Abstract
It was previously demonstrated that microRNA-199a (miR-199a) was down-regulated in testicular germ cell tumor (TGCT) partially caused by hypermethylation of its promoter. miR-199a is encoded by two loci in the human genome, miR-199a-1 on chromosome (Chr) 19 and miR-199a-2 on Chr 1. Both loci encode the same miR-199a. Another microRNA, microRNA-214 (miR-214), also locates on Chr 1. Previous study revealed that it is co-transcribed with miR-199a-2. However, the biological significance of the co-expression of miR-199a and miR-214 remains largely unknown. In this study, we determined that miR-199a and miR-214 were concordantly expressed in NT2 cells and TGCT patient tissues. After 5-aza treatment, miR-199-3p/5p and miR-214 expression was significantly increased. Silencing of DNMT1with siRNA restored the expression of miR-199a and miR-214, accompanied by de-methylation of the promoters of miR-199a-1/2. TP53 down-regulated the expression of DNMT1 in NT2 cells and overexpression of TP53 restored the expression of miR-199-3p/5p and miR-214. In addition, silencing of PSMD10 up-regulated the expression of TP53, while miR-214 over-expression resulted in PSMD10 down-regulation and TP53 up-regulation. Collectively, our findings highlighted a miR-199a/miR-214/PSMD10/TP53/DNMT1 self-regulatory network, which might be a potential therapeutic target in the treatment of TGCT.
As a key epigenetic modification, DNA methylation plays a crucial role in regulating gene expression in normal mammalian development. However, it was also observed that DNA methylation serves to modulate crucial growth regulators such as tumor suppressor genes (TSGs) and tumor suppressor microRNAs via promoter hypermethylation in cancer development1,2,3. When DNA is hypermethylated in the promoter region, genes or microRNAs (miRNAs) encoded are inactivated and silenced. DNA methylation is often dysregulated in tumor cells2. In the mammalian genome, DNA methylation is catalyzed by a family of DNA methyltransferases (DNMTs) that transfer a methyl group from S-adenyl-methionine (SAM) to the fifth carbon (C-5) of a cytosine residue to form 5mC. DNMT1 is primarily responsible for the maintenance, while DNMT3A and DNMT3B (de novo methyltransferases) are responsible for the establishment of genome DNA methylation patterns4,5.
Testicular germ cell tumor (TGCT) is the most frequent solid tumor of Caucasian adolescents and young adult males. It comprises a diverse group of neoplasms that can also be present in extragonadal sites, and is detrimental to male health and reproductive capacity6. Histologically, TGCTs are divided into seminomas, which resemble primordial germ cells (PGCs), and non-seminomas, which are either undifferentiated (embryonal carcinoma) or differentiated [embryonic (teratoma) or extra-embryonic (yolk sac choriocarcinoma)]. Embryonal carcinoma (EC) is the most frequent non-seminomatous tumor. It represents nearly 87% of non-seminoma7,8. Ntera2 (NT2) is one of the well-established pluripotent human testicular EC cell lines. This cell line has been extensively used in research on TGCT9,10,11,12. In this study, NT2 and normal human testis cell line Hs 1.Tes (HT, CRL-7002™) were used as cell models to study the tumorigenesis of TGCT.
miR-199a is a down-regulated miRNA caused by promoter hypermethylation in TGCT. miR-199a is encoded by two loci in the human genome, miR-199a-1 in Chr 19 and miR-199a-2 in Chr 1. Both loci encode miR-199a, which produces two mature miRNAs (miR-199a-3p and miR-199a-5p). Previous studies showed that the promoters of both miR-199a-1 and miR-199a-2 were hypermethylated in TGCTs12,13,14. However, the molecular mechanism underlying DNA hypermethylation in miR-199a promoter remains unknown. Previous study showed that DNMT3A did not regulate the expression of miR-199a in TGCT15. Whereas, it was reported that DNMT1 regulates miR-199a expression via mediating DNA methylation of miR-199a-1 promoter region16. Thus, it was suspected that DNMT1 also regulates miR-199a expression via mediating DNA methylation of miR-199a-2 promoter region in TGCT.
It was reported that the transcription of miR-199a-2 and miR-214 is regulated by the same promoter (miR-199a-2 promoter) as a single transcript in both human and mouse17,18. Co-expression of miR-199a and miR-214 was observed during normal development and in various diseases18,19,20,21,22,23. However, the significance of co-expression of miR-199a and miR-214 has not been fully elucidated. Besides, various studies showed that TP53 represses the transcription activity and expression of DNMT124,25,26,27. Interestingly, a more recent study reported that miR-214 regulates the expression of TP53 positively via directly targeting Gankyrin (also known as PSMD10), a negative regulator of tumor suppressor TP5328,29. Notably, the majority of clinical TGCTs express low levels of TP53, and TP53 mutations are rarely observed30,31. In addition, expression of DNMT1 was shown to be significantly upregulated in embryonal carcinoma32. These information together appear to suggest that miR-199a, miR-214, PSMD10, TP53 and DNMT1 may form a self-regulatory network in TGCT.
Results
Concordant expression of miR-199a and miR-214 in TGCT
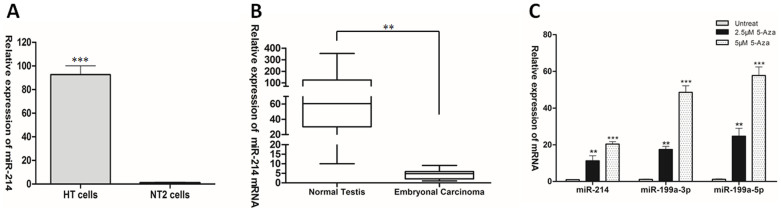
Since it was demonstrated that the promoters of miR-199a at both loci (Chr 1 and Chr 19) were hypermethylated, and the co-transcription of miR-199a-2 and miR-214 was directed by the miR-199a-2 promoter13,14,17,18, it is conceivable to propose that miR-214 showed similar expression pattern as miR-199a in TGCT. Indeed, qPCR results indicated that miR-214 was down-regulated in NT2 cells with more than 90-fold change when compared with HT cells (Fig. 1A). Moreover, the expression of miR-214 mRNA in clinical samples was also tested. miR-214 was significantly down-regulated in embryonal carcinoma compared to normal tissues (Fig. 1B). These results were consistent with the expression levels of miR-199a-3p and miR-199a-5p (two mature miRNAs of miR-199a) in embryonal carcinoma and NT2 cell lines14. Moreover, treatment with 5-aza restored the expression of miR-214, miR-199a-3p and miR-199a-5p in NT2 cells (Fig. 1C). These data suggested that miR-199a and miR-214 were concordantly expressed in TGCT, confirming the co-expression of miR-199a and miR-214.
