Abstract
Plasmodium falciparum alanine M1-aminopeptidase (PfA-M1) is a validated target for anti-malarial drug development. Presence of significant similarity between PfA-M1 and human M1-aminopeptidases, particularly within regions of enzyme active site leads to problem of non-specificity and off-target binding for known aminopeptidase inhibitors. Molecular docking based in silico screening approach for off-target binding has high potential but requires 3D-structure of all human M1-aminopeptidaes. Therefore, in the present study 3D structural models of seven human M1-aminopeptidases were developed. The robustness of docking parameters and quality of predicted human M1-aminopeptidases structural models was evaluated by stereochemical analysis and docking of their respective known inhibitors. The docking scores were in agreement with the inhibitory concentrations elucidated in enzyme assays of respective inhibitor enzyme combinations (r2≈0.70). Further docking analysis of fifteen potential PfA-M1 inhibitors (virtual screening identified) showed that three compounds had less docking affinity for human M1-aminopeptidases as compared to PfA-M1. These three identified potential lead compounds can be validated with enzyme assays and used as a scaffold for designing of new compounds with increased specificity towards PfA-M1.
Keywords: Drug designing, in silico screening, malaria, molecular docking, homology modeling
Background
The human malaria parasite Plasmodium falciparum infection leads to over two million deaths every year worldwide [1, 2]. During last six decades, considerable non-therapeutic malaria control measures have resulted in only limited success and with the limited success׳ of RTS,S/ASO1 in long term clinical trials, effective malaria vaccine is not in pipeline [3].The number of available antimalarial drugs are limited and Plasmodium has developed resistance against most of them, including second and third generation therapeutics such as artemisinin, antifolates and their derivatives [4, 5]. The problem of antimalarial drug resistance gets further aggravated by the existence of cross-resistance amongst drugs belonging to the same chemical series [4, 5]. Thus, it is essential to explore novel targets for antimalarial drug development.
Parasite specific hemoglobin degradation pathway is of special interest for development of antimalarial drugs [6]. During its intra-erythrocytic developmental stage Plasmodium catabolises >75% of the host cell haemoglobin inside its digestive vacuole, followed by terminal stage degradation in both parasite cytosol as well as vacuole [7, 8]. The free amino acids released from hemoglobin digestion are not only vital for parasite growth and development but also for maintaining osmotic integrity of the infected red blood cells and exchange of isoleucine with leucine from the RBC cytoplasm [9]. During this haemoglobin degradation process, two families of proteases- aspartic proteases (plasmepsins) and cysteine proteases (falcipains) degrade haemoglobin in small peptides which are subsequently digested by exopeptidases [10]. Plasmepsins and falcipains have not been much successful as antimalarial drug target possibly because of their overlapping functions [10]. On the other hand, out of eight available exopeptidases in Plasmodium, leucine and alanine exopeptidases are non-redundant and genetically essential [2, 7, 11]. These two metalloaminopeptidases, leucine aminopeptidase (PfA-M17) and alanine aminopeptidase (PfA-M1) are critical for parasite survival, because leucine (12.53 %) and alanine (13.23%) are not only most abundant amino acids in hemoglobin but also because plasmepsin and falcipainen prefer either of these two amino acids at their cleavage sites [12]. Thus, possibility of getting either alanine or leucine as N-terminal amino acids of hemoglobin derived peptide is very high. PfA-M1 further gets validated as drug target because inhibition of this enzyme by dipeptide analog bestatin leads to the parasite death [7, 13].
PfA-M1 belongs to M1-aminopeptidase family and has twelve homologues in human genome [14]. Available aminopeptidase inhibitors (bestatin and its derivatives) are non-specific in nature and inhibit almost all known aminopeptidases, including those from Plasmodium and human [15].
