Abstract
Inducible gene expression, which requires chromatin remodeling on gene promoters, underlies the epigenetically inherited differentiation program of most immune cells. However, chromatin-mediated mechanisms that underlie these events in T regulatory cells remain to be fully characterized. Here, we report that inducibility of FOXP3, a key transcription factor for the development of T regulatory cells, depends upon Kruppel-like factor 10 (KLF10) interacting with two antagonistic histone-modifying systems. We utilized chromatin immunoprecipitation, genome-integrated reporter assays, and functional domain KLF10 mutant proteins, to characterize reciprocal interactions between this transcription factor and either the Sin3-histone deacetylase complex or the histone acetyltransferase, p300/CBP-associated factor (PCAF). We characterize a Sin3-interacting repressor domain on the NH2 terminus of KLF10, which works to limit the activating function of this transcription factor. Indeed, inactivation of this Sin3-interacting domain renders KLF10 able to physically associate with PCAF as to induce FOXP3 gene transcription. We show that this biochemical data derived from studying our genome-integrated reporter cell system are recapitulated in primary murine lymphocytes. Collectively, these results advance our understanding of how a single transcription factor, namely KLF10, functions as a toggle to integrate antagonistic signals regulating FOXP3 and, thus, immune activation.
Keywords: T regulatory cell, KLF10, PCAF, Sin3, FOXP3
foxp3+ t regulatory (treg) cells may develop outside the thymus, generally in response to transforming growth factor β (TGFβ) and antigen to become critically important in intestinal immunologic homeostasis (adaptive Treg cells) (2, 6, 7, 22, 23, 26). Dysregulation of the transcription factor FOXP3, a key initiator of the Treg differentiation and functional program, leads to immune dysregulation in both mice (scurfy mouse) and humans (IPEX-immune polyendocrinopathy enteritis and X-linked- syndrome) (16, 38). While advances in T lymphocyte biology indicate the importance of TGFβ-induced activated T cells in both the induction (Th17 cells) and regulation (FOXP3+ Treg cells) of intestinal inflammation (3), the precise mechanistic events integrating the TGFβ signal to intracellular pathways that function as inducers or regulators of inflammation are not fully defined.
To identify these pathways, we recently characterized a role for a TGFβ-inducible Kruppel-like factor (KLF10) in silencing FOXP3 leading to enhanced colitis susceptibility, while providing mechanistic insights into how these functions require novel chromatin coupling events (40). These studies defined the importance of p300/CBP-associated factor (PCAF), a histone acetyltransferase (HAT) recruited by KLF10 to the FOXP3 transcriptional regulatory regions that are critical for the induction of this gene (40). In the experiments reported here, we build upon our previous discovery and demonstrate that KLF10 possesses the dual capacity to either positively or negatively regulate FOXP3 through its differential association with PCAF or the histone deacetylase binding protein Sin3, respectively. Collectively, these results increase our understanding of the chromatin pathways critical for mediating the inducibility of immune genes, such as FOXP3, in which alterations may cause human disease. Because drugs against these pathways are being used in clinical trials for several diseases, this new knowledge has both significant mechanistic value and potential biomedical relevance.
MATERIALS AND METHODS
Cloning of the FOXP3 promoter construct and the development of the genome integrated FLP cell line.
The cloning strategy has been previously described (40). Briefly, the human FOXP3 (NCBI AF235097) promoter locus, including the core promoter (−511 to +176) and the core promoter through the TGFβ-responsive enhancer region (Enhancer 1, −511 to 2738), was amplified by PCR using FOXP3 promoter sequence-specific primers. The genomic DNA extracted from CD4+ T cells of a healthy donor was used as template. The PCR product was subcloned initially into the pGL3 basic vector in frame with the luciferase reporter sequence (Promega, Madison, WI). We subsequently utilized the Flp-In system (Life Technologies, Grand Island, NY) for genome integration of the smaller FOXP3-core and larger FOXP3-core+enhancer 1 reporter constructs. First, we subcloned both FOXP3 promoter constructs with the luciferase reporter into the pcDNA5/FRT vector. Flp-In-Jurkat cells (Life Technologies) were cotransfected with either FOXP3-core or FOXP3-core+E1 in pcDNA5/FRT vector and the FLP-recombinase vector (pOG44) (FOXP3 core/pcDNA5/FRT ratio = 9:1), resulting in a stable integration of the gene of interest at the FLP recombination target (FRT) site in the genome. For the selective growth test, individual cells were grown in 24-well plates. The culture medium was supplemented with hygromycin at 250 μg/ml or 100 μg/ml. We subsequently refer to these cell lines as FLP-core or FLP-coreE1.
Mutation.
The KLF10 Sin3-interacting domain (SID) mutant was generated by using the Quick-Change site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). The binding domain fragment was mutated from GAAG to CCAC. The mutant construct was verified by sequencing.
The Human FOXP3 Core ChIP Primer is forward: 5′CAGATGACTCGTAAAGGGCAAAG-3′ and reverse: 3′CGATGAGTGTGTGCGCTGATAATC-5′.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) assays were performed using a ChIP isolation kit (EMD Millipore, Billerica, MA). One to two million Jurkat cells or primary lymphocytes were treated with 1% formaldehyde to cross-link histones to DNA. Fixed cells were sonicated to yield chromatin fragments of 200 to 1,000 bp. Antibodies used in the ChIP assays included Sin3a (no. 7691; Cell Signaling, Danvers, MA), His Tag (no. SC-7270; Santa Cruz Biotechnology, Dallas, TX), HDAC1 (no. 39534), and HDAC2 (no. 40967; Active Motif, Carlsbad, CA), acetyl-histone 4 (no. 06866; Millipore, Temecula, CA), and PCAF (no. Ab12188; Abcam, Cambridge, MA). DNA was recovered by phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation with the addition of an inert carrier. All ChIP results are presented controlled to FOXP3 expression of preimmunoprecipitated sample (input).
Quantitative real-time PCR.
Three microliters of above recovered DNA were used for each real-time PCR. PCR reactions were in 20 μl total volume that contained primers and 10 μl Express SYBR GreenER qPCR Supermaster mixes (Life Technologies). For control, we utilized preenriched chromatin (input).
For semiquantitative PCR, we amplified cDNA under the following conditions: initial denaturation, 94°C for 2 min, followed by 34 cycles with denaturation at 94°C for 30 s, annealing at 53°C for 30 s, and extension at 72°C for 60 s. All the PCR products were visualized by running 1.5% agarose gels electrophoresis and ethidium bromide staining for the pictures.
Transfection and luciferase assays.
Two million Jurkat cells were transfected using the Amaxa cell line nucleofector kit V for Jurkat cells, according to the optimized protocol provided with the kit (Lonza, Portsmouth, NH). Two micrograms of plasmid DNA for pcDNA 3.1, KLF10, KLF10-Sin3 mutation, and KLF family members were used in the nucleofection procedure. Luciferase assays were performed following the manufacturer's recommendations (Promega). For siRNA experiments, 300 nM siRNA was used (ON-TARGET plus siRNA, Thermo Scientific Dharmacon, Lafayette, CO). Cell stimulation conditions include plate-bound CD3 (2 μg/ml), soluble anti-CD28 (2 μg/ml; BD Biosciences, San Jose, CA), human IL-2 (100 ng/ml), and human TGF-β1 (5 ng/ml) in complete RPMI media (both cytokines; Peprotech, Rocky Hill, NJ).
Primary lymphocyte transduction and stimulation.
