased by 1°C per PCR cycle for first 10 cycles.
Exon-specific genomic PCR
Genomic DNA from the two tumours that did not express WWOX variant 1 on the basis of non-quantitative RT-PCR was screened for possible homo-zygous deletions by PCR amplification of individual WWOX exons. Reaction conditions as above and primers as in Table II.
Quantitative real-time RT-PCR
Quantitation of mRNA levels for β-actin, WWOX variant 1 (primers 8F2 and Z2) and WWOX variant 4 (primers Δ6-8F and Δ6-8R) was carried out in quadruplicate 20 μl reactions containing 1X SYBR Mastermix (Applied Biosystems), either 0.2 μl (β-actin and WWOX variant 1) or 1 μl (WWOX variant 4) first strand cDNA, and the relevant primer pair (listed in Table II). Final concentration of β-actin primers was 200 nM each, of variant 1 specific primers was 400 nM each, and of variant 4-specific primers was 200 nM (forward) and 50 nM (reverse). Reactions were run on the Rotorgene 2000 real-time PCR machine (Corbett Research, Australia), and fluorescence was detected using the FAM channel (source 470 nm; detector 510 nm). The PCR conditions for each primer pair are given in Table II, and all included a 15 min Taq activation step at 95°C before cycling, and a 4 min step at 72°C and a melt curve post-cycling. Specificity for variant 4 was obtained by designing the reverse primer to anneal across the exon 5-9 splice junction that is unique to the variant 4 transcript. Previous studies have used a similar reverse primer and found no cross-amplification from variant 1 (full length) cDNA (14). To ensure that our variant 4-specific primers did not misprime from variant 1 transcript, a mispriming control was included in all PCRs. This control was derived by transfecting a variant 1-over-expressing plasmid into PEOl cells, which express no endogenous variant 1 or variant 4. Variant 1 expression in this control line was in excess of that in the tumour samples (data not shown). A negative result in this control sample would thus indicate that the variant 4-specific PCR primers cannot misprime from the levels of variant 1 present in the tumours.
A standard curve was included in each run in triplicate and was prepared from four serial dilutions of cell line cDNA. For a standard curve to be accepted the best fit line had to have an R-value >0.99. Expression levels were extrapolated from the standard curve and normalised relative to β-actin expression for each sample. Selected products were run on a 2% agarose gel to confirm band size. To confirm sequence identity, selected samples were purified by treatment with Exonuclease I and Shrimp Alkaline Phosphatase (Amersham Pharmacia, UK), sequenced with ABI Prism® Big Dye and precipitated, according to manufacturer's instructions (Applied Biosystems).
Statistical analysis
Analyses for clinicopathological associations were conducted using Fisher's exact test, Mann-Whitney test and linear regression. These analyses were performed using SPSS Version 10 (SPSS Inc., USA) and the Analyse-it plug-in (Analyse-it Software Ltd., UK) for Microsoft Excel. Univariate analysis was performed comparing clinicopathological factors and WWOX variant expression to survival. All parameters found to be significant at the univariate level were included in the multivariate Cox regression analysis (forward stepwise likelihood ratio method; entry probability 0.05; removal probability 0.1).