Although key active site residues are conserved across aminopeptidases, but during the course of evolution and neofunctionalization, these M1 aminopeptidases have gone considerable sequence and structural changes, both in the active site cavity and rest of the protein structure. These sequence and structural differences between Plasmodium and human aminopeptidases can be exploited for the development of parasite specific aminopeptidase inhibitors. However, it is essential for any potential PfA-M1 specific inhibitor, identified either through ‘high throughput screening’, ‘virtual screening’, or any other method to be evaluated for their off-target activity. In silico structure based screening can be useful to predict offtarget binding of PfA-M1 inhibitors to human aminopeptidases. This approach has advantage of discarding compounds that show high affinity binding to targets human M1- aminopeptidases, at earlier stage of drug development. However, 3D-structures of only five out of twelve known human M1-aminopeptidases are available [16– 20]. Therefore, in the present study 3D-structures of remaining seven human M1- aminopeptidases were modeled using combination of homology modeling, threading and ab-initio modeling. After stereochemical and geometric evaluation of the modeled 3Dstructures were subjected to docking studies with their respective known inhibitor, to evaluate correlation between docking based predictions with enzyme assay experiments. Fifteen potential PfA-M1 inhibitors identified through virtual screening were further tested for their PfA-M1 specificity, out of which three compounds showed preferential binding towards PfA-M1 in comparison to the human M1- aminopeptidases.
Methodology
Selection of PfA-M1 human homologues:
The human homologues of PfA-M1 were selected from both literature [14, 21], and similarity search tools BLAST and PSIBLAST Table 1 (see supplementary material). The protein sequences of human M1-aminopeptidases, Aminopeptidase-Q (APQ) (Accession no. NP_776161.3), Placental LeucineAminopeptidase (PLAP) (Accession no. NP_005566.2), Puromycin Sensitive Aminopeptidase (PSA) (Accession no. NP_006301.3), Thyrotropin Releasing Hormone Degrading Ectoenzyme(TRHDE) (Accession no. NP_037513.1), Aminopeptidase-B (APB) (Accession no. NP_064601.3), Aminopeptidase-O (APO) (Accession no. NP_001180258.1) and Aminopeptidase B-like (APB) (Accession no. NP_060696.4), were retrieved from NCBI database (Table 1).
Modeling of 3D structure of PfA-M1 human homologs:
Reference structural templates for each protein sequence were identified through pairwise and multiple alignment using BLAST and CLUSTAL OMEGA. Modeling was done by using multiple templates through Phyre 2, Modeller 9.11 and Robetta server [22–24]. Ten models were generated for APQ, PLAP, TRHDE and PSA each by Phyre 2. For each APB, APB-L and APO, 5 models were generated using Modeller 9.11 and Robetta respectively [23, 24]. The generated models were energy minimized in water using OPLS force field with the convergence threshold of 0.05 by using Macromodel of Maestro – Schrodinger to remove steric clashes between atoms and to improve overall structural quality of predicted models [25].
Validation of Models:
3D- models were validated on the basis of stereochemical and geometric consideration and docking studies. The quality and stereochemistry of the models were evaluated using the Procheck, Whatcheck, Verify-3D, ERRAT, QMEANnorm and alignment with their respective template [26– 29]. The predicted protein structure models were ranked on the basis of QMEANnorm score, geometric and stereochemical considerations. The top ranked models were further validated and analyzed based on their Ramachandran plot, root mean square deviation values, Verify3D and ERRAT analysis.
Docking:
Docking studies were done in two stages using Grid based ligand docking with energetic (GLIDE) [30]. 1. The 3-D models of all seven human M1-aminopeptidases were docked with their known inhibitors retrieved from literature and Pubchem Bioassay search and most effective compounds were used for docking [31– 34]. 2. General aminopeptidase inhibitor Bestatin was docked against all seven human M1-aminopeptidases structural models and ligand striped PfA-M1 (3EBG). The receptor grid was generated using the metal binding sites as well asblind docking. The different conformations of the compounds were docked flexibly and maximum 1000 poses per compound were generated. The analysis of the poses, complexes and the binding affinities between the receptor and ligands was analyzed using Schrodinger׳s software [25] and correlation coefficient between Ki/IC50 and docking score was calculated using ‘CORREL’ function of MS Excel.
Screening of potential PfA-M1 Inhibitors for off target binding:
Fifteen potential PfA-M1 inhibitors, screened through virtual screening, were selected on the basis of their binding affinity and stability (as evident from docking score and binding energy). The selected compounds were evaluated for off-target binding by docking against human M1-aminopeptidases using the docking protocol as mentioned above.