Figure 1. Concordant expression of miR-199a and miR-214 in TGCT.
(A) miR-214 expression was assessed by RT-PCR in HT and NT2 cells. (B) miR-214 mRNA expression in normal testis tissues and embryonal carcinoma. (C) Effect of 5-aza treatment on expression of miR-199a and miR-214 in NT2 cells.
DNMT1 regulates the expression of miR-199a/miR-214 via promoter methylation in TGCT
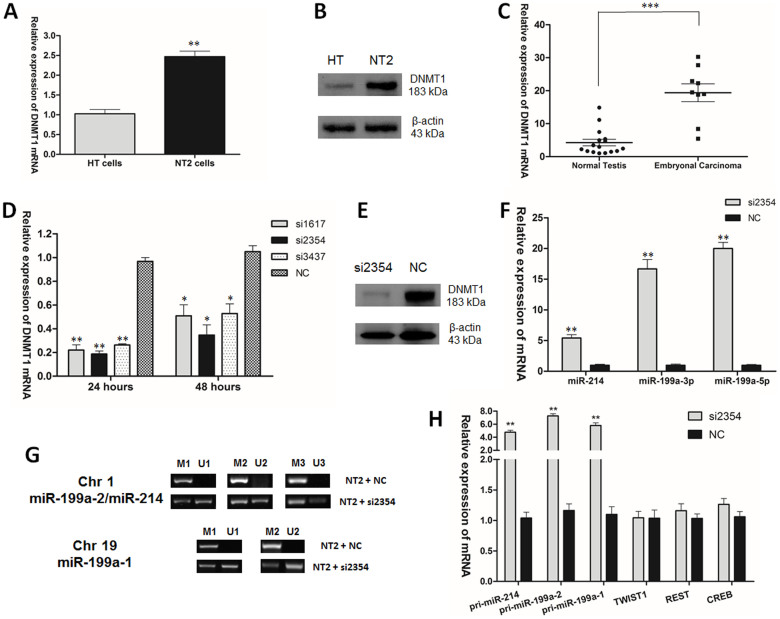
Consistent with the upregulation of DNMT1 in embryonal carcinoma32, DNMT1 was overexpressed in NT2 cells and embryonal carcinoma samples when compared with HT cells and normal testis tissues (Fig. 2A, B, C). Next, to select the siRNAs used to knock down DNMT1 in NT2 cells, the knock-down efficiencies of different siRNAs were compared. Results showed that si2354 had the best knock-down efficiency. Therefore, it was selected for subsequent experiments (Fig. 2D, E). When assessing the expression of miR-214 and miR-199a, it was found that knocking down DNMT1 increased the expression of mature miR-214, miR-199a-3p and miR-199a-5p, respectively, in NT2 cells (Fig. 2F). In addition, methylation-specific PCR (MSP) was used to examine the promoter methylation changes of miR-199a-1/2. Five sets of MSP primers were designed, of which two sets of primers focused on two CpG rich regions of miR-199a-1, and three sets of primers focused on three CpG rich regions of miR-199a-2. Each set of primers contained two pairs of primers to amplify either methylated or unmethylated alleles. miR-199a-1 is localized in Chr 19, with a size of 71bp (+1 to +71) in the human genome (10928102 to 10928172). The two regions selected for MSP analysis, based on their CpG content and being at the 5′ end of the gene33, are (−472 to −97) for M1/U1 and (−472 to −11) for M2/U2. miR-199a-2 is localized in Chr 1, with a size of 110 bp (+1 to +110) in the human genome (172113675 to 172113784). The three regions selected for MSP analysis are (−21 to +396) for M1/U1, (+65 to +374) for M2/U2 and (−21 to +100) for M3/U3. These three regions were previously identified to be in the potential promoter of miR-199a-214,18. MSP results showed that the promoter methylation levels of miR-199a in the two loci were correspondingly decreased after transfection with siDNMT1 (si2354) in NT2 cells (Fig. 2G). In addition, knocking down DNMT1 led to the elevation of pri-miR-199a-1/2 and pri-miR-214 (Fig. 2H). However, there are other transcription factors such as TWIST1, REST and CREB that are potentially involved in the regulation of miR-199a/miR-214 and would possibly affect the operation of the miR-199a/miR-214/PSMD10/TP53/DNMT1 network. The transcription factor TWIST1, a well-known positive regulator of the miR-199-2/miR-214 cluster transcription17, has been shown to be linked to DNMT1 in ovarian cancer34. Whereas the situation is different in TGCT, TWIST1 was most strongly expressed in seminoma and EC35, and the DNMT1 siRNA treatment led to no significant effect on the expression of TWIST1 in NT2 cells (Fig. 2H). REST, which preferentially bound to the methylated promoters of both miR-199a-1 and miR-199a-2/miR-214 had been shown to negatively regulate the expression of miR-199a13. CREB was previously reported to suppress the expression of miR-199a-3p during embryo implantation36. On the other hand, knocking down DNMT1 did not affect the expression of REST and CREB in NT2 cells significantly (Fig. 2H). These data indicated that the expression of miR-199/miR-214 could be affected by DNMT1 via promoter methylation, but not likely by the transcription factors TWIST1, REST or CREB.
Figure 2. DNMT1 regulated the expression of miR-199a/miR-214 via promoter methylation in TGCT.
(A) DNMT1 mRNA expression and (B) DNMT1 protein level in HT and NT2 cells. (C) DNMT1 mRNA expression in normal testis tissues and embryonal carcinoma. (D) DNMT1 mRNA expression and (E) DNMT1 protein level after transfection of siDNMT1 into NT2 cells. (F) miR-214, miR-199a-3p and miR-199a-5p mRNAs expression after transfection of siDNMT1 into NT2 cells. (G) Mature miR-199a-1/2 promoter methylation on Chr 1 and Chr 19 in NT2 cells after transfection of siDNMT1 into NT2 cells. (H) pri-miR-214, pri-miR-199a-1/2, TWIST1, REST and CREB mRNAs expression after transfection of siDNMT1 (si2354) into NT2 cells.