Naïve lymphocytes (CD4+CD62L+) were isolated using MACS bead magnetic sorting (no. 130–093-227; Miltenyi Biotech, Auburn, CA) from splenocytes of 4–6-wk-old mice. Ratio of viral particles to cell was MOI (multiplicity of infection) of 150. Cells were exposed for 48 h to adenoviral particles and subsequently washed prior to stimulation. For induction of FOXP3, cell-stimulating conditions included plate-bound CD3 (2 μg/ml), soluble anti-CD28 (2 μg/ml), human IL-2 (100 μg/ml), and human TGF-β1 (5 ng/ml) in complete RPMI media.
Immunoprecipitation and Western blot analysis.
Two micrograms of plasmid DNA for KLF10 in pcDNA3.1/His and KLF10-Sin3 mutation in pcDNA3.1/His were transfected into Jurkat cells by nucleofection, and 72 h later, cells were harvested. Protein was extracted from whole cell lysates derived from the harvested cells. The cells were lysed in lysis buffer (20 mM Tris·HCl, pH 8.0, 137 mM NaCl, 10% glycerol, 1% NP-40, and 2 mM EDTA). Protein was subjected to immunoprecipitation with His tag overnight and precipitated by protein A agarose beads. After three washes, we added 50-μl lysis buffer and boiled the eluant. The supernatants were run on 4–20% Bio-Rad gels. Upon transferral to nitrocellulose, the membrane was incubated with primary antibodies in 5% milk for 1 h. His tag and Sin3a proteins were detected with 1/25,000 dilution of anti-rabbit-horseradish peroxidase conjugate and peroxide solution and luminol enhancer solution detection kit (GE Health Care UK Limited).
For PCAF binding assays, 25 million pelleted Jurkat cells were lysed in 0.5 ml mammalian protein extraction reagent (MPER) lysis buffer (Thermo Scientific, Rockford, IL) supplemented with 0.5 M NaCl and Complete Inhibitor Cocktail (Roche Diagnostics, Indianapolis, IN), vortexed thoroughly, and incubated on ice for 5 min. 0.5 ml MPER buffer supplemented with Complete Inhibitor Cocktail without NaCl was then added for immunoprecipitation. Sixty microliters of glutathione S transferase (GST)-tagged protein was added. Sixty microliters of settled and washed glutathione beads (Sigma-Aldrich, St. Louis, MO) were incubated with lysate containing GST-tagged proteins. Reactions were rotated at 4°C for 3 h. After extensive washing, bound proteins were separated by SDS-PAGE and blotted with antibodies to PCAF (ab12188; Abcam). Three different antibodies were utilized to confirm non-cross-reactivity to other HATs (Anti-PCAF no. A-4012; Epigentek, Farmingdale, NY and no. AP12075b PCAF Antibody-c term; Abgent, San Diego, CA).
Molecular modeling of the KLF10 SID-Sin3 PHA2 complex with in silico mutational analyses.
The three-dimensional structure of the KLF10 SID complexed with the Sin3-paired amphipathic helix domain 2 (PAH2) domain was determined using a similar approach previously described for both KLF11 (41) and KLF16 (8). Briefly, we utilized the Sin3a-HBP1 complex (Protein Data Bank code 1s5r) as a template for KLF10-Sin3 SID homology-based modeling. After directly obtaining the three-dimensional structure of the Sin3a-PAH2 domain from the Sin3a-HBP1 complex, we modeled the structure of KLF10 SID bound with Sin3a-PAH2 domain (Protein Data Bank code 1G1E) by docking the KLF10 SID to the Sin3a-PAH2 domain to achieve maximal intermolecular interactions between the two partners using AutoDock 3.0.5 (27). In silico mutations were created using the builder module of Discovery Studio 4 (Accelrys, San Diego, CA), and the energy was minimized as previously described (14). Refinement of the complex involved a 2.0-ns (1-fs time step) molecular dynamics simulation using the CHARMm force field with an assignment of charges using the Momany-Rome method, as described previously (28). Using PROCHECK, the model was structurally verified and evaluated (21). For binding interphase analyses, complexes were energy minimized, and amino acid interactions were checked using the bond-monitoring function of Discovery Studio 4 (Accelrys) using a distance cutoff of 3.4 Å for conventional hydrogen bonds, 3.8 Å for weak hydrogen bonds, and 4 Å for salt bridges. The Ramachandran plot for the model showed 96.5% residues in most favored regions; thus, indicating that the model displays appropriate stereochemistry.
Chimera generation and colitis induction.
To make chimeras, 3- or 4-wk-old RAG-2−/− mice were total body-irradiated (10 Gy) before receiving 5 × 106 erythrocyte-depleted bone marrow cells that were extracted from KLF10−/− or wild-type mice, as previously described (9). Mice recovered for 2 wk, and flow analysis of peripheral blood demonstrated mature B- and T-cell populations (data not shown).
Mice were subsequently given water supplemented with 3% dextran sulfate sodium salt for 5 days. The water was then replaced with normal drinking water for 2 more days prior to the mice being killed for tissue removal for histology. The mice were weighed daily. The degree of colitis was quantified using three outcome variables: weight loss, colon histology, and a disease activity index. The disease activity index is an established clinical index of colitis severity encompassing clinical signs of colitis (wasting and hunching of the recipient mouse and the physical characteristics of stool) and an ordinal scale of colonic involvement (thickness and erythema) (13).
Statistical methodology.
Statistical analyses were performed using JMP version 9.0 (SAS institute, Cary, NC). Descriptive analyses, including means and standard errors were performed in normally distributed data using t-tests. For experiments with greater than two experimental groups, the Kruskal-Wallis nonparametric, single-sided ANOVA was used, and if variance were found to be significant, the Dunn's multiple comparison's test was utilized to demonstrate significant difference between selected data sets. A P value of <0.05 was considered as statistically significant.
Animal use.
All animal studies have been reviewed and approved by the Mayo Institutional Animal Care and Use Committee.
RESULTS
The TGFβ-inducible KLF10 represses the core FOXP3 promoter through physical association with the Sin3 corepressor protein.
Previous work has underscored a role for KLF10 in the chromatin-mediated silencing of FOXP3, the master transcription factor for Treg cells (40). However, the biochemical mechanisms that explain the functional interaction between these two transcription factors remain poorly understood. Consequently, we began this study utilizing Jurkat cell lines with genomic integration of relevant components of the FOXP3 core promoter in frame with a luciferase reporter construct (FLP Jurkat cells lines, see materials and methods). These cells lines are used to investigate KLF10-mediated transcriptional regulation of both the core promoter (FLP-core) and a larger regulatory region, including the downstream TGFβ-responsive enhancer (FLP-core E1) (34). Both sequences have been inserted into a single site of the Jurkat T-cell line genome using FLP recombinase, a model that circumvents the problems associated with the use of a nonepisomal reporter system, such as poor resolution of chromatin-mediated events (40). Basal luciferase production in these cell lines under the promotional control of cytomegalovirus (CMV) allows studies of transcriptional regulation in a manner similar to the widely used Gal4-based reporter, an episomal reporter system not optimal for epigenetic studies. We have previously demonstrated that the CMV promoter alone does not respond to KLF10-dependent regulatory mechanisms rendering the system appropriately controlled so as to draw useful inferences on how chromatin remodeling induced by this transcription factor affects the activity of the FOXP3 promoter (40).
