Abstract
Common chromosomal fragile sites are unstable genomic loci susceptible to breakage, rearrangement, and are highly recombinogenic. Frequent alterations at these loci in tumor cells led to the hypothesis that they may contribute to cancer development. The two most common chromosomal fragile sites FRA16D and FRA3B which harbor WWOX and FHIT genes, respectively, are frequently altered in human cancers. Here we report that environmental carcinogens, ultraviolet (UV) light, and Benzo[a]pyrene diol epoxide (BPDE), significantly downregulate expression of both genes. On the other hand, we observe that ionizing radiation (IR) does not affect expression of these genes, suggesting that the effect of repression exerted by UV and BPDE is not just a consequence of DNA damage but may be a result of different signaling pathways triggered by specific DNA lesions. Such downregulation correlates with an induction of an S-phase delay in the cell cycle. Treatment of UV-irradiated cells with caffeine abrogates the S-phase delay while concomitantly overcoming the repression phenomenon. This suggests the involvement of unique cell cycle checkpoint mechanisms in the observed repression. Therefore, it is hypothesized that protracted downregulation of the putative tumor suppressor genes WWOX and FHIT by environmental carcinogens may constitute an additional mechanism of relevance in the initiation of tumorigenesis.
Keywords: fragile sites, UV, BPDE, DNA damage, WWOX, FHIT
INTRODUCTION
Common chromosomal fragile sites are loci that exhibit gaps and breaks during metaphase in cells that have been cultured under conditions of replicative stress such as folate deficiency or treatment with aphidicolin [1]. These sites are normally stable in cultured human cells. However, following induction with replication inhibitors, these sites become hot spots for increased sister chromatid exchanges, translocations, and deletions [1,2]. The instability at such loci in tumor cells and their frequent alteration [3] led to the hypothesis that they may contribute to cancer development. Although the exact number of common fragile sites is a matter of interpretation, more than 70 aphidicolin-induced common fragile sites have been reported. Gaps and breaks at just 20 chromosomal fragile sites, however, represent over 80% of all fragile lesions observed in lymphocytes following treatment with low doses of aphidicolin [1]. FRA3B at 3p 14.2 stands out as the most active fragile site in the human genome, followed by 16q23 (FRA16D), 6q26 (FRA6E), 7q31.2 (FRA7G), and Xp22.3 (FRAXB) [4]. A recent study suggested that FRAXB, FRA3B, FRA7G, FRA7H, FRA16D, and their associated genes are unstable in cancer cells [5]. The frequency of gaps and breaks that occur after aphidicolin treatment varies in the population and appears to be determined by the combination of genetics and exposure to environmental carcinogens (reviewed in References [6,7]). This is exemplified by the fact that smokers show a higher level of expression of common chromosomal fragile sites than nonsmokers and by the association of allele losses at specific fragile sites (e.g., FRA3B) and smoking related cancer [8–11].
The fragile histidine triad (FHIT) gene was isolated from the FRA3B locus [12]. FHIT protein expression is reduced or absent overall in 60% of tumors, including lung, breast, esophagus, stomach, and bladder cancers (reviewed in Reference [13]) because of genomic deletions [10] or epigenetic changes, including 5’ CpG island methylation [14,15]. Recently, we have cloned a WW domain containing oxidoreductase (WWOX) gene from chromosome region 16q23.3–24.1 spanning the region of common fragile site FRA16D [16]. WWOX expression has been examined in a variety of tumors and cancer-derived cell lines and loss or reduced expression, aberrant transcripts as well as genomic deletions were observed in breast [16], ovary [17], esophageal [18], lung [19], liver cancers [20], and leukemias [21] among other tumor types. WWOX has been demonstrated to behave as a putative tumor suppressor [22,23]. In general, common chromosomal fragile sites do not have high sequence similarities and, therefore, specific sequences do not appear to be responsible for their fragile nature [6]. However, the two fragile genes FHIT and WWOX have common features; both span huge genomic loci of more than 1 Mb in size including the fragile regions, show frequent altered expression in cancers, and are suspected tumor suppressor genes [6,23]. A recent examination of the coordinate expression of the WWOX and FHIT genes in hematopoietic disorders provided evidence that loss or alteration of expression of the two genes can occur concordantly in hematopoietic tumors by epigenetic mechanisms such as histone modifications (methylation and deacetylation) at these fragile loci [21]. Additionally, a strong correlation was observed in loss of expression between WWOX and FHIT in invasive breast tumors [24].
Analyses of the mechanism of fragile site induction revealed that fragile sites are late replicating in S-phase and fragile site-inducers delay replication even more [25–27]. If replication is delayed at several different segments of a fragile region, cells would enter the G2/M phase of the cell cycle with such chromosomal segments still unreplicated leading to the formation of gaps and breaks, a phenomenon that would be enhanced by aphidicolin [6]. Of note is a recent demonstration that fragile site induction by aphidicolin does not inhibit expression of fragile site gene FHIT [28]. Loss of fragile site gene expression, in cancer, has been mostly attributed to allelic deletion, chromosomal translocation or gene inactivation as a result of exposure to environmental carcinogens [29]. However, no information is available on the regulation of expression of fragile site genes in cells following DNA damage by environmental carcinogens, some of which do not induce chromosomal fragility. Therefore, here we report on the expression pattern of WWOX and FHIT following DNA damage by ultraviolet (UV) light and benzo[a]pyrene diol epoxide (BPDE), two ubiquitous environmental carcinogens that induce bulky DNA adducts.