Hypothesis:
Multiple sequence alignment showed high sequence diversity amongst human (7.3 to 12.7% identities) as well as between human and P. falciparum aminopeptidases (6.5% to 12.7 % identity), with high variability at N-terminal region as compared to C-terminal catalytic domain (Supplementary data Figure 1). The signature ‘GAMEN’ motif is conserved in P. falciparum and six human M1-aminopeptidases (APN, APA, PLAP, ERAP1, ERAP2 and PSA). This motif is uniquely substituted by ‘HAMEN’ motif in APQ, ‘GGMEN’ motif in LTA4 and APB, ‘AAMEN’ motif in TRHDE, ‘VAMEN’ motif in APBLikeprotein and ‘LGMAS’ motif in APO. The Zn++ binding motif (HEXXHX18E) remains conserved across all the M1- aminopeptidases studied in the present work. Further, it was also observed that active site residues of PfA-M1 Glu 519, Tyr 580 are conserved in all its human homologues and Glu 463 and Ala 461 residues are conserved in 11 (except APO) and 9 (except APB, APO, LTA4) of the human M1-aminopeptidases respectively Supplementary data Figure 1.
Validation of predicted structural Models
Stereo chemical evaluation of models:
All the proteins show same four domain architecture but differ in arrangement of helices and sheets (Figure 1). The potential energy of the predicted human M1-aminopeidases ranged from 1.948e+05 to -1.750e+05 Kcal/mol, Qmean global score 0 to 1, the global Qmean6 score 0.544 to 0.625 and Qmean Z-scores - 1.95 to -2.43 and G-factor value -0.12 to -0.35, indicating absolute quality of models.The geometry of structural models were further evaluated using Ramachandran Plot and 98%-99% of residues were observed in favorable and allowed region Table 2 (see supplementary material). RMS Z-score were positive and near to 1, Verify-3D score ranged from 0.64 to 0.77, and ERRAT score ranged from 71.4 to 89.33 indicating primary structures compatibility with the environment of the residues in the 3D structure (Table 2).
Figure 1.
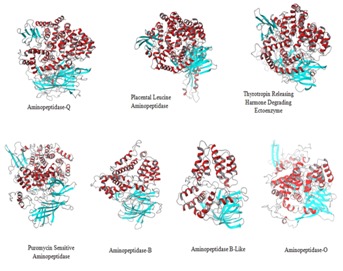
Ribbon representation of predicted 3D structural models of seven human M1 aminopeptidases. All the proteins show same four domain architecture but differ in arrangement of helices and sheets leading to differences in volume and accessibility of active site.
Molecular docking vs in vitro enzyme inhibition assays:
Results summarized in Table 3 (see supplementary material). Showed that most of the active compounds had good agreement between the docking score and experimental results (r2≈0.7). It suggests that the parameters of docking simulations and quality of structural models are good in reproducing experimental course of these compounds in all the modeled human M1- aminopeptidases. Further, bestatin docked in active site of all human M1 aminopeptidases and shoed interactions with active site residues (Figure 2) The observed difference among bestatin docking scores among PfA-M1 (- 6.0) and different human M1-amnopeptidases (-5.0 to 7.1) and binding energy (-35.30) and (-47.28 to -71.29) may be due to difference in binding affinity of bestatin towards different aminopeptidases as observed in inhibitory efficiency (Ki) of bestatin against different aminopeptidases [2, 35]. The bestatin PfA-M1 interactions observed with in silico docking and in the experimentally resolved structure were also in agreement [36].
Figure 2.
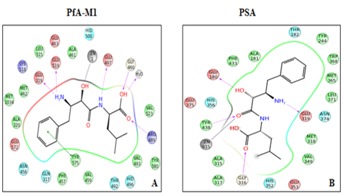
Ligand interaction diagrams of bestatin docking in PfA-M1 (A) and human PSA (B). Bestatin shows formation of hydrogen bond with active site residues and metal ion in both PfA-M1 and human PSA.