Results
Low level WWOX expression is detectable by real-time RT-PCR but not by immunoblotting
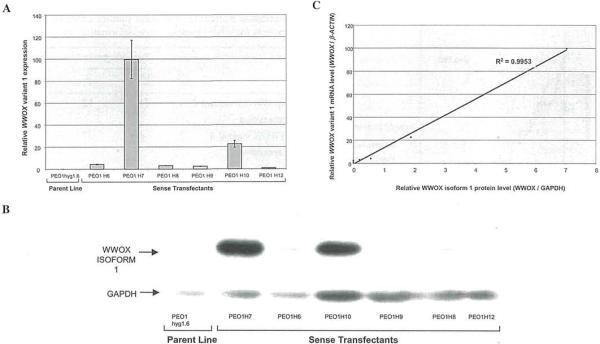
To assess the sensitivity of real-time quantitative RT-PCR and immunoblotting for measuring WWOX expression we used the PEOlhygl.6 cell line. PEOlhygl.6 cells are homozygously deleted for exons 4-8 of the WWOX gene, and thus express no full-length WWOX. We transfected these cells with a construct containing the WWOX variant 1 coding region, and quantified WWOX variant 1 mRNA and protein expression in the resultant transfectants by real-time RT-PCR and Western blot, respectively. Real-time RT-PCR detected variant 1 expression in 6 out of 6 of the transfectants but not in the parent line (Fig. 1 A). No WWOX variant 1 expression was detected in 3 vector-transfected controls (data not shown). Western blot analysis detected a single band corresponding to isoform 1 expression in 4 out of the 6 sense transfectants (Fig. 1B), and there was a strong linear correlation between WWOX mRNA and protein levels in this transfected cell line system (R2=0.995; Fig. 1C). The two transfectants in which protein was not detected were the lowest expressers of WWOX variant 1 mRNA by real-time RT-PCR (amplifying after 20-22 cycles when using 0.2 μl of first strand cDNA in the RT-PCR). This suggests that the sensitivity of the WWOX immunoblotting is less than the real-time RT-PCR. Due to its greater sensitivity we therefore used quantitative RT-PCR to investigate the WWOX expression profile in ovarian cancer.
Figure 1.
(A), Graph showing the expression of WWOX variant 1 mRNA in transfected cell lines. Relative WWOX variant 1 expression (calculated as the ratio of WWOX to β-ACTIN mRNA levels) was measured by quantitative real-time RT-PCR using the Rotorgene 2000. PEO1 hygl .6 cells were the parent cell line used for transfection, and express no variant 1. The sense transfectants (H6, H7, H8, H9, H10 and H12) were generated by transfection of PEOhygl.6 cells with a construct containing the WWOX variant 1 coding region, and express varying levels of variant 1 mRNA. (B), Graph showing the expression of WWOX isoform 1 protein in transfected cell lines. Protein lysate (40 μg) from each cell line was separated by electrophoresis on 10% SDS-PAGE gels, and was transferred to Trans-Blot membrane. WWOX isoform 1 was detected using a rabbit anti-WWOX antibody. GAPDH was detected using an anti-GAPDH antibody and was used as a loading control. No WWOX variant 1 protein was detectable in the parent line PEO 1 hygl.6 or the sense transfectants H9 and HI2. Variable levels of variant 1 expression were detected in the H6, H7, H8 and H10 transfectants. (C), Graph comparing WWOX variant 1 mRNA and protein levels in the transfected cell lines. Relative WWOX variant 1 mRNA expression (calculated as the ratio of WWOX to β-ACTIN mRNA levels) was measured by quantitative real-time RT-PCR using the Rotorgene 2000. Relative WWOX isoform 1 protein expression (calculated as the ratio of WWOX to GAPDH protein levels) was measured by densitometry from immunoblots. A strong linear correlation between variant 1 mRNA and protein levels (R=0.9953) is shown.
Full-length WWOX variant 1 is absent in two ovarian tumours
Non-quantitative RT-PCR using primers amplifying between exons 8 and 9 (primers 8F2 and Z2) revealed a product for all of the normal ovarian samples and 69 out of 71 tumour samples. The lack of full-length WWOX expression in the remaining two tumours was confirmed using two further primer sets amplifying between exons 7 and 8 (primers 7F2 and 8R2) and exons 4 and 9 (primers Ex4/4 and Z2). RT-PCR amplification across the whole open reading frame for these two tumours (using primers Ex 1/1 and CodR) revealed a truncated product of ~950 bp for one of the tumours and no product for the other tumour. We were unable to successfully clone the 950 bp product, and its size does not appear to match any known splice variants. Exon-specific PCR from genomic DNA revealed that all the exons were present in both cases, ruling out the possibility of homozygous deletion.