MATERIALS AND METHODS
Cell Lines and Treatments
MCF-7 and Saos-2 cells were maintained in complete IMEM (IMEM (Biosource, Rockville, MD) with 5% FBS and Gentamycin (50 µg/mL) and complete DMEM (Cambrex, Walkersville, MD) with 10% FBS Gentamycin (50 µg/mL), respectively. For UV irradiation, monolayers of 60%–70% confluent cells were washed once with phosphate buffered saline (PBS) and irradiated with UV-C emitted by five narrow band germicidal lamps emitting predominantly 254 nm. For protracted (three doses) UV-C exposure (10 J/m2 each time), cells were allowed to recover for 24 h between exposures. Cell survival was monitored by counting viable cells (Trypan blue-excluding) 24 h posttreatment. For ionizing (X-ray) irradiation, monolayers of cells at 60%–70% confluency were irradiated in RS2000 X-ray Irradiator (RadSource Technologies, Inc., Boca Raton, FL). Control cells were sham-irradiated in both the UV-C and ionizing radiation (IR) experiments. For BPDE treatment, (±)-7r,8t-dihydroxy-9,10t-oxy-7,8,9,10-tetrahydrobenzo[a] pyrene (BPDE) was obtained from ChemSyn Laboratories (Lenexa, KS), and stock solutions were prepared in tetrahydrofuran as previously described [30]. The integrity of BPDE stock solutions was checked immediately prior to use by a spectrophotometric assay [30]. Because BPDE is extremely labile in aqueous solution, the carcinogen was added to an aliquot of serum-free IMEM (at a dilution of 1/300) within 5 s of addition of the IMEM to the 60%–70% confluent monolayers of MCF-7 cells, which were previously rinsed once with PBS; final concentration of BPDE was 0.5 µM. Control cells were treated in 1/300 dilution of solvent in serum-free IMEM. Thirty minutes later, the serum-free medium was removed, the cells were rinsed once with PBS, and complete IMEM was added. For caffeine treatment, MCF-7 cells were pretreated for 1 h with complete IMEM containing 5 or 10 mM caffeine (Sigma, St. Louis, MO) prior to UV-C irradiation. The cells were maintained in the caffeine-containing complete IMEM after UV-C irradiation until harvested for RNA or for flow cytometry. Control cells were sham-treated in complete IMEM without caffeine. For cell cycle progression analysis, MCF-7 cells were grown in serum free IMEM for 96 h to arrest cells in G1/G0 phase. Cell growth stimulated by the addition IMEM containing 5% FBS for the indicated time points until harvested for RNA.
RNA Isolation and Northern Blot Analysis
Total RNA was isolated 24 h after treatments from the cells with TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Northern blots of total RNA were prepared with standard procedures. A 1200 bp BamHI-EcoRI restriction fragment of WWOX cDNA clone, spanning the whole amino acid-coding region, was used as probe. For FHIT, a 300 bp cDNA was used as probe. For p21, a 500 bp cDNA was used and for human GAPDH, 1200 bp cDNA was used as probe. The cDNA probes were labeled with [32P]dCTP with random priming (Prime It II; Stratagene, La Jolla, CA). The membranes were hybridized at 42°C overnight in hybridization buffer (10% Dextran Sulfate, 50% Formamide, 5× SSC, 1× Denhardts, 20 mM Tris (pH 7.5) 20 µg/mL herring sperm DNA) followed by washing once with 2x SSC/0.5%SDS and twice with 0.5 × SSC/0.5%SDS at 60°C. Washed membranes were exposed to X-ray film to develop the signals.
Western Blot Analysis
At the end of treatment protocols, cell monolayers were washed twice with PBS and lysed with RIPA buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl 0.5% Na-deoxycholate, 1% NP40, 0.1% SDS, and protease inhibitors (Roche, Indianapolis, IN). Protein samples were resolved in SDS-PAGE and transferred to PVDF membranes (Millipore, Billerica, MA) with standard protocols. Antibody treatment and signal detection were performed with Protein Detector™ Western blotting Kits (KPL, Inc., Gaithersburg, MD), as per manufacturer’s protocol. The affinity purified primary rabbit polyclonal antibody used against WWOX (pure concentration at 280 ng/mL) was developed at our laboratory [23] and was used at a 1:500 dilution and that against actin (ICN, Aurora, OH) was at 1:10 000 dilution. Appropriate Horseradish peroxidase-labeled secondary antibodies were used. The FHIT antibody used for Western analysis was a kind gift of Dr. Kay Huebner, Thomas Jefferson University, Kimmel Cancer Center.
[3H]-Thymidine Uptake
Control, caffeine-, and UV-treated MCF-7 cells were analyzed for DNA synthesis by adding 1 µCi/mL of [3H]-Thymidine (6.7 Ci/mmol; ICN) for a period of 4 h prior to the end of the time point. Cells were then washed twice with cold PBS and fixed and washed twice in ice-cold 10% Trichloroacetic acid for 5 min each. The fixed cells were lysed and solubilized in 0.5NNaOH. Aliquots were counted in scintillation counter.
Flow Cytometry
Cells were harvested with trypsin, pelleted, and washed with PBS. The cells were fixed with 70% ethanol at room temperature for 20 min and resuspended in PBS containing propidium iodide (25 µg/mL), RNAse-A (500 µg/mL), and Triton X-100 (0.5%). Cell cycle analysis was performed with Coulter® EPICS® Elite flow cytometer.
RESULTS
UV and BPDE but not Ionizing Radiation (IR) Downregulate Fragile Site Gene Expression
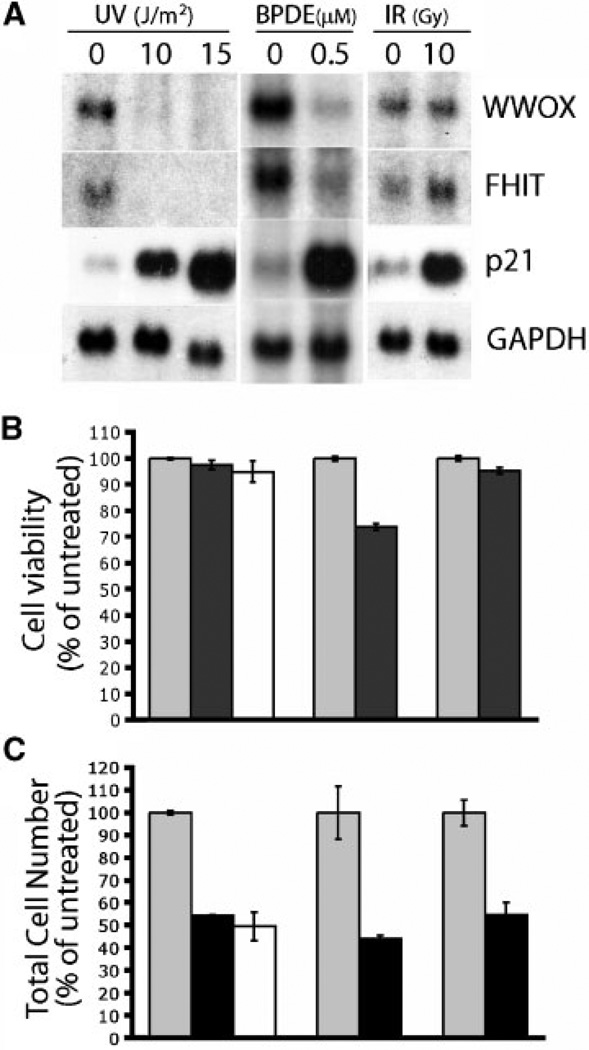
MCF-7 breast cancer cells were chosen as a model system for these studies because WWOX is abundantly expressed in this cell line. To determine the effect of UV-C irradiation on WWOX as well as FHIT expression, the cells were irradiated with UV-C at doses of 10 and 15 J/m2, total RNA was isolated and Northern blot hybridization carried out. Interestingly, as compared to control, WWOX and FHIT mRNA levels dramatically decreased by 24 h in the UV-irradiated samples (Figure 1A). At the moderate UV-C doses tested [31–34], cell number decreased about twofold at 24 h postirradiation (Figure 1B) without a significant decrease in cell viability (Figure 1C). As expected [31–33], the p21 mRNA level increased as a consequence of DNA damage by UV-C (Figure 1A). Thus, the decrease in cell number seen at 24 h is because of growth arrest rather than cell death (see below), which generally requires higher doses of UV-C [31,35]. Next we asked if IR, which causes DNA double strand breaks, also caused a similar effect on WWOX and FHIT expression. To test this, MCF-7 cells were irradiated with IR at a dose (10 Gy) approximately equitoxic to 10 J/m2 of UV-C radiation (Figure 1A). Northern analysis showed that IR-induced a DNA damage response as expected [32,36] as shown by the robust increase in p21 mRNA levels (Figure 1A), but did not cause a significant reduction of WWOX or FHIT transcripts. FHIT expression was observed to slightly increase in IR treated cells, however, the significance of this is unknown. Higher doses of IR (20, 30, and 40 Gy) were also ineffective in decreasing WWOX and FHIT mRNA levels, whereas increasing doses of UV (20 and 30 J/m2) caused downregulation of WWOX and FHIT transcripts similar to that obtained with 10 and 15 J/m2 (data not shown). Similar to UV, treatment of MCF-7 cells with BPDE, another environmental carcinogen which induces DNA bulky adduct formation, also caused a dramatic decrease of WWOX and FHIT transcripts at 24 h post-BPDE treatment (Figure 1A).