TP53 represses the expression of DNMT1 and increases the expression of miR-199a/miR-214 in TGCT
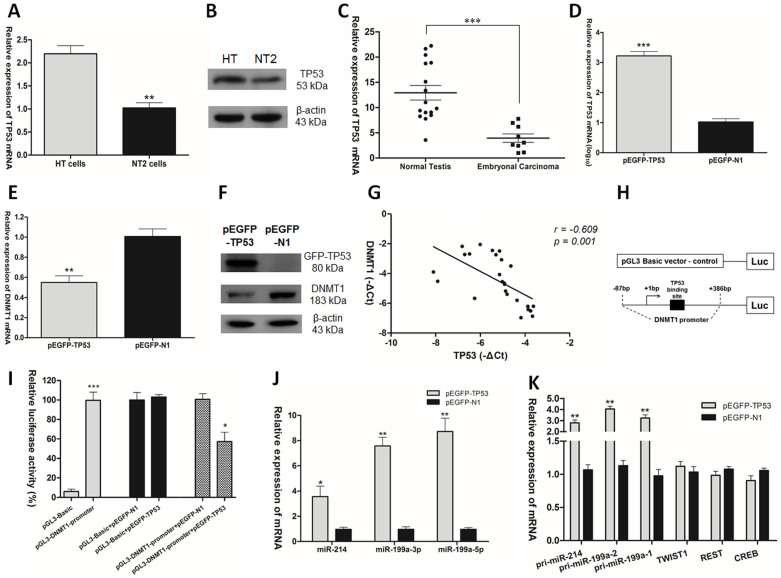
The expression of TP53 in HT cells and NT2 cells, as well as in clinical samples were compared by RT-qPCR. TP53 showed 2-fold down-regulation in NT2 cells (Fig. 3A). Results of Western blot analysis corroborated with this observation (Fig. 3B). In clinical samples, TP53 was significantly down-regulated in embryonal carcinoma compared to normal testis tissues (Fig. 3C). This is in agreement with the low expression of TP53 in the majority of TGCT samples30,31. When we overexpressed TP53 in NT2 cells with wide-type TP53 expression vector (pEGFP-TP53), we found that TP53 could inhibit the expression of DNMT1 (Fig. 3D, E, F). In addition, a statistically significant inverse correlation was observed between TP53 mRNA level and DNMT1 mRNA level in clinical samples (r = −0.609, p = 0.001. Pearson's correlation; Fig. 3G). To investigate whether TP53 regulates DNMT1 expression via modulating DNMT1 promoter activity, dual luciferase assay was employed to quantify the regulation of DNMT1 promoter activity gene by wild-type TP53 in NT2 cells. Fig. 3H showed the construct of pGL3-DNMT1 promoter luciferase reporter vector. Compared with pGL3-Basic vector, pGL3-DNMT1 promoter vector showed much higher luciferase activity. Co-transfection of pGL3-Basic vector with pEGFP-TP53 or pEGFP-N1 in NT2 cells, resulted in no significant difference between the two groups. However, co-transfection of pGL3- DNMT1 promoter vector with pEGFP-TP53 or pEGFP-N1 in NT2 cells showed that wild-type TP53 decreased DNMT1 promoter activity to 40% of the pEGFP-N1 control vector (Fig. 3I). Notably, overexpression of TP53 restored the expression of mature miR-214, miR-199a-3p and miR-199a-5p, respectively, in TGCT (Fig. 3J). On the other hand, TP53 is known to involve in posttranscriptional regulation of microRNA biogenesis37. To prove the elevation of mature miR-199a-3p/5p and miR-214 due to the regulation of TP53 on the transcription of pri-miR-199a-1/2 and pri-miR-214, the expression of pri-miR-199a-1/2 and pri-miR-214 were assessed after overexpression of TP53 in NT2 cells, and found that TP53 increased the primary transcription of miR-199a and miR-214 (Fig. 3K). In addition, to rule out the involvement of TWIST1, REST and CREB, the expression of TWIST1, REST and CREB were examined after overexpression of TP53 in NT2 cells. No significant change in the expression level of these transcription factors was observed (Fig. 3K). These findings revealed that TP53 repressed DNMT1 promoter activity and expression level, and enhanced the expression of miR-199a/miR-214. This effect of TP53 did not involve TWIST1, REST or CREB.
Figure 3. TP53 repressed the expression of DNMT1 and increased the expression of miR-199a/miR-214 in TGCT.
(A) TP53 mRNA expression and (B) TP53 protein level in HT and NT2 cells. (C) TP53 mRNA expression in normal testis tissues and embryonal carcinoma. (D) TP53 mRNA expression, (E) DNMT1 mRNA expression and (F) GFP-TP53 and DNMT1 protein levels after transfection of pEGFP-TP53 vector or pEGFP-N1 control vector into NT2. (G) Statistical analysis to evaluate correlation between TP53 and DNMT1 mRNA expression levels in clinical samples (r = −0.609, p = 0.001. Pearson's correlation). (H) Construct of pGL3-DNMT1 promoter luciferase reporter vector. (I) The vectors pGL3-Basic, pGL3-DNMT1 promoter, pGL3-Basic and pEGFP-TP53, pGL3-Basic and pEGFP-N1, pGL3-DNMT1 promoter and pEGFP-TP53, pGL3-DNMT1 promoter and pEGFP-N1, were co-transfected with internal control pRL-TK renilla luciferase vector into NT2 cells. Reporter luciferase activity was measured. Ratio of relative activity was calculated within group independent of each other. (J) Mature miR-214, miR-199a-3p and miR-199a-5p mRNAs expression after transfection of pEGFP-TP53 vector into NT2 cells. (K) The mRNAs expression of pri-miR-214, pri-miR-199a-1/2, TWIST1, REST and CREB after overexpression of TP53 in NT2 cells.
PSMD10 negatively regulates the expression of TP53 in TGCT
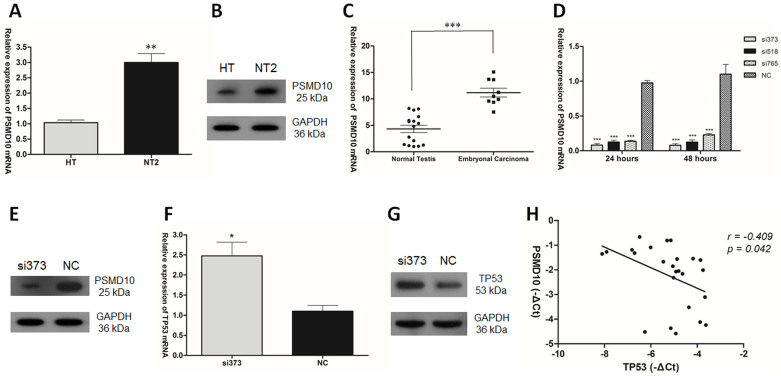
It was previously reported that PSMD10 is a negative regulator of tumor suppressor TP5329. To confirm the negative regulator role of PSMD10 on TP53, the expression of PSMD10 in HT cells and NT2 cells, as well as in clinical samples were examined. The expression level of PSMD10 showed 3-fold up-regulation in NT2 cells (Fig. 4A, B). The level of PSMD10 was also significantly higher in embryonal carcinoma (Fig. 4C). When si373 was used to knock down PSMD10 in NT2 cells, the expression of TP53 in NT2 cells increased (Fig. 4D, E, F, G). Moreover, correlation study showed that PSMD10 mRNA level was inversely correlated with TP53 mRNA level in clinical samples (r = −0.409, p = 0.042. Pearson's correlation; Fig. 4H).