Full-length WWOX variant 1 expression is reduced in ovarian tumours as compared to normal ovarian tissues
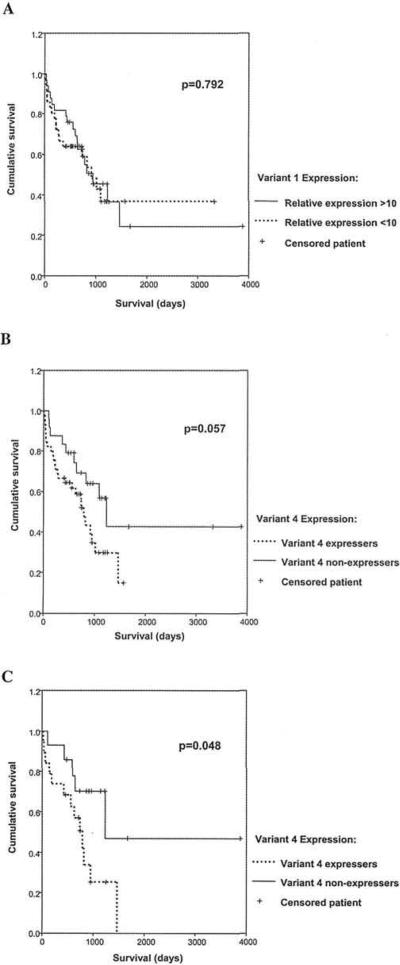
The relative WWOX variant 1 expression (WWOX/β-ACTIN), as determined by quantitative real-time RT-PCR, varied From 9.3 to 61.6 (median 22.9) in the 69 normal ovarian tissues and from 0 to 58.7 (median 9.57) in the ovarian tumours. Neither group of samples had a normal distribution of variant 1 expression (Fig. 2) but the median WWOX variant 1 expression was highly significantly reduced in the ovarian tumours compared to the normal tissues (p<0.0001; Mann-Whitney test). The level of WWOX variant 1 expression was not associated with any particular clinicopathological factor, but was decreased across all the tumour subtypes, suggesting that reduced WWOX variant 1 expression may be an early event in ovarian tumourigenesis. There was no significant difference in survival between high (actin-corrected level >10) and low (actin-corrected level <10) WWOX variant 1 expressers (Fig. 3A).
Figure 2.
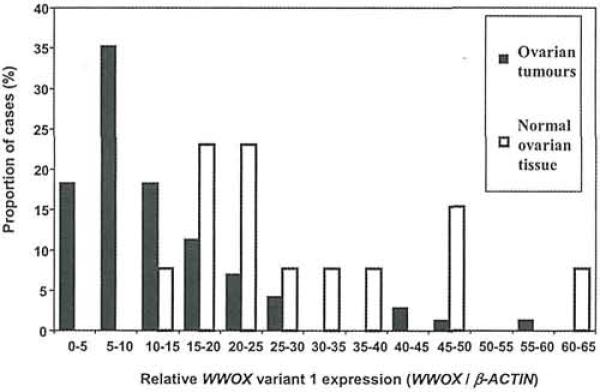
Graph showing WWOX variant 1 mRNA expression in 71 ovarian tumours (black bars) and 13 normal ovarian tissues (white bars). Relative WWOX variant 1 expression (calculated as the ratio of WWOX to β-ACTIN mRNA levels) was measured by quantitative real-time RT-PCR using the Rotorgene 2000. The median WWOX variant 1 expression is significantly lower in the tumours than in the normals (p<0.0001, Mann-Whitney test).
Figure 3.
Patient survival according to WWOX expression. Relative VKWOX variant 1 and variant 4 expression (calculated as the ratio of WWOX to β-ACTIN mRNA levels) was measured by quantitative real-time RT-PCR using the Rotorgene 2000. (A), Patients were divided into two groups with relative variant 1 expression levels >10 and <10. No significant difference in survival between the two groups was apparent. (B), Patients were divided into two groups, variant 4 expressers and variant 4 non-expressers. Patients expressing variant 4 showed a trend towards poorer survival, but this did not reach significance. (C), 33 patients whose tumours expressed high relative WWOX variant 1 levels (>10) were divided into two groups, variant 4 expressers and variant 4 non-expressers. Patients with high levels of WWOX variant 1 who co-expressed variant 4 showed a significantly poorer survival than those with no variant 4 (p=0.048).