Figure 1.
(A) UV and BPDE treatment but not IR downregulate expression of fragile site genes, WWOX and FHIT. MCF-7 cells were exposed to UV, BPDE, and IR, as described in Materials and Methods. Northern blots of total RNA (20 µg per lane) were hybridized with probes for WWOX, FHIT, p21, and GAPDH genes, as indicated. (B) Cell viability of treated cells. Cell viability was determined by trypan-blue exclusion and is expressed as a percentage of untreated controls. (C) UV, BPDE, and IR treatment resulted in cell growth arrest. Total viable cell number was determined by trypan-blue exclusion 24 h posttreatment. Each treatment was performed in triplicate and is represented as the average ±SD.
Downregulation of Expression of Fragile Site Genes Is p53-Independent
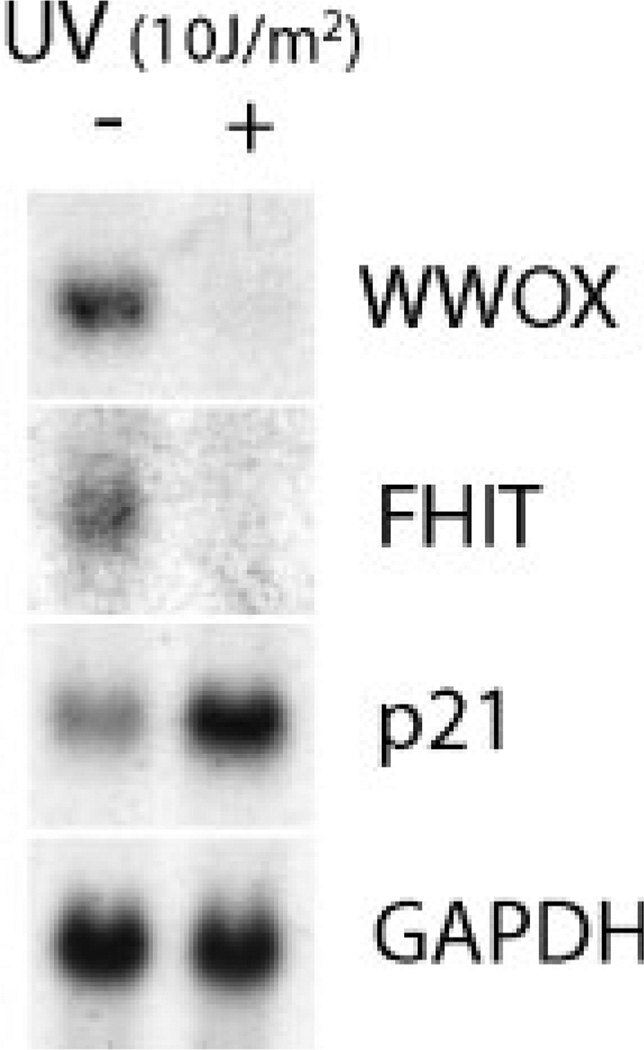
MCF-7 cells have endogenous wild-type p53 protein. In order to ascertain the role, if any, of p53 in the decreased expression of fragile site genes following UV treatment, we tested the p53-null Saos-2 cell line. Similarly to MCF-7 cells, UV irradiation downregulated the levels of FHIT and WWOX transcripts in Saos-2 cells, indicating that the repressive effect of UV on the fragile site genes was a p53-independent phenomenon (Figure 2). The increase in p21 mRNA level in Saos-2 can be attributed to a p53-independent mechanism as suggested by others [37].
Figure 2.
Downregulation of fragile site gene expression by UV is p53-independent. UV treatment of Saos-2 cells and Northern blot hybridization was carried out as mentioned for Figure 1. Note that WWOX and FHIT transcripts were decreased upon UV irradiation.
Repeated UV Irradiation Reduces the Level of WWOX Protein
In UV-irradiated MCF-7 cells, in spite of a dramatic reduction in the mRNA levels of WWOX, the protein expression level did not decrease considerably at 24 h. A considerable decrease in the protein expression was observed following multiple UV-C irradiations (24 h apart) at later time points (48 and 72 h), as shown by Western blot analysis (Figure 3). This suggests that the WWOX protein is relatively stable, and repeated genotoxic insults are required to cause a considerable decrease in protein levels. With the same Western blot membranes, we also tested the FHIT protein levels. However, we did not observe any significant reduction in levels of this protein (data not shown). The reason for a lack of reduction of FHIT levels, again could be because of a protein stability differences at the limited number of time points tested.
Figure 3.
Repeated UV irradiation decreases WWOX protein level. Total protein extracts were made from MCF-7 cells irradiated once (1×), twice (2×) each at 24 h intervals or thrice (3×) each at 24 h intervals or left untreated (−), at indicated time points, 50 µg of protein was resolved in 10% SDS-PAGE, transferred to PVDF membrane and probed with antibodies. Note a progressive decrease in WWOX protein levels upon repeated UV irradiations.