Previous studies have demonstrated that KLF10 is required to recruit P300/CBP-associated factor (PCAF; KAT2B) to the FOXP3 core promoter for gene induction (40). Congruent with these data, it has been found that in vivo, KLF10 is required for activation of the FOXP3 promoter during generation of adaptive Treg cells using episomal reporter systems (40). However, using improved, genomically integrated reporters, here, we found that KLF10 instead significantly repressed this promoter (Fig. 1). Using closely related KLF family members (KLF9, KLF11, KLF12, KLF13, and KLF16), we demonstrated FOXP3 repression to be specific to KLF10 in both stimulated and unstimulated FLP-core cell lines (second column in Fig. 1, A and B, respectively).
Fig. 1.
Kruppel-like factor 10 (KLF10) represses the core FOXP3 promoter in vitro. Luciferase reporter assays in the genome-integrated Jurkat cell lines demonstrating repression of core FOXP3 promoter function by KLF10. Relative luciferase units (RLU) upon overexpression of KLF family members into FLP-core cells with (A) or without (B) activating conditions (see materials and methods). KLF10 uniquely represses FOXP3 promoter function in both activating and resting conditions (0.63 ± 0.04 and 0.53 ± 0.10 RLU, second column Fig. 1, A and B, respectively). RLUs are normalized to EV control (1.00, white column). The data are expressed as means ± SE of three independent experiments with *P = 0.035 (A) and P = 0.004 (B). C: chromatin immunoprecipitation assay demonstrating binding of KLF10 to the FOXP3 core promoter locus. Inset: representative DNA gel for PCR reaction analysis of the expression of FOXP3 in cell fractions postimmunoprecipitation for His. Quantitative real-time PCR analysis of the expression of FOXP3 in cell fractions postimmunoprecipitation for His-tagged KLF10 in FLP-core cells transfected with KLF10-His expression vector demonstrates significant binding of KLF10 to the core promoter (3.81 ± 0.70-fold change over EV control). Results are presented controlled to FOXP3 expression of preimmunoprecipitated sample (input). EV, empty vector. The data are representative of three independent experiments with *P = 0.04.
To confirm the direct regulatory role of KLF10 upon the core FOXP3 promoter, we performed ChIP assays in Jurkat FLP-core cells. Overexpression of KLF10 resulted in significant binding of KLF10 to the FOXP3 core promoter (Fig. 1C). These experiments reveal that KLF10 functions as a direct transcriptional regulator of FOXP3, leading us to subsequently investigate the chromatin-mediated mechanisms underlying this effect.
Molecular modeling predicts association of KLF10 with the Sin3 corepressor complex.
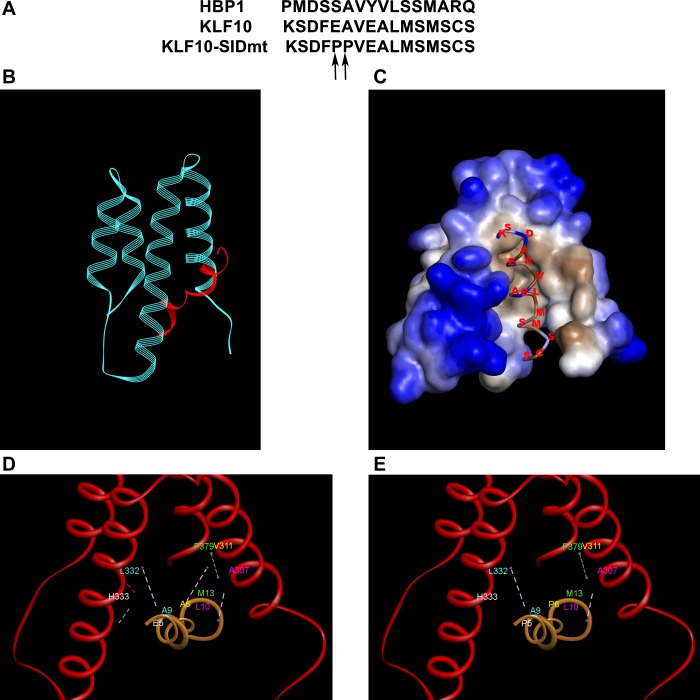
Previous studies have demonstrated that Sin3 interacts with several transcription factors through complexing of its second PAH with an α-helical repression motif present in the target protein, which accordingly has been termed the SID (Sin3 Interacting Domain) (28). Extensive biophysical and structural studies have demonstrated that critically located hydrophobic residues, which are embedded in an amphipathic α-helix, are critical for the interaction of transcription factors with Sin3. Interestingly, bioinformatics analyses using the linear motif searching software ELM (10) demonstrate that amino acids 36 to 51 within KLF10 contain a sequence that conforms to the SID consensus [FHYM].A[AV].[VAC]L[MV].[MI] previously identified in Sin3-interacting repressor proteins like HBP1 (Fig. 2A). Because the NMR structure of the HBP1-SID-Sin3 has been previously solved (32), this finding led us to use this complex to develop the first homology-based structural models for the putative KLF10 SID bound to the Sin3 PAH2 domain (4, 32) (Fig. 2), so as to further orient our subsequent functional studies using site-directed mutagenesis. Note that this protein complex adopts a five-helix-bundle fold, where the deep cleft of PAH2 is enriched in hydrophobic amino acids forming the docking pocket for the apolar upper surface of the KLF10 α-helix (Fig. 2B). The sidechains of V302, A307, I308, V311, L329, L332, Y335, V358, F376, and F379 from all four helices are sequestered from the hydrophilic surface to form the hydrophobic pocket of PAH2, thus, accommodating the KLF10 SID (Fig. 2C). Careful inspection of the protein-protein interaction interphase revealed that the KLF10 SID E5 forms a critical bond with Sin3 H333, KLF10 A6 with Sin3 V311, KLF10 A9 with Sin3 L332, KLF10 L10 with Sin3 A307, and KLF10 M13 with Sin3 F379 (Fig. 2D). Note KLF10 AA sequence numbering in Fig. 2 refers to the SID, not the full-length KLF10 protein. This information prompted us to evaluate the contributions of selected KLF10 SID residues toward the Sin3-dependent function by site-directed mutagenesis and immunoprecipitation. We sought to disrupt the critical binding between KLF10 E5 with Sin3 H333 and KLF10 A6 with Sin3 V311 by proline mutagenesis. The comparative binding interphase analyses depicted in Fig. 2, D and E predicts that these mutations would indeed impair Sin3 binding.
Fig. 2.
Putative structural model for the Sin3-PAH2 and KLF10-Sin3 interacting domain (SID) complex. Molecular modeling of KLF10 and Sin3 interaction utilizing HBP1 as a template was performed to predict disruptive domain mutations. A: sequence alignment of HBP1 SID, putative wild-type (wt) KLF10 SID, and E5P, A6P double KLF10 SID mutant. The established SID HBP1 was used as our modeling template. Consensus of the minimum SID is demonstrated in shaded gray. B: upper apolar surface of KLF10 SID alpha helix established hydrophobic interactions within the similarly charged pocket created by the four-helix bundle of the Sin3-PAH2. C: sequestration of hydrophobic sidechains (depicted as brown) away from hydrophilic surface (depicted as blue) to form the hydrophobic pocket, which accommodates the KLF10 SID. D: critical bonds predicted between KLF10SID and Sin3 upon examination of protein-protein interaction interphase. Pairs of interacting residues are labeled with the same color, while bonds are indicated by lines. E: predicted disruption of binding between KLF10 SID E5 with Sin3 H333 and KLF10 A6 with Sin3 V311 by proline mutagenesis. Pairs of interacting residues are labeled with the same color, while bonds are indicated by lines. Note the disappearance of lines indicating disruption of bonds upon mutations of E5 and A6 to P.