WWOX variant 4 expression in ovarian tumours is associated with stage, grade and poorer survival
To determine the expression of WWOX variant 4 in the 69 ovarian tumours, we performed non-quantitative RT-PCR on first strand cDNA using primers amplifying between exons 4 and 9 (primers Ex4/4 and Z2). This primer pair can amplify from both variant 1 and variant 4, producing products of 860 and 321 bp, respectively. The 321 bp variant 4 product lacking the exons 6-8 sequence was present in >50% of the tumour samples. In addition, two other PCR products, with sizes intermediate between the variant 1 and variant 4 products, were identified in a total of 12 out of 71 tumours (17%). Based on size criteria, these were assumed to be transcripts with deletions of exons 7 and 8 (~410 bp) and of exon 7 alone (~675 bp).
Thus, as was previously reported by us, WWOX variant 4 appears to be the most common alternate splice variant in ovarian tumours. However, to eliminate any possible interference in the PCR due to the competitive amplification of multiple variants, we developed a highly specific real-time RT-PCR for the detection of variant 4 using the Rotorgene 2000 (see Materials and methods). We utilised a primer pair (Δ6-8F and Δ6-8R) of which the reverse primer was designed to anneal to the exons 5-9 splice boundary that is unique to the variant 4 transcript. We routinely used a misprinting control to ensure that this variant 4-specific RT-PCR did not generate false positives by misprinting from variant 1 transcript (see Materials and methods). Expression of WWOX variant 4 was detected in 45 out of 71 (63%) ovarian tumour samples and was generally very low, amplifying after 32-39 cycles of PCR when using 1 μl of first strand cDNA per reaction. By comparison, variant 1 amplified significantly after 19-32 cycles, when using 0.2 μl of cDNA per reaction. The level of variant 4 expression in the tumours is lower than the standard curve generated for this RT-PCR, making quantitation of variant 4 levels unreliable. Nonetheless, this study accurately and sensitively detects the presence of variant 4 transcript in the tumours, and this expression pattern was analysed with respect to clinicopathological factors. There was a significant association between the presence of the variant 4 transcript and high grade ovarian cancer (p=0.006; Fisher's exact test; Table IIIA). The presence of the variant 4 transcript was also significantly associated with advanced stage disease (p=0.012; Fisher's exact test; Table IIIB). There was no correlation between expression of variant 4 and histology of ovarian tumour.
Table III.
A, Contingency tabic showing WWOX variant 4 expression according to ovarian tumour grade (for 66 of 69 patients for whom tumour grade information was available).
| Tumour grade |
Total | ||
|---|---|---|---|
| 1/2 | 3 | ||
| No WWOX variant 4 expression | 11 | 12 | 23 |
| WWOX variant 4 expression | 6 | 37 | 43 |
| Total | 17 | 49 | 66 |
| B, Contingency table showing WWOX variant 4 expression according to ovarian tumour stage (for 66 of 69 patients for whom tumour stage information was available). | |||
|---|---|---|---|
| Tumour stage |
Total | ||
| I/II | III/IV | ||
| No WWOX variant 4 expression | 10 | 14 | 24 |
| WWOX variant 4 expression | 5 | 37 | 42 |
| Total | 15 | 51 | 66 |
P=0.006, Fisher's exact test.
Variant 4 expression in the tumours was also compared with patient survival by Kaplan-Meier analysis. There was a trend towards worse survival in those patients whose tumours expressed WWOX variant 4 but this did not reach significance (p=0.057; Fig. 3B). However, in the patients expressing high levels of variant 1 (actin-corrected level >10), those that co-expressed variant 4 had significantly worse survival compared with those that did not (p=0.048; Fig. 3C). In contrast, expression of variant 4 did not significantly alter the survival in patients expressing low levels of variant 1 (data not shown). Analysis of variant 4 in a multivariate model demonstrated that expression of this variant was not an independent prognostic variable.
WWOX variant 4 is expressed in normal ovary tissue
We repeated the variant 4-specific real-time RT-PCR (primers Δ6-8F and Δ6-8R) on cDNA from the normal ovary samples. WWOX variant 4 expression was surprisingly detected in 9 out of 13 (69%) normal ovarian tissue samples, a frequency similar to that detected in the tumours. The relative WWOX variant 4 expression (WWOX/β-ACTIN) was low and appeared similar in the normal ovarian tissues (from 0 to 0.33) and in the ovarian tumours (from 0 to 3.65), although quantitation of such low expression is unreliable. Expression of variant 4 is thus not limited to malignant tissues.