S-Phase Checkpoint Mechanisms May be Responsible for the Downregulation of Expression of Fragile Site Genes
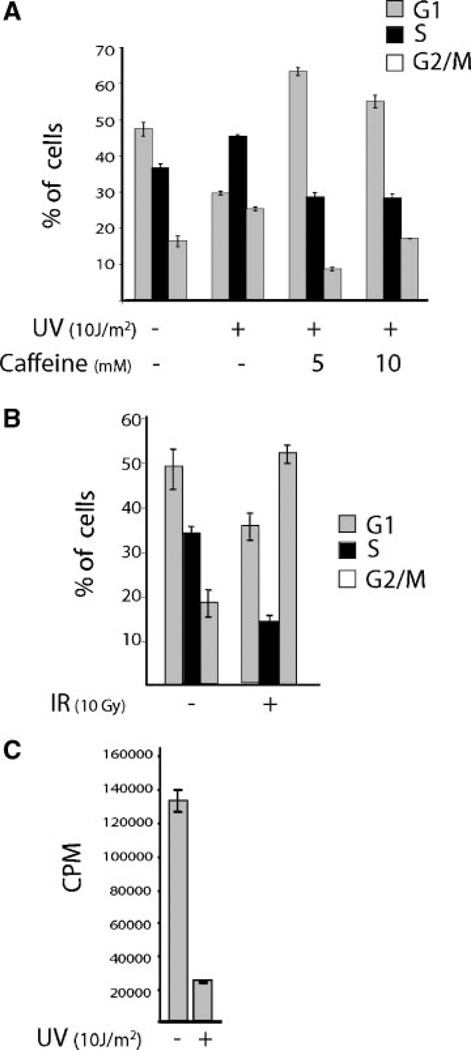
Different DNA damaging agents are known to alter the cell cycle profile of MCF-7 cells in different ways [38,39]. Flow cytometric analyses of UV-irradiated MCF-7 cells revealed that the fraction of cells in S-phase increased considerably in the total cell population (Figure 4A) indicating that UV-induced an S-phase checkpoint. On the other hand, IR-induced a G2 arrest in MCF-7 cells (Figure 4B). Similar to UV, BPDE treatment of MCF-7 cells also increased the S-phase fraction of the total cell population (data not shown). In order to confirm that the UV-induced increase in S-phase fraction was because of accumulation of cells in S-phase as a result of a delay and not because of increased cell proliferation, DNA synthetic rates in UV-C treated MCF-7 cells were monitored by uptake of [3H]-Thymidine. As shown in Figure 4C, a significant UV-C-induced reduction in the incorporation of [3H]-Thymidine was observed at 24 h posttreatment. This indicates that the increase in the fraction of S-phase cells shown in Figure 4A is, indeed, because of accumulation of cells at an S-phase checkpoint rather than to an increased rate of entry of cells into S-phase.
Figure 4.
(A) Caffeine abrogates S-phase delay. MCF-7 cells at 60%–70% confluency were pretreated or left untreated for 1 h with the indicated concentrations of caffeine and irradiated with UV. Caffeine treatment was continued for 24 h. The cells were harvested and processed for flow cytometry as mentioned in Materials and Methods. (B) IR causes primarily a G2/M arrest. MCF-7 cells at 60%–70% confluency were subjected to IR, and 24 h later harvested and processed for flow cytometry. (C) UV blocks DNA synthesis. MCF-7 cells were seeded in 12-well plate at 4 × 104 cells per well. Thirty-six hours later, the cells were irradiated with UV or left untreated for control. At 20-h post-UV irradiation, [3H]-Thymidine was added. Four hours later the cells were harvested and processed for counting radioactivity, counts per minute (CPM). Triplicate values for each treatment were plotted in the graphs.
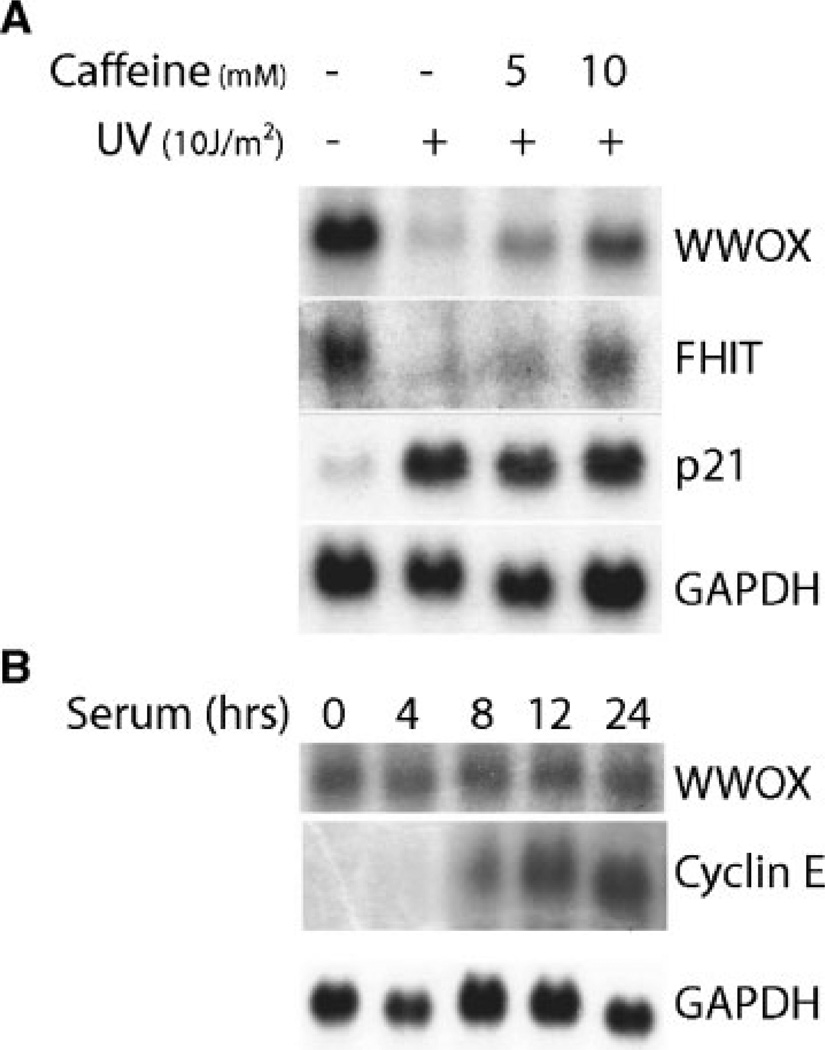
Caffeine is known to block the S-phase checkpoint response [40]. Hence, we investigated the effect of caffeine on the UV-induced S-phase delay and on the aforementioned decrease in transcript levels WWOX and FHIT. Interestingly, treatment of MCF-7 cells with caffeine abrogated the S-phase delay (Figure 4A) and significantly overcame the repression by UV of WWOX and FHIT expression (Figure 5A). Figure 4A also shows abrogation by caffeine of a small increase of G2/M checkpoint caused by UV that can be attributed to the known pleiotropic effects of caffeine. It is unlikely that the G2/M checkpoint response is involved in WWOX repression because IR causes a significant G2/M checkpoint response without affecting WWOX transcription. Therefore, the signaling pathways triggered at the S-phase checkpoint may play a role in the observed down-regulation.