Figure 4. PSMD10 negatively regulates the expression of TP53 in TGCT.
(A) PSMD10 mRNA expression and (B) PSMD10 protein level in HT and NT2 cells. (C) PSMD10 mRNA expression in normal testis tissues and embryonal carcinoma. (D) PSMD10 mRNA expression and (E) PSMD10 protein level after transfection of siPSMD10 into NT2 cells. (F) TP53 mRNA expression and (G) TP53 protein level after transfection of siPSMD10 into NT2 cells. (H) PSMD10 and TP53 mRNA levels inversely correlated in clinical samples (r = −0.409, p = 0.042). Statistical analysis to evaluate correlation was performed using Pearson's correlation analysis.
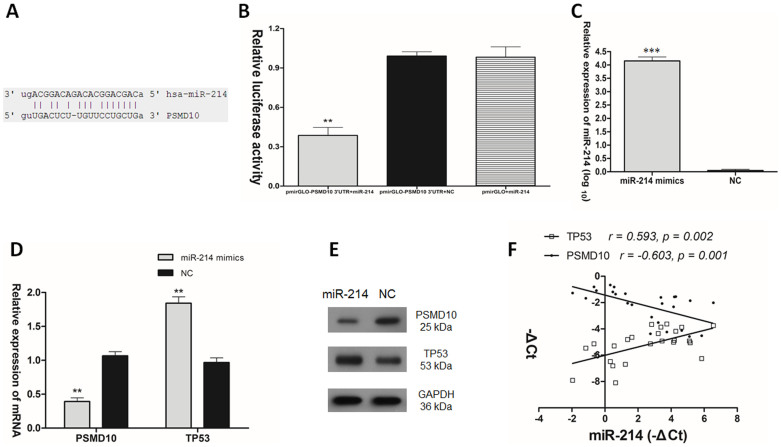
miR214 positively regulates the expression of TP53 via directly targeting PSMD10 in TGCT
It is known that PSMD10 was a direct target of miR-214 in myeloma28. To validate the direct targeting of miR-214 on PSMD10 in TGCT, a dual luciferase assay was performed. We constructed the pmirGLO-PSMD10 3′UTR reporter vector containing the putative miR-214 binding sites (Fig. 5A). The pmirGLO-PSMD10 3′-UTR reporter vector was co-transfected with miR-214 mimics or negative control RNA (NC), or empty pmirGLO with miR-214 mimics, into NT2 cells. As shown in Fig. 5B, when cotransfected with miR-214 mimics, the relative luciferase activities of pmirGLO-PSMD10 3′UTR luciferase reporter in NT2 cells were suppressed by almost 60% compared with the transfection with NC and empty pmirGLO vector. To investigate the effect of miR-214 on PSMD10 and TP53, miR-214 mimics was transfected into NT2 cells (Fig. 5C). RT-qPCR and Western blot analysis were used to evaluate the expression of PSMD10 and TP53 after transfection of miR-214. Overexpression of miR-214 greatly down-regulated PSMD10 expression and up-regulated TP53 expression (Fig. 5D, E). In addition, miR-214 mRNA level was inversely correlated with PSMD10 mRNA level (r = −0.603, p = 0.001. Pearson's correlation), while was positively correlated with TP53 mRNA level in clinical samples (r = 0.593, p = 0.002. Pearson's correlation) (Fig. 5F).
Figure 5. miR-214 regulated the expression of TP53 positively via directly targeting PSMD10 in TGCT.
(A) The putative human PSMD10 3′-UTR fragment containing miR-214 binding sequence was insert into the luciferase report vector pmirGLO downstream. (B) Dual luciferase assay of NT2 cells co-transfected with the luciferase constructs containing the PSMD10 3′-UTR as well as miR-214 mimics or scrambled oligonucleotides as the negative control. (C) miR-214 mRNA expression, (D) PSMD10 and TP53 mRNAs expression, and (E) PSMD10 and TP53 protein levels after transfection of miR-214 mimics into NT2 cells. (F) Statistical analysis to evaluate correlation between miR-214 and PSMD10 mRNA expression levels (r = −0.603, p = 0.001. Pearson's correlation), as well as between miR-214 and TP53 mRNA expression levels (r = 0.593, p = 0.002. Pearson's correlation) in clinical samples.
Discussion
Cancer was initially regarded as a disease of cell proliferation caused by mutations in genes that control proliferation and the cell cycle38. This is the Somatic Mutation Theory39. However, since the inception of epigenetics in the 1940s, discoveries implicating its role in cancer have been mounting continuously. More importantly, global changes in epigenetic regulation have been regarded as a hallmark of cancer. It is now realized that initiation and progression of cancer involves epigenetic abnormalities along with genetic alterations40,41. During the past decade, the epigenetics factors have changed from just one recognized marker, DNA methylation, to a variety of others, including a wide spectrum of histone modifications and microRNAs40. As the first identified epigenetic modification, DNA methylation has been widely studied. Dysregulation of DNA methylation has been found in cancer as a typical hallmark, and its consequence is the silencing of genes and noncoding RNAs40.
miR-199a is a typical example of silenced miRNA partially caused by promoter DNA hypermethylation in TGCT. Previous studies demonstrated that the promoters of miR-199a at both loci (Chr 1 and Chr 19) were almost completely methylated in TGCT when compared with normal testicular tissue13,14. More detailed analyses showed that miR-199a-5p, one of its two derivatives, suppressed TGCT invasion and proliferation via directly targeting PODXL and MAFB13,14. On the other hand, miR-199a-3p, its other derivative, was shown to negatively regulated DNA methylation in TGCT, partly through targeting DNMT3A. Overexpression of miR-199a-3p restored the expression of APC and MGMT tumor-suppressor genes in NT2 cells by affecting DNA methylation of their promoter regions15. Collectively, these data assigned a tumor suppressor role to miR-199a in TGCT. On the other hand, low expression of miR-214 has been observed in different types of cancer, such as myeloma28, hepatocellular carcinoma42,43, colorectal cancer44, breast cancer45, and primary central nervous system lymphomas (PCNSL)46. Moreover, it has been demonstrated that miR-214 acted as a tumor suppressor via downregulation of certain oncogenes [PSMD1028, β-catenin42,43, FGFR144, and Ezh245] in these different solid cancers. In our study, we demonstrated that in embryonal carcinoma and NT2 cells, miR-214 was also down-regulated. Inhibition of PSMD10 induced an increase of TP53 mRNA and protein levels. In addition, miR-214 was shown to directly targeted and down-regulated the expression of PSMD10, which subsequently up-regulated the expression of TP53. Moreover, a statistically significant inverse correlation between miR-214 mRNA level and PSMD10 mRNA level, while a statistically significant positive correlation between miR-214 mRNA level and TP53 mRNA level was observed in clinical samples. These data confirmed a tumor suppressor role for miR-214 in TGCT.