Indeed, the binding experiments shown in Fig. 3 experimentally confirm this prediction by showing that while the wild-type (wt) KLF10 interacts with Sin3, this binding is abrogated by the EA to PP mutations (Fig. 3A). Importantly, using ChIP assay, we determined that mutation of the SID does not affect the binding of KLF10 to DNA (Fig. 3B). These data indicate that the functional impact of this mutation is to selectively abolish chromatin coupling without affecting genomic binding by this transcription factor. This observation allows us to utilize the Jurkat FLP cell lines to define the role for the KLF10-SID in FOXP3 core promoter transcriptional activity. The KLF10-SID mutant (KLF10-SIDmt) was no longer capable of repressing FOXP3 core promoter function (Fig. 4A). Overexpression of the KLF10-SIDmt in the core-E1 FLP cell line produced equivalent results (Fig. 4B). Thus, these results identified an important mechanism for repressing FOXP3 through the coupling of KLF10 to the corepressor Sin3, via a defined HBP1-like SID.
Fig. 3.
Proline mutagenesis of the SID disrupts KLF10-Sin3 protein/protein interaction but not KLF10 DNA binding. A: immunoblot using antibody specific for Sin3 (top) or His tag (bottom) of Jurkat cell lysates after overexpression of His-tagged KLF10 wt (left lane) or KLF10 SID mutant construct (right lane). Note that as predicted in the molecular modeling experiments, wt KLF10 physically interacts with Sin3, and the proline mutagenesis completely disrupts this interaction. The data are representative of three independent experiments. B: KLF10 or KLF10 SID binding mutant constructs were expressed in Jurkat cells to demonstrate by chromatin immunoprecipitation the preserved competence of DNA binding by the KLF10 SID mutant construct. Inset, top: representative DNA gel for PCR reaction analysis of the expression of FOXP3 in cell fractions postimmunoprecipitation for His. Bottom: quantitative real-time PCR analysis of the expression of FOXP3 in cell fractions postimmunoprecipitation for His-tagged KLF10 demonstrates significant binding of both KLF10 (black column, 3.63 ± 0.3-fold change) and KLF10-SIDmt (gray column, 4.29 ± 0.2-fold change) constructs to the core promoter compared with EV control. Note the ability of the KLF10-SID mutant to bind DNA, equivalent to wt KLF10. The data are representative of three independent experiments; *P = 0.024.
Fig. 4.
KLF10 represses FOXP3 through association with Sin3. Luciferase reporter assays in genome-integrated Jurkat cell lines demonstrating disruption of repressor function upon mutation of the KLF10 SID. Relative luciferase counts upon overexpression of KLF10 or KLF10-SIDmt into FLP core (A) or FLP-core E1 cells (B), normalized to empty vector control (1.00, white column). Note the established repression of FOXP3 gene transcription by wt KLF10 is abrogated by proline mutagenesis of the KLF10-SID [0.97 ± 0.05 vs. 0.45 ± 0.07 RLU (A); and 1.14 ± 0.12 vs. 0.79 ± 0.03 RLU (B)]. The data are expressed as means ± SE of six independent experiments, *P = 0.003 (A); *P = 0.009 (B).
Members of the histone deacetylase subfamily I are critical for Sin3-mediated repression of FOXP3 by KLF10.
Given the established role for histone acetylation/deacetylation states and the transcriptional activity of FOXP3 (15, 33, 37), we next explored the role for a Sin3-deacetylase complex in this important regulatory process. We performed a series of ChIP assays to characterize the capacity for KLF10 to recruit Sin3 to the FOXP3 core promoter and the resultant effect on the resident histone acetylation state. Upon overexpression of the KLF10 wt construct, we observed a 2.7-fold increase of Sin3 at the FOXP3 core promoter (Fig. 5A) with a 0.44-fold change in histone 4 baseline acetylation state (Fig. 5B). Moreover, we found that the KLF10-SIDmt, a construct that does not bind Sin3 or repress transcriptional activity (Figs. 3A and 4A), failed to induce Sin3 recruitment to the core promoter (Fig. 5A) or lead to histone 4 deacetylation (Fig. 5B). Concordant with the Sin3-dependent loss of histone 4 acetylation at the FOXP3 core promoter, ChIP assay specific to histone deacetylase 1 (HDAC1) demonstrated a 1.25-fold increase of this enzyme at the core promoter upon expression of the wt KLF10 but not the KLF10-SIDmt (Fig. 5C). Thus, this line of experimentation demonstrated that a chromatin-mediated mechanism of FOXP3 silencing through KLF10 requires Sin3 and the corepressor complex member, HDAC1.
Fig. 5.
KLF10 recruits HDAC chromatin-modifying complexes to the FOXP3 promoter. Chromatin immunoprecipitation assay demonstrating that recruitment of the Sin3-HDAC repressor complex to the FOXP3 core promoter locus is dependent upon the KLF10 SID. Quantitative real-time PCR analysis of the expression of FOXP3 in cell fractions postimmunoprecipitation for Sin3 (A), H4 polyacetylation (B), or HDAC1 (C) in FLP-core-E1 cells transfected with KLF10 or KLF10-SIDmt expression vectors. Note the capacity of wt KLF10 but not the KLF10-SIDmt protein to recruit Sin3 (2.717 ± 0.35 vs. 1.63 ± 0.09-fold change; *P = 0.024) and histone deacetylase HDAC1 (1.25 ± 0.12-fold change vs. 0.43 ± 0.05-fold change; *P = 0.024) to the core promoter resulting in loss of baseline histone 4 acetylation state (0.44 ± 0.15 vs. 0.82 ± 0.14-fold change; *P = 0.034). The data represent means ± SE of three independent experiments. Inset: representative DNA gel for PCR reaction analysis of the expression of FOXP3 in cell fractions postimmunoprecipitation for Sin3 (A) and poly H4-Ac (B).
Evidence for the existence of a histone acetyltransferase pathway mediated by PCAF with the ability to antagonize KLF10-Sin3-HDAC-dependent FOXP3 repression.
KLF family members can frequently regulate both gene activation and repression (5, 14, 24). Indeed, we have previously identified a key role for PCAF-mediated KLF10 functions in the activation of both murine and human FOXP3 promoters (40). For this purpose, we again utilized the FOXP3 core-E1 promoter genome integrated cell line to directly test the ability of PCAF, KLF10, and Sin3 to competitively regulate gene transcription. Consistent with previous data, overexpression of the wt KLF10 construct significantly represses FOXP3 core promoter function (black columns 2 and 3, Fig. 6), and FOXP3 repression depends upon the SID (KLF10-SIDmt, fourth column, Fig. 6). To characterize the role for PCAF in the derepression evident upon mutation of the SID, we performed identical experiments upon knockdown of PCAF through siRNA methodology. Notably, through an apparent dominant effect of the SID-Sin3-HDAC repressor mechanism, the knockdown of PCAF does not affect KLF10 repression of FOXP3 gene transcription (middle black column, Fig. 6). In contrast, PCAF knockdown significantly abrogates the activity of the FOXP3 promoter when the interaction of KLF10 with the Sin3-HDAC complex is disrupted by mutation of its SID (furthest right column vs. fourth column, Fig. 6).
Fig. 6.