Figure 5.
(A) Caffeine significantly overcomes downregulation of expression of fragile site genes. MCF-7 cells were subjected to UV-irradiation and caffeine treatment as indicated in Figure 4A. Northern blots of total RNA (30 µg per lane) from these treated cells were hybridized as indicated in Figure 1. Note the increase in the intensity of WWOX and FHIT signals upon caffeine treatments. (B) WWOX expression is not cell cycle regulated. MCF-7 cells were grown in serum-free medium for 4 d to synchronize the cells in G1/G0 phase. Total RNA was isolated from cells incubated in 5% serum containing medium for the indicated times and analyzed by Northern blotting with WWOX, Cyclin E, and GAPDH probes.
It was a possibility that the observed repression of WWOX and FHIT expression was a consequence of DNA damage induced alterations in the cell cycle. It has been previously shown that FHIT expression does not change during cell cycle progression [41]. To determine whether WWOX expression was cell cycle regulated, MCF-7 cells were arrested in G1-phase by serum starvation and WWOX expression levels were determined following serum stimulation. We observed normal levels of WWOX mRNA after serum starvation (G1 arrest) that remained unchanged as the cells progressed through the S- and G2/M-phases (Figure 5B). This was in sharp contrast to cyclin E mRNA that was undetectable in the G1-phase and dramatically increased as the cells went through S- and G2/M-phases. We conclude that WWOX expression is not dependent on cell cycle-specific signaling in normal growing cells but can be controlled by DNA damage induced S-phase checkpoint mechanisms.
DISCUSSION
Humans are daily exposed to an enormous variety of DNA-damaging agents. Therefore, it is not surprising that elaborate molecular regulatory systems exist to maintain cellular genomic integrity. UV causes DNA lesions at sites not occupied by histones by producing cyclobutane pyrimidine dimer, which accounts for nearly three-quarters of the damage to DNA induced by this carcinogen. Other types of UV-induced lesions include the pyrimidine-(6-4)pyrimidine dimers and a small fraction of pyrimidine hydrates and glycols [42]. The polycyclic aromatic hydrocarbon (PAH) benzo [a] pyrene (BP) is another ubiquitous environmental carcinogen. BP is product from combustion of fuel and tobacco and is one of the most potent carcinogens to which humans are frequently exposed [43,44]. A metabolic pathway mediated by cytochrome P450 and epoxide hydrolase converts BP to BPDE (benzo [a] pyrene diol epoxide) [45], the ultimate carcinogenic metabolite responsible for virtually all stable DNA adducts derived from BP. Cis or trans opening of the epoxide ring of BPDE by the exocyclic amino group N2 of guanine or N6 of adenine results in covalent DNA adducts [46]. IR, on the other hand, produces primarily double strand breaks and thymine glycols. Cells respond to DNA damage by undergoing cell cycle arrest or apoptosis mostly mediated by the tumor suppressor, p53. The first of these two pathways, involves blocking cell cycle progression to allow the DNA repair machinery of the affected cell to remove or repair the damage site prior to replication [47–49]. Nucleotide excision repair is the main pathway involved in the repair of bulky DNA lesions in DNA such as those induced by UV and BPDE [50,51]. The alternative pathway, apoptosis, is considered as a way to eliminate cells that have DNA so extensively damaged that mutation-free recovery is unlikely. During such cell cycle “checkpoints” for DNA repair or apoptosis, certain set of genes are activated whose protein products participate in these pathways whereas certain other genes, whose products may not be required are repressed [52,53]. The kind of response viz., arrest at different phases of cell cycle, or activation of apoptotic pathway is dependent on the nature of the damaging agents (carcinogens) and the type of cells [38,54–56].
The difference in the expression response at the FHIT and WWOX loci to damage induced by IR or U V is of interest. Both damaging agents produce a response that includes the phosphorylation and stabilization of p53, the activation of MAPK pathways, dramatic changes in transcription of damage response genes, and the establishment of one or more cell cycle checkpoints. However, these damage responses are mediated by different pathways. The ATM kinase pathway is primarily responsible for the IR-dependent response, whereas the related ATR kinase pathway is activated by UV. There is some evidence that BPDE-induced S-phase arrest is also mediated by the ATR kinase pathway [57]. Although similar in many ways, these damage response pathways are qualitatively distinct; note, for example, the differential cell cycle response to IR and UV shown in Figure 4. Thus, the downregulation of FHIT and WWOX expression by UV and BPDE but not by IR could therefore be a consequence of qualitative differences between the ATR- and ATM-mediated response pathways. Additionally, it has been demonstrated that the in vivo kinase activities of ATR and ATM remain unaffected upon caffeine treatment [40,58]. Within this context, our observation that caffeine is able to significantly block the repressive effect of UV on WWOX and FHIT expression, suggests that the ATR-Chk1 signaling pathway is unlikely to be the major mechanism behind the observed repressive phenomenon.
Alternatively, it is possible to speculate that the observed differences may be a consequence of the type of DNA damage produced by the different agents. As stated above, IR produces primarily double strand breaks and thymine glycols, the latter being a nondistortive DNA lesion. UV, on the other hand, produces cyclobutane pyrimidine dimers and 6-4 photoproducts both of which are classified as “bulky” lesions. The 6-4 photoproducts produce especially large distortions in DNA structure. The major BPDE-DNA adduct, formed by addition of this PAH to the exocyclic amino group of deoxyguanine, is also quite bulky, and has recently been shown to induce large kinks in adducted DNA [59]. These bulky adducts represent strong blocks to both transcription and replication, and could directly affect the expression of genes such as FHIT and WWOX, which produce extremely long transcripts. Additionally, BPDE-DNA adducts have been shown to directly affect the function of several transcription factors, including Sp1 and E2F1, by a hijacking mechanism in treated cells [60,61]. This effect is thought to be specific for DNA adducts that distort the DNA, mimicking the binding sites for transcription factors that bend DNA when bound. It is therefore possible that alterations in transcription factor availability at the promoters for the FHIT and WWOX genes, produced by the distortive DNA adducts in UV- or BPDE-treated cells but not in IR-treated cells are involved in the differential response observed.