Discussion
The WWOX gene was identified from a study of human breast cancer, and has subsequently been suggested as a tumour suppressor gene for a number of different tumour types. This is supported by the observations that WWOX is capable of tumour suppression in vivo in a breast cancer cell line model, and its apparent role in apoptosis in murine fibroblast studies. However, inactivating mutation or epigenetic silencing of WWOX in primary tumours is rare, which questions the clinical significance of this gene. Because of this contradiction between the strong functional evidence of WWOX tumour suppression and rare gene inactivation in tumours, we decided to investigate potential alternative mechanisms of WWOX dysfunction in ovarian cancer - namely decreased gene expression and production of alternate WWOX variant splice-forms.
Until very recently, the available antibodies directed against WWOX protein only recognised the full-length variant 1 protein, not any of the shorter forms. Two groups have now generated antibodies that detect WWOX short-form proteins. Watanabe et al (22) could only detect expression of short-form proteins if the proteasome was blocked. They interpreted this as meaning that these isoforms were targeted for destruction and were not biologically active. While this may be true, we show here that the sensitivity of the real-time RT-PCR assay is superior to immunoblotting as evidenced by PEOl WWOX transfectants that express low level WWOX variant 1 mRNA but no detectable protein. We also show that expression of the alternate transcripts is generally orders of magnitude lower than that of variant 1. Thus it may be that the short-form proteins are expressed at levels below the sensitivity of the available antibodies, and that blocking the proteasome could result in the alternative isoforms rising to a level that is detectable by immunoblotting.
We demonstrated a tight linear correlation between WWOX mRNA and protein levels in transfected cells. Based on this, and the greater sensitivity of RT-PCR, we chose to use quantitative real-time RT-PCR to investigate the expression profile of WWOX variant 1 and variant 4 (the predominant alternate variant in ovarian tumours) in a panel of normal ovary tissues and ovarian tumours. We have demonstrated that WWOX variant 1 mRNA expression is significantly reduced in human ovarian tumours compared to normal ovarian tissues, supporting its role as a tumour suppressor gene. In contrast to the rarity of point mutation in the WWOX gene, we found decreased expression of WWOX variant 1 to be highly frequent in ovarian cancer. Tumours in our study showed 87% (60/69) variant 1 expression levels lower than the median expression level of the normal ovaries, and 49% (34/69) of the tumours exhibited less variant 1 expression than the lowest expressing normal ovary sample. Thus reduced WWOX variant 1 expression was apparent in the majority of the ovarian tumours.
In addition, 2/71 tumours expressed no full length variant 1. This frequency is consistent with previous reports describing the absence of variant 1 in 1/36 oesophageal cancers (17), in 2/27 non-small cell lung cancers (18) and in 1/20 breast cancers (20). The latter example, like our two cases, demonstrated no evidence of exonic homozygous deletion.
We have also demonstrated the expression of the variant 4 transcript in 63% of our panel of human ovarian tumours. This compares to a frequency of only 5.5% in oesophageal squamous cell carcinoma (17) and 11.1% in non-small cell lung carcinoma (18), although a further 14.8% of these lung tumours showed other alternate transcripts. It is unlikely that this discrepancy in variant 4 expression frequency is related to the different methods of detection, since we also detected a high frequency of variant 4 expression in ovarian tumours (50%) using a competitive, non-quantitative PCR similar to that used in the earlier studies. The expression of variant 4 mRNA was significantly associated with high tumour grade and advanced stage ovarian carcinoma. There was a trend towards adverse survival in patients who expressed this variant and significantly worse survival in those patients with robust variant 1 expression who co-expressed variant 4.