Derepression depends upon the histone acetyltransferase (HAT), p300/CBP-associated factor (PCAF). Luciferase reporter assay demonstrates that abrogation of repressor function evident upon mutation of the KLF10 SID is dependent upon the HAT, PCAF. Relative luciferase counts, normalized to empty vector control (1.00; white column), upon overexpression of KLF10 (black columns), or KLF10-SIDmt (columns 3–5) into FLP-core cells with concomitant knockdown of PCAF (PCAF siRNA) or scramble siRNA (scr) control. KLF10 represses FOXP3 promoter function [0.62 ± 0.03 relative light units (RLU), second column from the left] only when the SID domain is intact (1.65 ± 0.34 RLU KLF10-SIDmt, second column from the right). Note the derepression evident upon proline mutagenesis of the KLF10-SIDmt (second column from the right) entirely depends upon the expression of PCAF (1.65 ± 0.34 RLU, second column from the right vs. 0.58 ± 0.13 RLU, furthest right column; P = 0.005). The data express the means ± SE of 3 independent experiments.
These data suggest that Sin3 repression is independent of PCAF activity (black columns); however, PCAF derepression depends upon abrogation of Sin3-KLF10 interaction. These data are congruent with our previously published report that PCAF is recruited to the FOXP3 core promoter in wt murine lymphocytes, but not in KLF10-deficient lymphocytes (40). However, it remains to be established whether the access of PCAF to the FOXP3 promoter, although facilitated by KLF10, involves a direct protein-protein interaction with this transcription factor or occurs through the recruitment by other proteins. Therefore, we next explored the potential for physical association between KLF10 and PCAF. For this purpose, we generated GST fusion proteins of either full-length KLF10 or truncation mutants. We initially performed binding assays using purified recombinant PCAF that demonstrated, by Western blot analysis, putative interaction in this cell-free system to binding domains in both the NH2 (KLF10 1–210) and COOH terminus (KLF10 210–350) (Fig. 7A). Extending these results to an in vivo system, we repeated GST binding assays using Jurkat cell lysates. Using this system, we find that differently from other related transcription factors, such as KLF16 and KLF13, KLF10 couples to PCAF not through the zinc finger domain but rather via the NH2-terminal region of the protein (1–210), Fig. 7B. Thus, combined, our biochemical and transcriptional assays demonstrated that KLF10 interacts with both PCAF and Sin3, forming complexes that mediate FOXP3 activation and repression, respectively. To confirm physiological relevance, we subsequently explored this differential interaction in primary murine lymphocytes.
Fig. 7.
KLF10 associates with PCAF. Assays of protein-protein interaction demonstrate physiologically relevant interaction between PCAF and the N-terminus domain of KLF10. A: immunoblot for PCAF in cell-free association assay of purified PCAF and GST constructs indicated. Note in this cell-free assay, PCAF associates with both the NH2 and COOH terminus of KLF10. B: immunoblot for PCAF using lysates of Jurkat lymphocytes incubated with GST fusion proteins (GST control, GST-KLF10 1–210, or GST-KLF10 210–350). Purified PCAF run on the left lane represents the positive control. Note that in a physiologically relevant system using Jurkat cell lysates, PCAF clearly interacts with the NH2 terminus, but not the COOH terminus of KLF10.
We systematically evaluated the role for KLF10 in primary lymphocytes beginning with the whole animal and culminating in mechanistic single-cell experiments. As we have previously demonstrated colitis susceptibility in the KLF10-deficient mouse (39), we began by confirming that the colitis susceptibility transfers with the immune compartment in bone marrow chimera models. Indeed, recipient mice of KLF10-deficient bone marrow demonstrate enhanced colitis, as assessed by weight loss, clinical disease activity, and histology in response to dextran sodium sulfate compared with wild-type bone marrow recipients (Fig. 8). Having demonstrated the relevance of KLF10 to intestinal immunoregulation in vivo, we focused more deeply on the function of KLF10 in primary CD4+ lymphocytes.
Fig. 8.
Rag2−/− mice reconstituted with KLF10−/− bone marrow exhibit more severe colitis than Rag2−/− mice reconstituted with wt bone marrow (BM). Data are presented as means ± SE (n = 6 mice per group). A: weight loss at the time of death. The values of body weight are expressed as a percentage of initial body weight on day 0. Rag2−/− reconstituted with KLF10−/− BM demonstrated significantly enhanced weight loss compared with mice reconstituted with wt BM (6.89 ± 0.53 vs. 3.64 ± 1.31%, P = 0.04). B: macroscopic disease score based on the presence of clinical signs of colitis (wasting and hunching of the recipient mouse and the physical characteristics of stool) and an ordinal scale of colonic involvement (thickness and erythema) shows Rag2−/− reconstituted with KLF10−/− BM had significantly enhanced disease activity compared with mice reconstituted with Wt lymphocytes (4.31 ± 0.38 vs. 2.64 ± 0.33; P = 0.01). Levels of cytokines IL-6 (C) and IL-10 (D) were determined in serum using mouse cytometric bead array cytokine assay (BD Biosciences, San Jose, CA, USA) and analyzed using FCAP array version 3 software (Soft Flow Hungary). Rag2−/− reconstituted with KLF10−/− lymphocytes demonstrated higher levels of proinflammatory cytokine IL-6 (166.3 ± 41.34 vs. 65.65 ± 27.60 pg/ml; P = 0.11) and lower levels of anti-inflammatory cytokine IL-10 than Rag2−/− mice reconstituted with Wt bone marrow (0.56 ± 0.56 vs. 6.60 ± 3.82 pg/ml; P = 0.08). E: representative histologic sections of mouse colon upon death from the two experimental conditions. Note a higher degree of crypt loss and diffuse eosinophilic infiltration in the KLF10−/− recipient animals compared with WT recipient controls.
We subsequently investigated endogenous FOXP3 gene regulation utilizing a C57/BL6 transgenic mouse line. In this mouse line, the transgenic expression of the Coxsackievirus and adenovirus receptor allows efficient gene transduction with adenoviral packaged expression vectors in resting lymphocytes (36, 40). Wild-type KLF10 or KLF10-SIDmt adenoviral constructs were packaged and transduced into resting CD4+ naïve murine lymphocytes. Greater than five-fold expression of transduced constructs was evident at 6–48 h (Fig. 9). The trend to enhanced KLF10 expression upon mutation of the SID likely relates to the established capacity of wt KLF10 to repress its own gene transcription (18). After a 48-h resting period, cells were stimulated for 5 days to induce FOXP3 through TCR activation and exogenous TGFβ (see materials and methods). Through a series of ChIP assays, we characterized chromatin changes that occur at the native FOXP3 core promoter upon overexpression of wild-type or SID binding mutant KLF10. Congruent with the cell line experiments, overexpression of wt KLF10 resulted in 2.4-fold increased recruitment of Sin3 to the native FOXP3 core promoter compared with the KLF10-SID binding mutant (Fig. 10, A and B). In the absence of Sin3 binding, overexpression of the KLF10-SID binding mutant led to 1.5-fold increased acetylation of histone 4 compared with wt KLF10 (Fig. 10, C and D), and an ∼7.5-fold increased recruitment of the HAT, PCAF at the native FOXP3 core promoter (Fig. 10, E and F). We repeated the above assay and performed alternative quantitative methodology in primary lymphocytes to confirm physiological relevance. By quantitative real-time PCR, we demonstrate KLF10 to recruit Sin3 (Fig. 10B) and result in loss of histone 4 acetylation at the FOXP3 core promoter in primary T cells transduced with wt KLF10 and activated to induce FOXP3 (Fig. 10D). Upon transduction with KLF10-SIDmt, PCAF is recruited to the core promoter with resultant restoration of H4 acetylation (Fig. 10, D and F).