Common chromosomal fragile sites contribute to genomic instability by their propensity as prime sites for the occurrence of chromosomal translocations, deletions, gene amplifications, and integration of oncogenic viruses, thereby contributing to cancer development and progression [4,6,7]. In this study, we have clearly demonstrated that the two most commonly altered fragile site genes WWOX/FRA16D and FHIT/FRA3B are downregulated in a coordinated manner by the common environmental carcinogens, UV and BPDE. Such a decrease in expression level occurred following induction of S-phase checkpoint in an asynchronously cycling cell population. Caffeine is known to inhibit, among others, the S-phase checkpoint and the DNA repair process [40,62]. In this study, caffeine treatment of UV-irradiated MCF-7 cells abrogated the S-phase delay and significantly prevented the decrease of the WWOX and FHIT transcripts, indicating that the S-phase checkpoint mechanisms contribute to the observed downregulation. It can be hypothesized that DNA damage repair mechanism contributes to the repression of WWOX and FHIT transcription following UV and BPDE exposure. In the event of DNA damage, elongating RNA Polymerase II is blocked by many DNA lesions in the transcribed strand [63], resulting in transcriptional downregulation of the genes. In stark contrast to the fragile site genes, the observed p21 upregulation, the unaffected GAPDH expression as well as the failure of IR to reduce the expression of WWOX and FHIT indicate that decrease in mRNA abundance following DNA damage is not a global phenomenon. Additionally, such a decrease in WWOX transcripts caused by a single moderate (nonlethal) UV dose (10 J/m2) is transient. In time-course experiments, such reduction was noticeable as early as 3 h postirradiation and dramatically reduced by 18 and 24 h, however, at 48 h the WWOX transcripts re-appeared (data not shown). This also implies that the observed repression is unlikely because of gene inactivation by fragile site induction and also speaks for the viability of the cells (Figure 1B). In addition, UV has not been reported to induce chromosomal fragility. It is worth noting that no deleterious effects were noted by the UV treatment (10 J/m2) because examination by phase contrast microscopy and trypan-blue exclusion proved that the cells remained viable and healthy.
At present, no information is available on specific regulatory transcription factor(s) of the two genes that could be affected by these carcinogens. However, their chromosomal localization at fragile regions and their being easy targets for environmental carcinogens could be reasoned for their concordant downregulation. Moreover, the structural nature of both fragile sites spanning huge genomic regions (up to 2 Mb) combined with the presence of large introns may pose considerable delay in the repair/recovery processes through specific pathways following DNA damage leading to the downregulation of expression of these genes.
Therefore, the observed decrease in expression also appears to be a mechanism for loss of expression of fragile site genes, in addition to the previously described gross genomic abnormalities. It has been recently suggested that chromosomal breaks on the DNA at fragile sites represent regions that have escaped the ATR-CHK1 DNA damage checkpoint mechanisms [2]. Thus, one could imagine a scenario in which one allele of a fragile site gene could be inactivated by deletion or fragile site induction while the other allele could be subject to downregulation (probably, in transcription) by environmental carcinogens as observed in this study, leading to complete silencing of the gene. Interestingly, supporting this concept and our findings, recently Iida et al. [64] reported that the genotoxic carcinogen methyleugenol induces a marked reduction of Wwox and Fhit expression in mouse liver while the noncarcinogenic congener eugenol has no effect. Importantly, methyleugenol forms bulky DNA adducts making it probable that the observed “in vivo” transcriptional repression could be mediated by the same mechanisms induced by UV and BPDE damaged DNA reported here. We hypothesize that such a continuous (transcriptional) silencing of fragile site associated putative tumor suppressor genes such as WWOX and FHIT by protracted exposure to environmental carcinogens may play a significant role in the initiation and development of cancer.
Since the submission of our manuscript, Ishii et al. [65] reported that UV irradiation resulted in the reduction of WWOX and FHIT gene expression and suggested a role for the G1-S checkpoint.
ACKNOWLEDGMENTS
We thank Dr. David G. Johnson for providing the p21 and cyclin E cDNA, and Charles Claypool for FACS analysis. This study was supported by NCI grant RO-1 CA 102444-01, and NIEH center grant ES 07784.
Abbreviations
- UV
ultraviolet
- BPDE
benzo[a]pyrene diol epoxide
- IR
ionizing radiation
- FHIT
fragile histidine triad
- WWOX
WW domain containing oxidoreductase
- PBS
phosphate buffered saline
- BP
benzo [a] pyrene
REFERENCES
- 1.Glover TW, Berger C, Coyle J, Echo B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum Genet. 1984;67:136–142. doi: 10.1007/BF00272988. [DOI] [PubMed] [Google Scholar]
- 2.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 3.Yunis JJ, Soreng AL. Constitutive fragile sites and cancer. Science. 1984;226:1199–1204. doi: 10.1126/science.6239375. [DOI] [PubMed] [Google Scholar]
- 4.Smith Dl, Huang H, Wang L. Common fragile sites and cancer (review) Int J Oncol. 1998;12:187–196. [PubMed] [Google Scholar]
- 5.Arlt MF, Miller DE, Beer DG, Glover TW. Molecular characterization of FRAXB and comparative common fragile site instability in cancer cells. Genes Chromosomes Cancer. 2002;33:82–92. doi: 10.1002/gcc.10000. [DOI] [PubMed] [Google Scholar]
- 6.Huebner K, Croce CM. FRA3B and other common fragile sites: The weakest links. Nat Rev Cancer. 2001;1:214–221. doi: 10.1038/35106058. [DOI] [PubMed] [Google Scholar]
- 7.Popescu NC. Genetic alterations in cancer as a result of breakage at fragile sites. Cancer Lett. 2003;192:1–17. doi: 10.1016/s0304-3835(02)00596-7. [DOI] [PubMed] [Google Scholar]