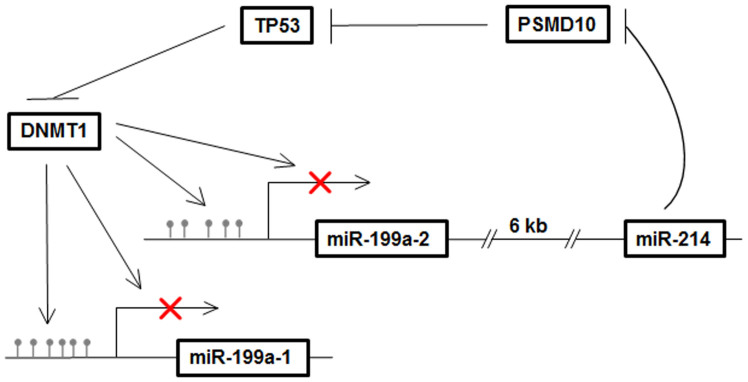
As a master regulator, miRNA is involved in multiple cellular processes during normal development and diseases47. Previous studies mainly focused on the elucidation of miRNA functions48. However, the question of how microRNA expression is regulated was rarely asked. In the present study, we explored the mechanism of the down-expression of miR-199a/miR-214 in TGCT. Early studies indicated that miR-199a-2 and miR-214 were co-transcribed with the promoter of miR-199a-2, and co-expression of miR-199a and miR-214 has been observed in various systems17,18,19,20,21,22,23. Notably, concordant expression of miR-199a and miR-214 was also observed in TGCT. Since it was reported that silencing of miR-199a-1/2 was partially caused by promoter DNA hypermethylation, the expression of miR-214, miR-199a-3p, and miR-199a-5p, respectively, were correspondingly up-regulated after treatment with 5-aza. Moreover, we showed that DNMT1, the maintenance DNA methyltransferase, inhibited the expression of primary and mature miR-199a/miR-214 via affecting the promoter DNA methylation of miR-199a-1 and miR-199a-2/miR-214 in TGCT. In unraveling the regulatory mechanisms of miR-199a/miR-214 expression in greater detail, we further found that TP53 repressed the transcription activity and expression of DNMT1 as previously reported, which suggested that TP53 is a potential positive regulator of miR-199a/miR-214. Indeed, over-expressed TP53 would up-regulate the expression of primary and mature miR-199a/miR-214, respectively, in TGCT. Therefore, a new miR-199a/miR-214/PSMD10/TP53/DNMT1 self-regulatory network was suggested in TGCT progression, in which TP53 was activated by miR-214 and participated in the positive regulation of miR-199a/miR-214 via repressing DNMT1 (Fig. 6). However, previous study reported that DNMT1 was not expressed in seminoma, but upregulated in embryonal carcinoma32,49. Thus the regulatory network suggested in the current study might only operate in non-seminoma and not in seminoma. Several well-known regulators of miR-199a and miR-214 expression in systems other than TGCT, such as TWIST117, REST13 and CREB36, were not affected after DNMT1 siRNA (si2354) treatment and overexpression of TP53 in NT2 cells. These results suggested that the regulation of miR-199a and miR-214 is highly complex and the regulatory network is tissue and/or tumor dependent. The miR-199a/miR-214/PSMD10/TP53/DNMT1 self-regulatory network identified in the current study could regulate the expression of miR-199a/miR-214 in TGCT. However, the existence of other regulatory pathway cannot be ruled out.
Figure 6. Summary diagram describes the miR-199a/miR-214 self-regulatory network via TP53 and DNMT1 in TGCT.
It suggested that TP53 was activated by miR-214 and participated in the positive regulation of miR-199a/miR-214 via repressing DNMT1.
The miR-199a/miR-214/PSMD10/TP53/DNMT1 regulatory network contributed to the down-regulation of miR-199a, miR-214, and TP53, respectively, as well as the up-regulation of DNMT1 in TGCT, and partly explains the mechanism involved in DNA hypermethylation of miR-199a promoter in TGCT. However, DNMT3B and DNMT3L, the other DNA methyltransferases, were not examined in this study. DNMT3L specifically expressed in TGCTs and EC cell lines and was essential for the growth of human EC50. DNMT3B was also strongly expressed in EC samples51. Since DNMT3A did not affect DNA methylation of the miR-199a promoter regions15 and knocking down DNMT1 did not cause complete demethylation of the miR-199a-1/2 promoter regions, it is reasonable to assume that DNMT3B, DNMT3L, or both might also act as a regulator of miR-199a/miR-214 expression. Elucidation of the role of DNMT3B and DNMT3L in miR-199a/miR-214 silencing will help us understand better the mechanism of DNA methylation and involvement of DNMTs in the regulation of miR-199a/miR-214 in TGCT.
In summary, our findings highlight a miR-199a/miR-214/PSMD10/TP53/DNMT1 self-regulatory network, the dysfunction of which may contribute to tumor survival and progression of TGCT. Our study offers new insight into the biological significance conferred by the co-expression of miR-199a and miR-214, and provide a potential therapeutic approach in targeting miR-199a/miR-214/PSMD10/TP53/DNMT1 regulatory network for the treatment of TGCT.
Methods
Primary TGCT specimens
Genomic RNA of 9 embryonal carcinoma (EC) obtained from TGCT patients were purchased from Oncomatrix (San Marcos, CA, USA). Normal testicular RNA (16 cases) were purchased from Zyagen (San Diego, CA, USA). Each RNA samples was isolated from a single individual.