Fig. 9.
Quantitative real-time PCR analysis of the expression of KLF10 in primary naïve CD4+ murine lymphocyte cells transduced with KLF10 or KLF10-SIDmt expression vectors. Note that overexpression of KLF10 adenoviral vectors results in 5–10-fold increased expression over the empty adenoviral vector. Results are presented controlled to KLF10 expression of empty vector-transduced cells.
Fig. 10.
: KLF10 associates alternatively with Sin3 or PCAF to repress or activate FOXP3 gene transcription in primary naïve CD4+ lymphocytes. Using adenoviral constructs in primary murine lymphocytes, chromatin immunoprecipitation demonstrates recruitment of alternatively Sin 3 or PCAF to the FOXP3 core promoter locus to depend upon the KLF10 SID. A, C, E: Inset: DNA gel for PCR reaction analysis of the expression of FOXP3 in cell fractions postimmunoprecipitation for Sin3 (A), poly histone 4 (H4)-Ac (C), and PCAF (C) in primary lymphocytes posttransduction with KLF10 or KLF10-SIDmt expression vectors. Histogram represents densitometry units normalized by input for associated gels. Note that recapitulating the data from cell lines, wt KLF10, but not the KLF10-SIDmt protein, recruits Sin3 to the core promoter (2.43 ± 0.18 vs. 1.00 ± 0.1 densitometry units normalized to input), resulting in a decrease of the histone 4 acetylation state (52.45 ± 4.45 vs. 77.95 ± 7.14 densitometry units normalized to input; C). KLF10-SIDmt recruits PCAF to the core promoter (11.43 ± 1.58 vs. 2.51 ± 1.09 densitometry units normalized to input). The data are expressed as means ± SE of three independent experiments. B, D, F: quantitative real-time PCR analysis of the expression of FOXP3 in cell fractions postimmunoprecipitation for Sin3 (B), H4 polyacetylation (D), or PCAF (F) in primary T cells transfected with EV, KLF10, or KLF10-SIDmt expression vectors. Note the capacity of wt KLF10 but not the KLF10-SIDmt protein to recruit Sin3 (2.93 ± 0.25 vs. 1.23 ± 0.30-fold change; P = 0.04). In the absence of Sin3, the KLF10-SIDmt recruits PCAF (4.56 ± 1.22-fold change vs. 0.51 ± 0.03-fold change) to the core promoter resulting in enhancement of the histone 4 acetylation state (0.27± 0.15 vs. 2.59 ± 1.59-fold change). Data are expressed as means ± SE of three independent experiments.
Importantly, we assessed the functional relevance of these KLF10-dependent chromatin modifications through native FOXP3 transcriptional activity. Five days after adenoviral transduction of wt or SID binding mutant KLF10 into primary murine CD4+ lymphocytes activated to induce FOXP3, mRNA was isolated, and quantitative RT-PCR was performed to measure FOXP3 gene transcriptional activity. Upon overexpression of the KLF10-SIDmt compared with wt KLF10, a seven-fold increase in FoxP3 mRNA was evident (Fig. 11A).
Fig. 11.
KLF10 overexpression drives FOXP3 protein production upon disruption of binding with Sin3. A: quantitative real-time PCR analysis of the expression of FoxP3 mRNA in primary CD4+ T-cell lysates posttransduction with wt KLF10 (white column) or KLF10-SIDmt (black column). Consistent with the chromatin landscape, KLF10-SIDmt results in enhanced native FOXP3 gene transcription in primary cells (7.02 ± 1.15 vs. 0.95 ± 0.12 fold change; *P = 0.03, means ± SE). Data are expressed as means ± SE of four independent experiments. Inset gel upper left demonstrates DNA gel for PCR reaction for the expression of Foxp3 in primary CD4+ T-cell lysates. B: flow cytometry for FOXP3 protein in primary KLF10-deficient CD4+ T lymphocytes induced with TGFβ posttransduction with wt KLF10, KLF10-SIDmt, or EV adenoviral constructs. Note the established block in FOXP3 transduction in the absence of KLF10 (EV, 6.8% left upper dot blot) and the significant enhancement in FOXP3 protein production upon reconstitution of cells with KLF10-SIDmt but not wt KLF10 (10.7% lower left dot plot vs. 1.7% upper right dot plot). The experiment was repeated three times and mean ± SE for FOXP3 protein expression in EV transduced (white column), KLF10 transduced (black column), and KLF10-SIDmt transduced (gray column) demonstrate significant enhancement in FOXP3 protein in KLF10-SIDmt transduced but not wt KLF10-transduced cells compared with EV (1.57 ± 0.07 vs. 0.57 ± 0.16; *P = 0.02).
Subsequently, we assessed the physiological relevance of these KLF10-dependent events by assay of FOXP3 protein production upon transduction of wt or SIDmt KLF10 constructs. Primary KLF10-deficient lymphocytes were transduced with KLF10 or KLF10-SIDmt adenovirus and activated for 12 days in conditions to induce FOXP3. Note, as demonstrated previously, KLF10-deficient CD4+ lymphocytes produce minimal FOXP3 protein upon TGFβ stimulation (40). The transduction of KLF10-SIDmt, but not wt KLF10, into KLF10-deficient CD4+ T cells leads to partial rescue of FOXP3 protein production (Fig. 11B). Taken together, these results demonstrate that KLF10 has the ability to silence or activate the FOXP3 core promoter through either association with the histone deacetylase corepressor complex, Sin3, or the histone acetyltransferase, PCAF. This observation has biological relevance as several intracellular signaling cascades have the ability to selectively activate or inactivate the interaction of many transcription factors with HAT or HDAC pathways which, in the case of KLF10, should function as a physiological switch for the regulation of FOXP3 by this protein (Fig. 12).
Fig. 12.
KLF10 dichotomous regulation of FOXP3. A model depicting how KLF10 functions as a switch integrating cell signaling cascades to alternatively activate or silence the FOXP3 core promoter through interaction with the HAT, PCAF (top) or the Sin3/HDAC complex (bottom), respectively.
DISCUSSION
In this report, we characterize a novel mechanism of reversible chromatin-dependent silencing of the key immunoregulatory gene FOXP3. We demonstrate coupling of the DNA-binding protein KLF10 with the Sin3/HDAC repressor complex. In the absence of this Sin3 binding event, KLF10 may couple with the HAT, PCAF to facilitate the activation state of FOXP3. While literature describing epigenetic regulation of FOXP3 to date has focused on less reversible DNA methylation events (19, 39), this report is the first to characterize a rapidly reversible, chromatin-dependent pathway, integrating key signaling events critical to immunoregulation. As persistent activation of FOXP3 is critical to Treg function (39), the chromatin-dependent events regulating induction or silencing of the FOXP3 promoter locus in humans are critically important. The importance of understanding these mechanisms is evident in the disease-inducing potential of self-reactive “ex-Treg cells” identified through cell fate mapping as to have extinguished FOXP3 expression (42); yet mechanistic insights into purported silencing events are lacking.
Epigenetic regulation of FOXP3.