- 8.Stein CK, Glover TW, Palmer JL, Glisson BS. Direct correlation between FRA3B expression and cigarette smoking. Genes Chromosomes Cancer. 2002;34:333–340. doi: 10.1002/gcc.10061. [DOI] [PubMed] [Google Scholar]
- 9.Ban S, Cologne JB, Neriishi K. Effect of radiation and cigarette smoking on expression of FUdR-inducible common fragile sites in human peripheral lymphocytes. Mutat Res. 1995;334:197–203. doi: 10.1016/0165-1161(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 10.Sozzi G, Huebner K, Croce CM. FHIT in human cancer. Adv Cancer Res. 1998;74:141–166. doi: 10.1016/s0065-230x(08)60766-6. [DOI] [PubMed] [Google Scholar]
- 11.Tseng JE, Kemp BL, Khuri FR, et al. Loss of Fhit is frequent in stage I non-small cell lung cancer and in the lungs of chronic smokers. Cancer Res. 1999;59:4798–4803. [PubMed] [Google Scholar]
- 12.Ohta M, Inoue H, Cotticelli MG, et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 13.Ishii H, Dumon KR, Vecchione A, et al. Potential cancer therapy with the fragile histidine triad gene: Review of the preclinical studies. Jama. 2001;286:2441–2449. doi: 10.1001/jama.286.19.2441. [DOI] [PubMed] [Google Scholar]
- 14.Zochbauer-Muller S, Fong KM, Maitra A, et al. 5’ CpG island methylation of the FHIT gene is correlated with loss of gene expression in lung and breast cancer. Cancer Res. 2001;61:3581–3585. [PubMed] [Google Scholar]
- 15.Tanaka H, Shimada Y, Harada H, et al. Methylation of the 5’ CpG island of the FHIT gene is closely associated with transcriptional inactivation in esophageal squamous cell carcinomas. Cancer Res. 1998;58:3429–3434. [PubMed] [Google Scholar]
- 16.Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60:2140–2145. [PubMed] [Google Scholar]
- 17.Paige AJ, Taylor KJ, Taylor C, et al. WWOX: A candidate tumor suppressor gene involved in multiple tumor types. Proc Natl Acad Sci USA. 2001;98:11417–11422. doi: 10.1073/pnas.191175898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroki T, Trapasso F, Shiraishi T, et al. Genetic alterations of the tumor suppressor gene WWOX in esophageal squamous cell carcinoma. Cancer Res. 2002;62:2258–2260. [PubMed] [Google Scholar]
- 19.Yendamuri S, Kuroki T, Trapasso F, et al. WW domain containing oxidoreductase gene expression is altered in non-small cell lung cancer. Cancer Res. 2003;63:878–881. [PubMed] [Google Scholar]
- 20.Park S-W, Ludes-Meyers J, Zimonjic DB, Durkin ME, Popescu NC, Aldaz CM. Frequent downregulation and loss of WWOX gene expression in human hepatocellular carcinoma. Br J Cancer. 2004;91:753–759. doi: 10.1038/sj.bjc.6602023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishii H, Vecchione A, Furukawa Y, et al. Expression of FRA16D/WWOX and FRA3B/FHIT genes in hematopoietic malignancies. Mol Cancer Res. 2003;1:940–947. [PubMed] [Google Scholar]
- 22.Bednarek AK, Keek-Waggoner CL, Daniel RL, et al. WWOX, the FRA16D gene, behaves as a suppressor of tumor growth. Cancer Res. 2001;61:8068–8073. [PubMed] [Google Scholar]
- 23.Ludes-Meyers JH, Bednarek AK, Popescu NC, Bedford M, Aldaz CM. WWOX, the common chromosomal fragile site, FRA16D, cancer gene. Cytogenet Genome Res. 2003;100:101–110. doi: 10.1159/000072844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guler G, Uner A, Guler N, et al. The fragile genes FHIT and WWOX are inactivated coordinately in invasive breast carcinoma. Cancer. 2004;100:1605–1614. doi: 10.1002/cncr.20137. [DOI] [PubMed] [Google Scholar]
- 25.Le Beau MM, Rassool FV, Neilly ME, et al. Replication of a common fragile site, FRA3B, occurs late in S phase and is delayed further upon induction: Implications for the mechanism of fragile site induction. Hum Mol Genet. 1998;7:755–761. doi: 10.1093/hmg/7.4.755. [DOI] [PubMed] [Google Scholar]
- 26.Hellman A, Rahat A, Scherer SW, Darvasi A, Tsui LC, Kerem B. Replication delay along FRA7H, a common fragile site on human chromosome 7, leads to chromosomal instability. Mol Cell Biol. 2000;20:4420–4427. doi: 10.1128/mcb.20.12.4420-4427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palakodeti A, Han Y, Jiang Y, Le Beau MM. The role of late/slow replication of the FRA16D in common fragile site induction. Genes Chromosomes Cancer. 2004;39:71–76. doi: 10.1002/gcc.10290. [DOI] [PubMed] [Google Scholar]
- 28.Michael D, Rajewsky MF. Induction of the common fragile site FRA3B does not affect FHIT expression. Oncogene. 2001;20:1798–1801. doi: 10.1038/sj.onc.1204243. [DOI] [PubMed] [Google Scholar]
- 29.Pekarsky Y, Zanesi N, Palamarchuk A, Huebner K, Croce CM. FHIT: From gene discovery to cancer treatment and prevention. Lancet Oncol. 2002;3:748–754. doi: 10.1016/s1470-2045(02)00931-2. [DOI] [PubMed] [Google Scholar]
- 30.MacLeod MC, Adair G, Humphrey RM. Differential efficiency of mutagenesis at three genetic loci in CHO cells by a benzo[a]pyrene diol epoxide. Mutat Res. 1988;199:243–254. doi: 10.1016/0027-5107(88)90252-7. [DOI] [PubMed] [Google Scholar]
- 31.Koch-Paiz CA, Amundson SA, Bittner ML, Meltzer PS, Fornace AJJ. Functional genomics of UV radiation responses in human cells. Mutat Res. 2004;549:65–78. doi: 10.1016/j.mrfmmm.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 32.Amundson SA, Bittner M, Chen Y, Trent J, Meltzer P, Fornace AJJ. Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotixic stress responses. Oncogene. 1999;18:3666–3672. doi: 10.1038/sj.onc.1202676. [DOI] [PubMed] [Google Scholar]
- 33.Froesch BA, Aime-Sempe C, Leber B, Andrews D, Reed JC. Inhibition of p53 transcriptional activity by Bcl-2 requires its membrane-anchoring domain. J Biol Chem. 1999;274:6469–6475. doi: 10.1074/jbc.274.10.6469. [DOI] [PubMed] [Google Scholar]
- 34.Mori T, Anazawa Y, liizumi M, Fukuda S, Nakamura Y, Arakawa H. Identification of the interferon regulatory factor 5 gene (IRF-5) as a direct target of p53. Oncogene. 2002;21:2914–2918. doi: 10.1038/sj.onc.1205459. [DOI] [PubMed] [Google Scholar]
- 35.Oda K, Arakawa H, Tanaka T, et al. p53AIP1, a potential mediator of p53-dependent apoptosis and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- 36.Fujiuchi N, Aglipay JA, Ohtsuka T, et al. Requirement of IFI16 for the maximal activation of p53 induced by ionizing radiation. J Biol Chem. 2004;79:20339–20344. doi: 10.1074/jbc.M400344200. [DOI] [PubMed] [Google Scholar]