Cell culture and transfection
Ntera 2 (NT2) and Hs 1.Tes (HT) cell lines were purchased from ATCC (Manassas, VA, USA) supplemented with 10% FBS and incubated in a 37°C humidified incubator supplied with 5% CO2. Synthetic double-stranded miR-214 mimics, scramble oligonucleotides used as negative control (NC) (GenePharma, Shanghai, China) at a final concentration of 20 nM were introduced into NT2 cells by Lipofectamine 2000 kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Cells were harvested at 48 hours after transfection. For demethylation analysis, 1 × 105 NT2 cells were seeded for 24 h and treated with 2.5 μm or 5 μm of 5-aza-2-deoxycytidine (5-aza) (Sigma, St Louis, MO, USA) for 72 h.
Inhibition of DNMT1, PSMD10 expression and overexpression of TP53
The small interfering RNAs (siRNAs) specifically targeting DNMT1 (siDNMT1), namely, si1617, si2354 and si3437, respectively, and PSMD10 (siPSMD10), namely, si373, si518 and si765, respectively, were designed and synthesized by GenePharma, Shanghai, China. The synthetic siDNMT1, siPSMD10, or negative control siRNA at a final concentration of 20 nM were introduced into NT2 cells by using Lipofectamine 2000 kit (Invitrogen, Carlsbad, CA) as previously described under Cell Culture and Transfection. Wild-type TP53 expression vector (GFP-p53, Plasmid 12091, GeneBank ID: AAD28628.1), bought from Addgene (Cambridge, MA, USA), produced the GFP-TP53 fusion protein (about 80 kDa). In addition, we re-named GFP-p53 as pEGFP-TP53, which was consistent with its vector backbone pEGFP-N1. The pEGFP-N1 vector, used as negative control, was purchased from Clontech Laboratories, Inc. (Mountain View, CA, US). The siRNAs or vectors were introduced into NT2 cells by Lipofectamine 2000 kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Transfected cells were harvested at 24 or 48 hours to test the knock-down efficiency of siDNMT1 and siPSMD10. For downstream examination after transfection of siRNAs (siDNMT1 or siPSMD10) or vectors (pEGFP-TP53 or pEGFP-N1) in NT2 cells, the transfected cells were harvested at 48 hours.
Construction of luciferase reporter vector
The fragment (nucleotides -11bp to +285bp) of PSMD10 3′-untranslated region (3′-UTR) containing an intact miR-214 recognition sequence was amplified by PCR from genomic DNA and inserted into the Firefly/Renilla dual reporter vector pmirGLO (Promega, Madison, WI) at the 3′-end of the firefly luciferase gene. The DNMT1 promoter (nucleotides −87bp to +386bp), containing the TP53 binding site24, was amplified by PCR from genomic DNA and inserted into the pGL3-Basic luciferase reporter vector to construct the pGL3-DNMT1 promoter luciferase reporter vector. The pRL-TK renilla luciferase vector was used as internal control. The primers for the construction of luciferase reporter vector were as follows: pmirGLO-PSMD10 3′UTR vector, forward 5′- CGAGCTCTGGAAGGTTAAACAGCTTGGA-3′ and reverse 5′- GCTCTAGAAGACTCACAACAGCCACAGAA-3′; pGL3-DNMT1 promoter vector, forward 5′-CTAGCTAGCCGTGGAGCTTGGACGA-3′ and reverse 5′- CCCAAGCTTCCGCACCACCCCACGCAT-3′.
Reverse-Transcription (RT) reaction and quantitative polymerase chain reaction (qPCR)
The harvested cells were placed in TRIzol® Reagent (Invitrogen, Carlsbad, CA), and total RNAs were extracted according to the TRIzol manufacturer's instructions. Total RNA (1 μg) was primed by random hexamers and converted into cDNA using SuperScript III (Invitrogen). The SYBR green-based real-time PCR was performed in an Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA), and GAPDH was used as an endogenous control to normalize the amount of total mRNA in each sample. The primer sequences for qPCR analysis were as follows: DNMT1, forward 5′-GTGGGGGACTGTGTCTCTGT-3′ and reverse 5′-TGAAAGCTGCATGTCCTCAC-3′; TP53, forward 5′- TCCACTACAACTACATGTGTAAC-3′ and reverse 5′- GTGAAATATTCTCCATCCAGTG-3′; PSMD10, forward 5′- GGTGTCCCAAGGAGCAAGTA-3′ and reverse 5′- ACACTGGGGACAACAACACA-3′; TWIST1, forward: 5′- CGGGAGTCCGCAGTCTTA-3′ and reverse 5′-GCTTGAGGGTCTGAATCTTG-3′; REST, forward: 5′-GAGGCCACATAACTGCACTG-3′ and reverse 5′-TGTCCTTACTCAAGTTCTCAGAAGA-3′; CREB, forward: 5′- GTGTTACGTGGGGGAGAGAA-3′ and reverse 5′- GGCTCCAGATTCCATGGTC-3′; pri-miR-199a-1, forward 5′- CCGCTCTGTCCCTTCTGACG-3′ and reverse 5′-AAACCCTGCCTCCTGCTCC-3′; pri-miR-199a-2, forward 5′-TGCCCAGTCTAACCAATGTGC-3′ and reverse 5′-AGCTGAATGCAACCCCTGG-3′; pri-miR-214, forward 5′- GTATCTGTCTATGAGCAAAGGAAACC-3′ and reverse 5′-GGTGTAGATGCTATGGTGTGAGGGC-3′; GAPDH, forward 5′- TGCACCACCAACTGCTTAGC-3′ and reverse 5′- GGCATGGACTGTGGTCATGAG-3′. The expression of mature miR-199a-3p, miR-199a-5p and miR-214 were measured using the well-established stem-loop RT-qPCR method as previously described52, and the level of miRNAs expression were normalized by U6 RNA. Real-time PCR was performed with a standard SYBR-Green PCR kit protocol on ABI 7900HT Fast Real Time PCR system (Applied Biosystems, Foster City, CA). The qPCR primer sequences for miR-199a-3p/5p have been previously described15, while the sequence-specific primers for miR-214 and U6 were as follows: Stem loop primer for miR-214, GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGA.
GCCAACACTGCC; miR-214, forward 5′-GGTGCAGGGTCCGAGGTAT-3′ and reverse 5′- CCGACAGCAGGCACAGACA-3′; U6, forward 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse 5′- CGCTTCACGAATTTGCGTGTCAT-3′.