A consistent feature of Treg cells with stable FOXP3 expression is relative hypomethylation of the conserved noncoding sequence 2 (CNS2) within intron 1 of the FOXP3 promoter locus (Treg-specific demethylated region, TSDR) (29). The hypomethylated CNS2 region has been demonstrated to bind the transcription factors CREB (cyclic adenosine monophosphate response element binding protein) and Ets-1 (E26-AMV virus oncogene cellular homolog 1) stabilizing expression, and this DNA methylation pattern appears to be most frequently associated with thymus-derived, “natural” Treg cells (20). Comparatively little is known about the chromatin-dependent events regulating adaptive Treg cells, the mechanisms of which we address here. Histone acetylation states, altered through the competitive recruitment of HAT and HDAC histone-modifying complexes are key mechanisms of chromatin dynamic regulation of inducible genes, particularly within the immune pathways (31). Indeed, recruitment of SMAD proteins to conserved nucleotide sequence 1 has previously been demonstrated to lead to histone acetylation at the core promoter; however, further mechanistic details were lacking (34). We have previously deduced indirectly through the study of KLF10-deficient lymphocytes that KLF10 likely plays a role in the recruitment of PCAF to the core promoter and induction of FOXP3 in adaptive Treg cells (40), yet the direct role for KLF10 to alternatively induce or repress FOXP3 expression through the regulation of acetylation/deacetylation of regional nucleosomes is the novel discovery outlined in this report. Our data would suggest that Sin3 repressor function appears to be dominant over PCAF activation; however, we do not formally exclude the possibility that Sin3 binding normally functions as a rheostat of PCAF-induced activation as has been demonstrated for KLF11 and the recruitment of heterochromatin protein 1 (25, 30). In this system, PCAF titration experiments do not affect FOXP3 gene activation (data not shown), and PCAF knockdown does not affect Sin3-mediated repression; thus, we favor the interpretation that Sin3 repressor function is dominant over PCAF activation. One would expect the Sin3-mediated mechanism to be inactivated in order for KLF10 to express PCAF-induced activation. Inactivation may occur through KLF10 posttranslational modifications downstream of lymphocyte cell signaling cascades in a similar manner to Sin3-binding disruption to KLF11 (12) and KLF16 (8).
Posttranslational modification of KLF10.
Key functionally relevant, posttranslational modifications, including serine/threonine phosphorylation and ubiquitination of KLF10 have been previously observed (1, 35). Several key pieces of data are important to consider within the context of this report. First, the SID of the closely related family member KLF16 bears a tyrosine residue (Y10) that is a Src-family tyrosine kinase target and upon which phosphorylation clearly regulates function (8). Second, ERK-induced phosphorylation of KLF11, the most closely related family member to KLF10, has been clearly demonstrated to disrupt Sin3 binding and repressor function (11). Finally, specific kinase inhibitor therapy has recently been shown to modulate Treg function (17). Thus, given the key role for TCR-dependent Src-family tyrosine kinases in lymphocytes, further experiments as to functionally relevant phosphorylation events on KLF10 are ongoing.
Perspectives and Significance
Thus, we report a novel mechanism of reversible chromatin-dependent silencing of the key immunoregulatory gene FOXP3. Through differential coupling with the Sin3/HDAC repressor complex or the HAT-PCAF, the DNA-binding protein KLF10 functions as a switch mediating FOXP3 repression or activation, respectively (model, Fig. 12). These insights have broad relevance to TGFβ-induced regulatory T cells; yet further investigation is required to draw conclusions on the role for KLF10 in maintenance of FOXP3 expression in stable Treg cells. Further insight into lymphocyte receptor signaling events leading to the modification of key functional domains of KLF10 will likely lead to novel therapeutic targets for induction or repression of FOXP3 and Treg cells.
GRANTS
This research is supported by grants from the National Institute of Allergy and Infectious Disease to W. A. Faubion, Jr. (NIH Grant AI89714-R01) and NIDDK-52913 and P30DK-084567 to R. A. Urrutia. Molecular modeling experiments were supported by the Epigenomics Translational Program of the Mayo Clinic Center for Individualized Medicine.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: Y.X., P.A.S., O.O.S., M.D., S.K., R.A.U., and W.A.F. performed experiments; Y.X., P.A.S., O.O.S., T.C.S., M.D., S.K., G.A.L., R.A.U., and W.A.F. analyzed data; Y.X., P.A.S., O.O.S., T.C.S., M.D., S.K., R.A.U., and W.A.F. prepared figures; Y.X., P.A.S., O.O.S., G.A.L., and W.A.F. drafted manuscript; Y.X., P.A.S., O.O.S., G.A.L., R.A.U., and W.A.F. edited and revised manuscript; Y.X., P.A.S., O.O.S., T.C.S., M.D., G.A.L., R.A.U., and W.A.F. approved final version of manuscript; P.A.S., O.O.S., G.A.L., R.A.U., and W.A.F. interpreted results of experiments; G.A.L., R.A.U., and W.A.F. conception and design of research.
REFERENCES
- 1.Alemu EA, Sjottem E, Outzen H, Larsen KB, Holm T, Bjorkoy G, Johansen T. Transforming growth factor-β-inducible early response gene 1 is a novel substrate for atypical protein kinase Cs. Cell Mol Life Sci 68: 1953–1968, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ana Faria HLW Oral tolerance MC. Immunol Rev 206: 232–259, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441: 235–238, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Brubaker K, Cowley SM, Huang K, Loo L, Yochum GS, Ayer DE, Eisenman RN, Radhakrishnan I. Solution structure of the interacting domains of the Mad-Sin3 complex: implications for recruitment of a chromatin-modifying complex. Cell 103: 655–665, 2000 [DOI] [PubMed] [Google Scholar]
- 5.Buttar NS, DeMars CJ, Lomberk G, Rizvi S, Bonilla-Velez J, Achra S, Rashtak S, Wang KK, Fernandez-Zapico ME, Urrutia R. Distinct role of Kruppel-like factor 11 in the regulation of prostaglandin E2 biosynthesis. J Biol Chem 285: 11433–11444, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen W, Jin W, Hardegen N, Lei Kj Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J Exp Med 198: 1875–1886, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curotto de Lafaille MA, Lafaille JJ. Natural and adaptive Foxp3+ regulatory T cells: more of the same or a division of labor. Immunity 30: 626–635, 2009 [DOI] [PubMed] [Google Scholar]
- 8.Daftary GS, Lomberk GA, Buttar NS, Allen TW, Grzenda A, Zhang J, Zheng Y, Mathison AJ, Gada RP, Calvo E, Iovanna JL, Billadeau DD, Prendergast FG, Urrutia R. Detailed structural-functional analysis of the Kruppel-like factor 16 (KLF16) transcription factor reveals novel mechanisms for silencing Sp/KLF sites involved in metabolism and endocrinology. J Biol Chem 287: 7010–7025, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Jong YP, Abadia-Molina AC, Satoskar AR, Clarke K, Rietdijk ST, Faubion WA, Mizoguchi E, Metz CN, Alsahli M, ten Hove T, Keates AC, Lubetsky JB, Farrell RJ, Michetti P, van Deventer SJ, Lolis E, David JR, Bhan AK, Terhorst C. Development of chronic colitis is dependent on the cytokine MIF. Nat Immunol 2: 1061–1066, 2001 [DOI] [PubMed] [Google Scholar]