There are several possible explanations for the association of WWOX variant 4 with adverse clinical parameters. It could represent: i) a surrogate marker of cancer-associated disruption at FRA16D, ii) a surrogate marker of cancer-associated disruption of splicing fidelity, iii) an oncogenic, gain of function WWOX isoform, or iv) a dominant negative WWOX iso-form. WWOX variant 4 predicts a protein lacking the enzymatic oxidoreductase function, but retaining the protein binding WW-domains, and thus suggests possible function as a dominant negative isoform, sequestering binding partners of WWOX isoform 1 and inhibiting its putative tumour suppressor role. In this regard, it is important to note that WWOX iso-form 1 and WWOX isoform 4 have different intracellular locations. While WWOX isoform 4 is located in the nucleus, WWOX isoform 1 is located normally in the cytoplasm, but translocates to the nucleus following TNF-α treatment (14,15). This suggests that low levels of WWOX isoform 4 do not preclude it having a dominant negative effect on the function of WWOX isoform 1, as competition for binding partners may not be on an equal basis.
However, it is also known that alternative splicing variants are commonly associated with cancer. It has been estimated that in normal cells spliceosome errors occur in 2-3% of transcripts (23), and this rises to 10-20% of transcripts in cancer cells (24). Cancer-associated splice variants have been reported for a number of genes including EGFR, CD44 and NER (25). Some of these alternate transcripts have the potential to play a role in tumourigenesis e.g., by inhibiting apoptosis (CD79) (26) or by blocking tumour suppressor activity (BIN 1) (27). Wang et al (28) performed a genome-wide computational screen identifying 26,258 alternative splicing variants, of which 845 were significantly associated with human cancer. It thus seems that cancer is associated with a generalised relaxation of splicing fidelity, perhaps due to mutation or altered expression of components of the splicing apparatus. Our results here reveal low levels of WWOX variant 4 expression in 69% of non-malignant ovarian tissues, a frequency similar to that detected in ovarian tumours. Alternate WWOX transcripts have been noted in normal tissues previously but this has been somewhat understated (20,21). This perhaps argues against a dominant negative hypothesis, and suggests that the variant may be an infrequently produced splice form, even in normal cells.
The role of WWOX variant 4 is clearly speculative at this time and progress is reliant on the demonstration that it is actually translated. In addition, the possible dominant negative role of variant 4 could be tested using the following models: i) transfection of variant 4 into immortalised human ovarian surface epithelial cells that express normal WWOX variant 1, or ii) RNAi knockdown of variant 4 expression in cell lines that are abundant expressers of both WWOX variant 1 and variant 4. These models can then be used to determine whether regulation of variant 4 expression can i) abrogate, or ii) reconstitute WWOX-mediated suppression of tumourigenicity in nude mice.
In summary, we show that the expression of full-length WWOX (variant 1) is significantly and frequently reduced in ovarian tumours compared to normal ovarian tissue. This decreased variant 1 expression is not associated with any particular clinicopathological factor, but is decreased across all the tumour subtypes, suggesting that reduced WWOX variant 1 expression is a frequent and early event in ovarian tumourigenesis. We also demonstrate that the predominant WWOX alternate transcript (variant 4) is significantly associated with advanced ovarian cancer. We report the first evidence that this variant splice form is frequently expressed in non-malignant ovarian tissue, arguing against its involvement in tumourigenesis. These findings taken together strengthen the case for the WWOX gene and full length expression as demonstrating a tumour suppressor role.
Acknowledgements
The authors would like to acknowledge the contribution of Agnes Gallacher (sequencing) and Syvia Rye (clinical data retrieval) to this study. A.J.W.P. and H.G. are supported by a project grant from the Scottish Hospitals Endowments Research Trust (SHERT). The work of the other UK-based authors was funded by Cancer Research UK.