Dual luciferase reporter assay
3 × 104 NT2 cells were plated in each well of a 24-well plate the day before transfection. NT2 cells were co-transfected with pmirGLO-PSMD10 3′UTR construct and miR-214 mimics or a negative control, or co-transfected with empty pmirGLO plasmid and miR-214 mimics by Lipofectamine 2000 (Invitrogen, Carlsbad, CA) as previously described by cell culture and transfection methods. Moreover, the plasmids pGL3-Basic, pGL3-DNMT1 promoter, pGL3-Basic and pEGFP-TP53, pGL3-Basic and pEGFP-N1, pGL3-DNMT1 promoter and pEGFP-TP53, pGL3-DNMT1 promoter and pEGFP-N1, which were co-transfected with internal control pRL-TK renilla luciferase vector into NT2 cells by Lipofectamine 2000. Four wells for each group in a single experiment. A luciferase activity assay was performed 48 hours after transfection with the dual luciferase reporter assay system (Promega). The relative luciferase activity was normalized with Renilla luciferase activity.
Western blot analysis
Whole cell lysates were prepared with RIPA lysis buffer. Protein concentration was determined for each sample with the Thermo Scientific™ Pierce™ BCA Protein Assay. The protein samples were 1:1 diluted in 2x sample buffer containing β-mercaptoethanol (BME) and the mixture was boiled at 95-100°C for 5 minutes. Equal amounts of cell lysate proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. After incubation with antibodies specific for either DNMT1 (ab13537, Abeam), TP53 (sc-126, Santa Cruz Biotechnology), PSMD10 (sc-101498, Santa Cruz Biotechnology), β-Actin (A1978, SIGMA-ALDRICH) or GAPDH (Abeam), the blots were developed with HRP (horseradish peroxidase)-conjugated anti-mouse IgG (Santa Cruz Biotechnology, Santa Cruz, CA) and visualized with enhanced chemiluminescence.
Methylation specific PCR (MSP)
DNA was extracted using Invitrogen™ genomic DNA (gDNA) extraction kits. The bisulfite conversion of gDNA (400 ng) was conducted with EZ DNA Methylation-Gold™ Kit (Zymo Research Corporation, USA). MSP was performed as previously described53. The bisulfite-treated DNA (80–100 ng) was amplified with the methylation-specific primers or the unmethylated-specific primers, and the amplified products were analyzed on agarose gel. In this study, MSP assay was performed 72 hours after transfection with siDNMT1 (si2354) in NT2 cells. The primer sequences for miR-199a-1/2 used in MSP assays were described in our previous study15.
Statistical analysis
The expressions of miR-214, TP53, PSMD10 and DNMT1 in embryonal carcinoma and normal testis tissues were compared by the unpaired 2-tailed t test. The relationship between miR-214 and PSMD10 mRNA expression levels, miR-214 and TP53 mRNA expression levels, TP53 and PSMD10 mRNA expression levels, TP53 and DNMT1 mRNA expression levels were analyzed by Pearson's correlation. The differences between samples analyzed by luciferase assay and RT-qPCR were determined by two-tailed Student's t test. Data were expressed as means and standard deviations (SD) from at least three independent experiments. All p values were two-sided and were obtained with the SPSS 16.0 software package (SPSS, Chicago, IL). A p value < 0.05 was considered statistically significant (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Author Contributions
B.F.C. planned the research, performed the experiments and drafted the manuscript. Y.K.S., S.G. and L.L. offered help in the entire project and carefully read the manuscript. W.Y.C. supervised the entire project, discussed the results, edited and proof read the manuscript.
Acknowledgments
This work was supported by funds provided by the Chinese University of Hong Kong, Vigconic Ltd, International, and the CUHK-Shandong University Joint Laboratory.
References
- Chen C. Z. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med 353, 1768–1771 (2005). [DOI] [PubMed] [Google Scholar]
- Baylin S. B. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol 2 Suppl 1, S4–11 (2005). [DOI] [PubMed] [Google Scholar]
- Li W. & Chen B. F. Aberrant DNA methylation in human cancers. J Huazhong Univ Sci Technolog Med Sci 33, 798–804 (2013). [DOI] [PubMed] [Google Scholar]
- Lechner M., Boshoff C. & Beck S. Cancer epigenome. Adv Genet 70, 247–276 (2010). [DOI] [PubMed] [Google Scholar]
- Chen B. F. & Chan W. Y. The de novo DNA methyltransferase DNMT3A in development and cancer. Epigenetics 9, 669–677 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert D., Rapley E. & Shipley J. Testicular germ cell tumours: predisposition genes and the male germ cell niche. Nat Rev Cancer 11, 278–288 (2011). [DOI] [PubMed] [Google Scholar]
- Bosl G. J. & Motzer R. J. Testicular germ-cell cancer. N Engl J Med 337, 242–253 (1997). [DOI] [PubMed] [Google Scholar]
- Oosterhuis J. W. & Looijenga L. H. Testicular germ-cell tumours in a broader perspective. Nat Rev Cancer 5, 210–222 (2005). [DOI] [PubMed] [Google Scholar]
- Burger H., Nooter K., Boersma A. W. M., Kortland C. J. & Stoter G. Expression of p53, Bcl-2 and Bax in cisplatin-induced apoptosis in testicular germ cell tumour cell lines. Br J Cancer 77, 1562–1567 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch S., Mayer F., Honecker F., Schittenhelm M. & Bokemeyer C. Efficacy of cytotoxic agents used in the treatment of testicular germ cell tumours under normoxic and hypoxic conditions in vitro. Br J Cancer 89, 2133–2139 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotheim R. I. et al. Differentiation of human embryonal carcinomas in vitro and in vivo reveals expression profiles relevant to normal development. Cancer Res 65, 5588–5598 (2005). [DOI] [PubMed] [Google Scholar]
- Cheung H. H. et al. Genome-wide DNA methylation profiling reveals novel epigenetically regulated genes and non-coding RNAs in human testicular cancer. Br J Cancer 102, 419–427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S. et al. Molecular mechanisms of regulation and action of microRNA-199a in testicular germ cell tumor and glioblastomas. PLoS One 8, e83980 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung H. H. et al. Methylation of an intronic region regulates miR-199a in testicular tumor malignancy. Oncogene 30, 3404–3415 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. F., Gu S., Suen Y. K., Li L. & Chan W. Y. microRNA-199a-3p, DNMT3A, and aberrant DNA methylation in testicular cancer. Epigenetics 9, 119–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J. et al. Reactive oxygen species regulate ERBB2 and ERBB3 expression via miR-199a/125b and DNA methylation. EMBO Rep 13, 1116–1122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. B. et al. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res 37, 123–128 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K. et al. MicroRNAs miR-199a-5p and -3p target the Brm subunit of SWI/SNF to generate a double-negative feedback loop in a variety of human cancers. Cancer Res 71, 1680–1689 (2011). [DOI] [PubMed] [Google Scholar]