- 37.Haapajarvi T, Kivinen L, Heiskanen A, et al. UV radiation is a transcriptional inducer of p21(Cip1/Waf1) cyclin-kinase inhibitor in a p53-independent manner. Exp Cell Res. 1999;24:8272–8279. doi: 10.1006/excr.1999.4403. [DOI] [PubMed] [Google Scholar]
- 38.Khan QA, Dipple A. Diverse chemical carcinogens fail to induce G1 arrest in MCF-7 cells. Carcinogenesis. 2000;21:1611–1618. [PubMed] [Google Scholar]
- 39.Janicke RU, Engels IH, Dunkern T, Kaina B, Schulze-Osthoff K, Porter AG. Ionizing radiation but not anticancer drugs causes cell cycle arrest and failure to activate the mitochondrial death pathway in MCF-7 breast carcinoma cells. Oncogene. 2001;20:5043–5053. doi: 10.1038/sj.onc.1204659. [DOI] [PubMed] [Google Scholar]
- 40.Kaufmann WK, Heffernan TP, Beaulieu LM, et al. Caffeine and human metabolism: The magic and the mystery. Mutat Res DNA. 2003;532:85–102. doi: 10.1016/j.mrfmmm.2003.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo Z, Vishwanatha JK. Effect of regulated expression of the fragile histidine triad gene on cell cycle and proliferation. Mol Cell Biochem. 2000;204:83–88. doi: 10.1023/a:1007068823848. [DOI] [PubMed] [Google Scholar]
- 42.Sage E. Distribution and repair of photolesions in DNA: Genetic consequences and the role of sequence context. Photochem Photobiol. 1993;57:163–174. doi: 10.1111/j.1751-1097.1993.tb02273.x. [DOI] [PubMed] [Google Scholar]
- 43.Phillips DH. Polycyclic aromatic hydrocarbons in the diet. Mutat Res. 1999;443:139–147. doi: 10.1016/s1383-5742(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 44.Phillips DH. Fifty years of benzo(a)pyrene. Nature. 1983;303:468–472. doi: 10.1038/303468a0. [DOI] [PubMed] [Google Scholar]
- 45.Sims P, Grover PL, Swaisland A, Pal K, Hewer A. Metabolic activation of benzo(a)pyrene proceeds by a diol-epoxide. Nature. 1974;252:326–328. doi: 10.1038/252326a0. [DOI] [PubMed] [Google Scholar]
- 46.Jerina DM, Chadha A, Cheh AM, Schurdak ME, Wood AW, Sayer JM. Covalent bonding of bay-region diol epoxides to nucleic acids. Adv Exp Med Biol. 1991;283:533–553. doi: 10.1007/978-1-4684-5877-0_70. [DOI] [PubMed] [Google Scholar]
- 47.Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 48.Sionov RV, Haupt Y. The cellular response to p53: The decision between life and death. Oncogene. 1999;18:6145–6157. doi: 10.1038/sj.onc.1203130. [DOI] [PubMed] [Google Scholar]
- 49.Vousden KH. p53: Death star. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 50.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 51.Yang LL, Maher VM, McCormick JJ. Relationship between excision repair and the cytotoxic and mutagenic effect of the 'anti' 7,8-diol-9,10-epoxide of benzo[a]pyrene in human cells. Mutat Res. 1982;94:435–447. doi: 10.1016/0027-5107(82)90306-2. [DOI] [PubMed] [Google Scholar]
- 52.Ko U, Prives C. p53: Puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 53.Mirza A, Wu Q, Wang L, et al. Global transcriptional program of p53 target genes during the process of apoptosis and cell cycle progression. Oncogene. 2003;22:3645–3654. doi: 10.1038/sj.onc.1206477. [DOI] [PubMed] [Google Scholar]
- 54.Kuerbitz SJ, Plunkett BS, Walsh WV, Kastan MB. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc Natl Acad Sci USA. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nelson WG, Kastan MB. DNA strand breaks: The DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol. 1994;14:1815–1823. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fan S, Twu NF, Wang JA, et al. Down-regulation of BRCA1 and BRCA2 in human ovarian cancer cells exposed to adriamycin and ultraviolet radiation. Int J Cancer. 1998;77:600–609. doi: 10.1002/(sici)1097-0215(19980812)77:4<600::aid-ijc21>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 57.Guo N, Faller DV, Vaziri C. Carcinogen-induced S-phase arrest is Chk1 mediated and caffeine sensitive. Cell Growth Differ. 2002;13:77–86. [PubMed] [Google Scholar]
- 58.Cortez D. Caffeine inhibits checkpoint responses without inhibiting the ataxia-telangiectasia-mutated (ATM) and ATM- and Rad3-related (ATR) protein kinases. J Biol Chem. 2003;278:37139–37145. doi: 10.1074/jbc.M307088200. [DOI] [PubMed] [Google Scholar]
- 59.Pietrasanta LI, Smith BL, MacLeod MC. A novel approach for analyzing the structure of DNA modified by Benzo[a]pyrene diol epoxide at single-molecule resolution. Chem Res Toxicol. 2000;13:351–355. doi: 10.1021/tx9902035. [DOI] [PubMed] [Google Scholar]
- 60.MacLeod MC, Powell KL, Tran N. Binding of the transcription factor, Sp1, to non-target sites in DNA modified by benzo[a]pyrene diol epoxide. Carcinogenesis. 1995;16:975–983. doi: 10.1093/carcin/16.5.975. [DOI] [PubMed] [Google Scholar]
- 61.Butler AP, Johnson DG, Kumar AP, Narayan S, Wilson SH, MacLeod MC. Disruption of transcription in vitro and gene expression in vivo by DNA adducts derived from a benzo[a]pyrene diol epoxide located in heterologous sequences. Carcinogenesis. 1997;18:239–244. doi: 10.1093/carcin/18.2.239. [DOI] [PubMed] [Google Scholar]
- 62.Deplanque G, Ceraline J, Lapouge G, Dufour P, Bergerat JP, Klein-Soyer C. Conflicting effects of caffeine on apoptosis and clonogenic survival of human K1 thyroid carcinoma cell lines with different p53 status after exposure to cisplatin or UVc irradiation. Biochem Biophys Res Commun. 2004;314:1100–1106. doi: 10.1016/j.bbrc.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 63.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 64.lida M, Anna CH, Holliday WM, et al. Unique patterns of gene expression changes in liver after treatment of mice for 2 weeks with different known carcinogens and non-carcinogens. Carcinogenesis. 2005;26:689–699. doi: 10.1093/carcin/bgi005. [DOI] [PubMed] [Google Scholar]
- 65.Ishii H, Mimori K, Inageta T, et al. Components of DNA damage checkpoint pathway regulate UV exposure-dependent alterations of gene expression of FHIT and WWOX at chromosome fragile sites. Mol Cancer Res. 2005;3:103–108. doi: 10.1158/1541-7786.MCR-04-0209. [DOI] [PubMed] [Google Scholar]