Footnotes
References
- 1.Iwabuchi H, Sakamoto M, Sakunaga H, Ma YY, Carcangiu ML, Pinkel D, Yang-Feng TL, Gray JW. Genetic analysis of benign, low-grade and high-grade ovarian tumors. Cancer Res. 1995;55:6172–6180. [PubMed] [Google Scholar]
- 2.Cleton-Jansen AM, Moerland EW, Kuipers-Dijkshoorn NJ, Callen DF, Sutherland GR, Hansen B, Devilee P, Cornelisse CJ. At least two different regions are involved in allelic imbalance on chromosome arm 16q in breast cancer. Genes Chromosomes Cancer. 1994;9:101–107. doi: 10.1002/gcc.2870090205. [DOI] [PubMed] [Google Scholar]
- 3.Aldaz CM, Chen T, Sahin A, Cunningham J, Bondy M. Comparative allelotype of in situ and invasive human breast cancer: high frequency of microsatellite instability in lobular breast carcinomas. Cancer Res. 1995;55:3976–3981. [PubMed] [Google Scholar]
- 4.Driouch K, Dorion-Bonnet F, Briffod M, Champeme MH, Longy M, Lidereau R. Loss of heterozygosity on chromosome arm 16q in breast cancer metastases. Genes Chromosomes Cancer. 1997;19:185–191. doi: 10.1002/(sici)1098-2264(199707)19:3<185::aid-gcc8>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki H, Komiya A, Emi M, Kuramochi H, Shiraishi T, Yatani R, Shimazaki J. Three distinct commonly deleted regions of chromosome arm 16q in human primary and metastatic prostate cancers. Genes Chromosomes Cancer. 1996;17:225–233. doi: 10.1002/(SICI)1098-2264(199612)17:4<225::AID-GCC4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 6.Nishida N, Fukuda Y, Kokuryu H, Sadamoto T, Isowa G, Honda K, Yamaoka Y, Ikenaga M, Imura H, Ishizaki K. Accumulation of allelic loss on arms of chromosomes 13q, 16q and 17p in the advanced stages of human hepatocellular carcinoma. Int J Cancer. 1992;51:862–868. doi: 10.1002/ijc.2910510605. [DOI] [PubMed] [Google Scholar]
- 7.Chen T, Sahin A, Aldaz CM. Deletion map of chromosome 16q in ductal carcinoma in situ of the breast: refining a putative tumor suppressor gene region. Cancer Res. 1996;56:5605–5609. [PubMed] [Google Scholar]
- 8.Paris PL, Witte JS, Kupelian PA, Levin H, Klein EA, Catalona WJ, Casey G. Identification and fine mapping of a region showing a high frequency of allelic imbalance on chromosome 16q23.2 that corresponds to a prostate cancer susceptibility locus. Cancer Res. 2000;60:3645–3649. [PubMed] [Google Scholar]
- 9.Latil A, Cussenot O, Fournier G, Driouch K, Lidereau R. Loss of heterozygosity at chromosome 16q in prostate adenocarcinoma: identification of three independent regions. Cancer Res. 1997;57:1058–1062. [PubMed] [Google Scholar]
- 10.Li C, Berx G, Larsson C, Auer G, Aspenblad U, Pan Y, Sundelin B, Ekman P, Nordenskjold M, van Roy F, Bergerheim US. Distinct deleted regions on chromosome segment 16q23-24 associated with metastases in prostate cancer. Genes Chromosomes Cancer. 1999;24:175–182. [PubMed] [Google Scholar]
- 11.Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60:2140–2145. [PubMed] [Google Scholar]
- 12.Paige AJ, Taylor KJ, Stewart A, Sgouros JG, Gabra H, Sellar GC, Smyth JF, Porteous DJ, Watson JE. A 700-kb physical map of a region of 16q23.2 homozygously deleted in multiple cancers and spanning the common fragile site FRA16D. Cancer Res. 2000;60:1690–1697. [PubMed] [Google Scholar]
- 13.Mangelsdorf M, Ried K, Woollatt E, Dayan S, Eyre H, Finnis M, Hobson L, Nancarrow J, Venter D, Baker E, Richards RI. Chromosomal fragile site FRA16D and DNA instability in cancer. Cancer Res. 2000;60:1683–1689. [PubMed] [Google Scholar]
- 14.Bednarek AK, Keck-Waggoner CL, Daniel RL, Laflin KJ, Bergsagel PL, Kiguchi K, Brenner AJ, Aldaz CM. WWOX, the FRA16D gene, behaves as a suppressor of tumor growth. Cancer Res. 2001;61:8068–8073. [PubMed] [Google Scholar]