- Wienholds E. et al. MicroRNA expression in zebrafish embryonic development. Science 309, 310–311 (2005). [DOI] [PubMed] [Google Scholar]
- Yi R. et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet 38, 356–362 (2006). [DOI] [PubMed] [Google Scholar]
- van Rooij E. et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A 103, 18255–18260 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer L. et al. Differential micro-RNA expression in primary CNS and nodal diffuse large B-cell lymphomas. Neuro Oncol 13, 1090–1098 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhakumar D. et al. Combined agonist-antagonist genome-wide functional screening identifies broadly active antiviral microRNAs. Proc Natl Acad Sci U S A 107, 13830–13835 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. K. et al. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res 70, 5807–5817 (2010). [DOI] [PubMed] [Google Scholar]
- Kwok. HPV-16 E6 upregulation of DNMT1 through repression of tumor suppressor p53. Oncol Rep 24, 1599–1604 (2010). [DOI] [PubMed] [Google Scholar]
- Cheng J. C., Auersperg N. & Leung P. C. Inhibition of p53 represses E-cadherin expression by increasing DNA methyltransferase-1 and promoter methylation in serous borderline ovarian tumor cells. Oncogene 30, 3930–3942 (2011). [DOI] [PubMed] [Google Scholar]
- Cheng J. C., Auersperg N. & Leung P. C. Inhibition of p53 induces invasion of serous borderline ovarian tumor cells by accentuating PI3K/Akt-mediated suppression of E-cadherin. Oncogene 30, 1020–1031 (2011). [DOI] [PubMed] [Google Scholar]
- Misiewicz-Krzeminska I. et al. Restoration of microRNA-214 expression reduces growth of myeloma cells through positive regulation of P53 and inhibition of DNA replication. Haematologica 98, 640–648 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W. et al. Retinoblastoma protein modulates gankyrin-MDM2 in regulation of p53 stability and chemosensitivity in cancer cells. Oncogene 27, 4034–4043 (2008). [DOI] [PubMed] [Google Scholar]
- Bauer S. et al. Therapeutic potential of Mdm2 inhibition in malignant germ cell tumours. Eur Urol 57, 679–687 (2010). [DOI] [PubMed] [Google Scholar]
- Kersemaekers A. M. et al. Role of P53 and MDM2 in treatment response of human germ cell tumors. J Clin Oncol 20, 1551–1561 (2002). [DOI] [PubMed] [Google Scholar]
- Omisanjo O. A. et al. DNMT1 and HDAC1 gene expression in impaired spermatogenesis and testicular cancer. Histochem Cell Biol 127, 175–181 (2007). [DOI] [PubMed] [Google Scholar]
- Antequera F. Structure, function and evolution of CpG island promoters. Cell Mol Life Sci 60, 1647–1658 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu Y. W. et al. Double RNA interference of DNMT3b and DNMT1 enhances DNA demethylation and gene reactivation. Cancer Res 63, 6110–6115 (2003). [PubMed] [Google Scholar]
- Vare P. & Soini Y. Twist is inversely associated with claudins in germ cell tumors of the testis. Apmis 118, 640–647 (2010). [DOI] [PubMed] [Google Scholar]
- Sun X., Guo J. H., Chen H., Ruan Y. C. & Chan H. C. Regulation of miR101/miR-199a-3p targeting cyclooxygenase-2 by the epithelial sodium channel during embryo implantation (911.5). FASEB J 28 (2014). [DOI] [PubMed] [Google Scholar]
- Siomi H. & Siomi M. C. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell 38, 323–332 (2010). [DOI] [PubMed] [Google Scholar]
- Soto A. M. & Sonnenschein C. The somatic mutation theory of cancer: growing problems with the paradigm? Bioessays 26, 1097–1107 (2004). [DOI] [PubMed] [Google Scholar]
- Dawson M. A., Kouzarides T. & Huntly B. J. Targeting epigenetic readers in cancer. N Engl J Med 367, 647–657 (2012). [DOI] [PubMed] [Google Scholar]
- Rodriguez-Paredes M. & Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med 17, 330–339 (2011). [DOI] [PubMed] [Google Scholar]
- Sharma S., Kelly T. K. & Jones P. A. Epigenetics in cancer. Carcinogenesis 31, 27–36 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Ooi L. L. & Hui K. M. MiR-214 targets beta-catenin pathway to suppress invasion, stem-like traits and recurrence of human hepatocellular carcinoma. PLoS One 7, e44206 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. J. et al. MiR-214 inhibits cell growth in hepatocellular carcinoma through suppression of beta-catenin. Biochem Biophys Res Commun 428, 525–531 (2012). [DOI] [PubMed] [Google Scholar]
- Chen D. L. et al. Identification of miR-214 as a negative regulator of colorectal cancer liver metastasis via regulation of FGFR1 expression. Hepatology 60, 598–609 (2014). [DOI] [PubMed] [Google Scholar]
- Derfoul A. et al. Decreased microRNA-214 levels in breast cancer cells coincides with increased cell proliferation, invasion and accumulation of the Polycomb Ezh2 methyltransferase. Carcinogenesis 32, 1607–1614 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer L. et al. Differential micro-RNA expression in primary CNS and nodal diffuse large B-cell lymphomas. Neuro Oncol 13, 1090–1098 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Julie Li Y. S., Huang H. D., Shyy J. Y. & Chien S. microRNA: a master regulator of cellular processes for bioengineering systems. Annu Rev Biomed Eng 12, 1–27 (2010). [DOI] [PubMed] [Google Scholar]
- Bushati N. & Cohen S. M. microRNA functions. Annu Rev Cell Dev Biol 23, 175–205 (2007). [DOI] [PubMed] [Google Scholar]
- Netto G. J. et al. Global DNA hypomethylation in intratubular germ cell neoplasia and seminoma, but not in nonseminomatous male germ cell tumors. Mod Pathol 21, 1337–1344 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami K. et al. DNMT3L Is a Novel Marker and Is Essential for the Growth of Human Embryonal Carcinoma. Clin Cancer Res 16, 2751–2759 (2010). [DOI] [PubMed] [Google Scholar]
- Arai E., Nakagawa T., Wakai-Ushijima S., Fujimoto H. & Kanai Y. DNA methyltransferase 3B expression is associated with poor outcome of stage I testicular seminoma. Histopathology 60, E12–E18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33, e179 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman J. G., Graff J. R., Myohanen S., Nelkin B. D. & Baylin S. B. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 93, 9821–9826 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]