- 10.Dinkel H, Van Roey K, Michael S, Davey NE, Weatheritt RJ, Born D, Speck T, Krüger D, Grebnev G, Kuban M, Strumillo M, Uyar B, Budd A, Altenberg B, Seiler M, Chemes LB, Glavina J, Sánchez IE, Diella F, Gibson TJ. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res 42: D259–D266, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellenrieder V, Buck A, Harth A, Jungert K, Buchholz M, Adler G, Urrutia R, Gress TM. KLF11 mediates a critical mechanism in TGF-β signaling that is inactivated by Erk-MAPK in pancreatic cancer cells. Gastroenterology 127: 607–620, 2004 [DOI] [PubMed] [Google Scholar]
- 12.Ellenrieder V, Zhang JS, Kaczynski J, Urrutia R. Signaling disrupts mSin3A binding to the Mad1-like Sin3-interacting domain of TIEG2, an Sp1-like repressor. EMBO J 21: 2451–2460, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faubion WA, De Jong YP, Molina AA, Ji H, Clarke K, Wang B, Mizoguchi E, Simpson SJ, Bhan AK, Terhorst C. Colitis is associated with thymic destruction attenuating CD4+25+ regulatory T cells in the periphery. Gastroenterology 126: 1759–1770, 2004 [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Zapico ME, van Velkinburgh JC, Gutiérrez-Aguilar R, Neve B, Froguel P, Urrutia R, Stein R. MODY7 gene, KLF11, is a novel p300-dependent regulator of Pdx-1 (MODY4) transcription in pancreatic islet β cells. J Biol Chem 284: 36482–36490, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang HD, Bopp T, Schmitt E, Klein-Hessling S, Serfling E, Hamann A, Huehn J. Epigenetic control of the Foxp3 locus in regulatory T cells. PLoS Biol 5: e38, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godfrey VL, Wilkinson JE, Russell LB. X-linked lymphoreticular disease in the scurfy (sf) mutant mouse. Am J Pathol 138: 1379–1387, 1991 [PMC free article] [PubMed] [Google Scholar]
- 17.Graham JA, Fray M, de Haseth S, Lee KM, Lian MM, Chase CM, Madsen JC, Markmann J, Benichou G, Colvin RB, Cosimi AB, Deng S, Kim J, Alessandrini A. Suppressive regulatory T cell activity is potentiated by glycogen synthase kinase 3β inhibition. J Biol Chem 285: 32852–32859, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guillaumond F, Grechez-Cassiau A, Subramaniam M, Brangolo S, Peteri-Brunback B, Staels B, Fievet C, Spelsberg TC, Delaunay F, Teboul M. Kruppel-like factor KLF10 is a link between the circadian clock and metabolism in liver. Mol Cell Biol 30: 3059–3070, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janson PC, Winerdal ME, Marits P, Thorn M, Ohlsson R, Winqvist O. FOXP3 promoter demethylation reveals the committed Treg population in humans. PLoS One 3: 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HP, Leonard WJ. CREB/ATF-dependent T-cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med 204: 1543–1551, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8: 477–486, 1996 [DOI] [PubMed] [Google Scholar]
- 22.Li MO, Sanjabi S, Flavell RA. Transforming growth factor-β controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity 25: 455–471, 2006 [DOI] [PubMed] [Google Scholar]
- 23.Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-β1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity 26: 579–591, 2007 [DOI] [PubMed] [Google Scholar]
- 24.Lomberk G, Grzenda A, Mathison A, Escande C, Zhang JS, Calvo E, Miller LJ, Iovanna J, Chini EN, Fernandez-Zapico ME, Urrutia R. Kruppel-like factor 11 regulates the expression of metabolic genes via an evolutionarily conserved protein-interaction domain functionally disrupted in maturity onset diabetes of the young. J Biol Chem 15: 15, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lomberk G, Mathison AJ, Grzenda A, Seo S, DeMars CJ, Rizvi S, Bonilla-Velez J, Calvo E, Fernandez-Zapico ME, Iovanna J, Buttar NS, Urrutia R. Sequence-specific recruitment of heterochromatin protein 1 via interaction with Kruppel-like factor 11, a human transcription factor involved in tumor suppression and metabolic diseases. J Biol Chem 287: 13026–13039, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marie JC, Letterio JJ, Gavin M, Rudensky AY. TGF-β1 maintains suppressor function and Foxp3 expression in CD4+CD25+ regulatory T cells. J Exp Med 201: 1061–1067, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comp Chem 30: 2785–2791, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang YP, Kumar GA, Zhang JS, Urrutia R. Differential binding of Sin3 interacting repressor domains to the PAH2 domain of Sin3A. FEBS Lett 548: 108–112, 2003 [DOI] [PubMed] [Google Scholar]
- 29.Polansky JK, Kretschmer K, Freyer J, Floess S, Garbe A, Baron U, Olek S, Hamann A, von Boehmer H, Huehn J. DNA methylation controls Foxp3 gene expression. Eur J Immunol 38: 1654–1663, 2008 [DOI] [PubMed] [Google Scholar]
- 30.Seo S, Lomberk G, Mathison A, Buttar N, Podratz J, Calvo E, Iovanna J, Brimijoin S, Windebank A, Urrutia R. Kruppel-like factor 11 differentially couples to histone acetyltransferase and histone methyltransferase chromatin remodeling pathways to transcriptionally regulate dopamine D2 receptor in neuronal cells. J Biol Chem 287: 12723–12735, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suarez-Alvarez B, Baragano Raneros A, Ortega F, Lopez-Larrea C. Epigenetic modulation of the immune function: A potential target for tolerance. Epigenetics 8: 7, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swanson KA, Knoepfler PS, Huang K, Kang RS, Cowley SM, Laherty CD, Eisenman RN, Radhakrishnan I. HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nat Struct Mol Biol 11: 738–746, 2004 [DOI] [PubMed] [Google Scholar]
- 33.Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, Li B, Turka LA, Olson EN, Greene MI, Wells AD, Hancock WW. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med 13: 1299–1307, 2007 [DOI] [PubMed] [Google Scholar]
- 34.Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol 9: 194–202, 2008 [DOI] [PubMed] [Google Scholar]
- 35.Venuprasad K, Huang H, Harada Y, Elly C, Subramaniam M, Spelsberg T, Su J, Liu YC. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat Immunol 9: 245–253, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan YY, Leon RP, Marks R, Cham CM, Schaack J, Gajewski TF, DeGregori J. Transgenic expression of the coxsackie/adenovirus receptor enables adenoviral-mediated gene delivery in naive T cells. Proc Natl Acad Sci USA 97: 13784–13789, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Tao R, Hancock WW. Using histone deacetylase inhibitors to enhance Foxp3+ regulatory T-cell function and induce allograft tolerance. Immunol Cell Biol 87: 195–202, 2009 [DOI] [PubMed] [Google Scholar]
- 38.Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, Levy-Lahad E, Mazzella M, Goulet O, Perroni L, Bricarelli FD, Byrne G, McEuen M, Proll S, Appleby M, Brunkow ME. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet 27: 18–20, 2001 [DOI] [PubMed] [Google Scholar]
- 39.Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol 8: 277–284, 2007 [DOI] [PubMed] [Google Scholar]
- 40.Xiong Y, Khanna S, Grzenda AL, Sarmento OF, Svingen PA, Lomberk GA, Urrutia RA, Faubion WA., Jr Polycomb antagonizes p300/CREB-binding protein-associated factor to silence FOXP3 in a Kruppel-like factor-dependent manner. J Biol Chem 287: 34372–34385, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang JS, Moncrieffe MC, Kaczynski J, Ellenrieder V, Prendergast FG, Urrutia R. A conserved alpha-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol Cell Biol 21: 5041–5049, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou X, Bailey-Bucktrout SL, Jeker LT, Penaranda C, Martinez-Llordella M, Ashby M, Nakayama M, Rosenthal W, Bluestone JA. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat Immunol 10: 1000–1007, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]