- 15.Chang NS, Pratt N, Heath J, Schultz L, Sieve D, Carey GB, Zevotek N. Hyaluronidase induction of a WW domain-containing oxidoreductase that enhances tumor necrosis factor cytotoxicity. J Biol Chem. 2001;276:3361–3370. doi: 10.1074/jbc.M007140200. [DOI] [PubMed] [Google Scholar]
- 16.Paige AJ, Taylor KJ, Taylor C, Hillier SG, Farrington S, Scott D, Porteous DJ, Smyth JF, Gabra H, Watson JE. WWOX: a candidate tumor suppressor gene involved in multiple tumor types. Proc Natl Acad Sci USA. 2001;98:11417–11422. doi: 10.1073/pnas.191175898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuroki T, Trapasso F, Shiraishi T, Alder H, Mimori K, Mori M, Croce CM. Genetic alterations of the tumor suppressor gene WWOX in esophageal squamous cell carcinoma. Cancer Res. 2002;62:2258–2260. [PubMed] [Google Scholar]
- 18.Yendamuri S, Kuroki T, Trapasso F, Henry AC, Dumon KR, Huebner K, Williams NN, Kaiser LR, Croce CM. WW domain containing oxidoreductase gene expression is altered in non-small cell lung cancer. Cancer Res. 2003;63:878–881. [PubMed] [Google Scholar]
- 19.Ried K, Finnis M, Hobson L, Mangelsdorf M, Dayan S, Nancarrow JK, Woollatt E, Kremmidiotis G, Gardner A, Venter D, Baker E, Richards RI. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation break-points in cancer cells. Hum Mol Genet. 2000;9:1651–1663. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- 20.Driouch K, Prydz H, Monese R, Johansen H, Lidereau R, Frengen E. Alternative transcripts of the candidate tumor suppressor gene, WWOX, are expressed at high levels in human breast tumors. Oncogene. 2002;21:1832–1840. doi: 10.1038/sj.onc.1205273. [DOI] [PubMed] [Google Scholar]
- 21.Ishii H, Vecchione A, Furukawa Y, Sutheesophon K, Han SY, Druck T, Kuroki T, Trapasso F, Nishimura M, Saito Y, Ozawa K, Croce CM, Huebner K. Expression of FRA16D/WWOX and FRA3B/FHIT genes in hematopoietic malignancies. Mol Cancer Res. 2003;1:940–947. [PubMed] [Google Scholar]
- 22.Watanabe A, Hippo Y, Taniguchi H, Iwanari H, Yashiro M, Hirakawa K, Kodama T, Aburatani H. An opposing view on WWOX protein function as a tumor suppressor. Cancer Res. 2003;63:8629–8633. [PubMed] [Google Scholar]
- 23.Skandalis A, Ninniss PJ, McCormac D, Newton L. Spontaneous frequency of exon skipping in the human HPRT gene. Mutat Res. 2002;501:37–44. doi: 10.1016/s0027-5107(02)00013-1. [DOI] [PubMed] [Google Scholar]
- 24.LeHir H, Charlet-Berguerand N, De Franciscis V, Thermes C. 5′-End RET splicing: absence of variants in normal tissues and intron retention in phaeochromocytomas. Oncology. 2002;63:84–91. doi: 10.1159/000065725. [DOI] [PubMed] [Google Scholar]
- 25.Xu Q, Lee C. Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucleic Acids Res. 2003;31:5635–5643. doi: 10.1093/nar/gkg786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cragg MS, Chan HT, Fox MD, Tutt A, Smith A, Oscier DG, Hamblin TJ, Glennie MJ. The alternative transcript of CD79b is overexpressed in B-CLL and inhibits signaling for apoptosis. Blood. 2002;100:3068–3076. doi: 10.1182/blood.V100.9.3068. [DOI] [PubMed] [Google Scholar]
- 27.Ge K, DuHadaway J, Du W, Herlyn M, Rodeck U, Prendergast GC. Mechanism for elimination of a tumor suppressor: aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proc Natl Acad Sci USA. 1999;96:9689–9694. doi: 10.1073/pnas.96.17.9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Lo HS, Yang H, Gere S, Hu Y, Buetow KH, Lee MP. Computational analysis and experimental validation of tumor-associated alternative RNA splicing in human cancer. Cancer Res. 2003;63:655–657. [PubMed] [Google Scholar]