Human M1-aminopeptiadases structural models:
Structural alignment of predicted human M1-aminopeptidase structural models with their respective templates show an alignment score ranged from 0.07 to 0.20 and an RMSD score from 1.44 to 2.18 respectively. While superimposition of the Cα- backbone of predicted models with PfA-M1 shows RMSD value of ranged from 3.04-3.95 Å (Figure 3A-B) and RMSD value 1.47 - 3.96 Å among different human M-aminopeptidases. Indicating high level of structural difference among different human M1-aminopeptidases and between that of human and Plasmodium M1-aminopeptidases. Volume of active site cavity also varied among PfA-M1 (2132.77 Å3) and human M1- aminopeptidases (347.80 -2684.66 Å3). These differences have possibly evolved to accommodate substrates of different shape and size in their respective enzyme active site cavity viz antigenic peptide and cytokine receptors for ERAP1 and ERAP2, Leukotriene A4 for LTA4, oxytocin and vasopressin for PLAP.
Figure 3.
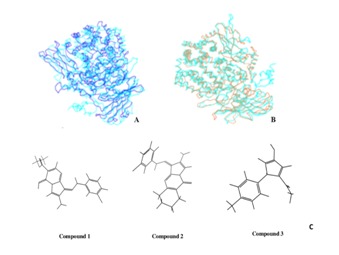
Backbone superimposition of PfA-M1 (in dark blue) with PSA (in turquoise) with (A) and (B) APN (in brown). C Molecular structureof three PfA-M1 inhibitors that shows high affinity binding towards P. falciparum PfA-M1 than most of the human M1 aminopeptidases.
Screening of potential PfA-M1 inhibitors for off target binding:
Out of fifteen compounds selected for high binding affinity towards PfA-M1, three compounds (Compound 1, Compound 2 and Compound 3) showed weak binding affinity towards human M1-aminopeptidases as compared to PfA-M1 (Figure 3C). Compound 1 had strong binding affinity for PfA-M1 (docking score -11.25 and binding energy -84.71 Kcal/mol) and forms very stable complex with it as compared to human M1- aminopeptidases. Strong H-bond interactions were formed with Glu 497, Arg 489 and Glu319 and hydrophobic interactions observed with His 500, Met 571, Thr 305, Val 459 and Met 1034. Docking result shows that human LTA4 (docking score -10.80) has strong affinity for compound1 but in terms of stability LTA4 does not form a stable complex,as indicated by less binding energy (-54.10 Kcal/mol) than PfAM1 (-84.71 Kcal/mol). APQ and APB-L having docking score of -5.56 and 4.86 respectively had weak affinity for Compound 1. In APB-L the ligand formed hydrogen bond with Phe 217and had major hydrophobic interactions with Ala 318, Val 218, Ala 175, Leu 314andAla 317. Interestingly no interactions were observed in the conserved metal binding site region. Other enzymes that appeared to be sensitive to Compound1 are APN, ERAP2 and PSA.
Compound 2 showed strong binding affinity for PfA-M1 (docking score -11.07 and binding energy -54.98 Kcal/mol). The ligand formed coordination bond with Zn and hydrogen bonds with residues Ala 461, Glu 463 and Glu 319 of active site (Figure 4A). Human M1 aminopeptidases LTA4, ERAP1, APO, APB, APB-L, PSA, APQ and APA had weak affinity for Compound 2 (docking score ranging from -8.3 to -4.9) as compared to PfA-M1. Human M1-aminopeptidases that have better docking score than PfA-M1 are ERAP2, TRHDE, PLAP and APN (docking score ranging between -10.421 to -9.18) but none forms a stable complex with Compound 2 as indicated by low binding energy ranging from -50.15 to -41.0 kcal/mol (Figure 4B).
Figure 4.
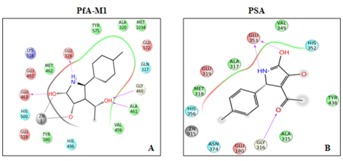
Ligand interaction diagrams of Compound 2 docking in PfA-M1 A) and human PSA; B) Compound 2 docking shows metal ion interaction with PfA-M1 but not with PSA. Number of hydrogen bonds formed by Compound 2 with PfA-M1 are more than with human PSA.
Compound 3 also had high binding affinity for PfA-M1 (docking score -10.27) and forms very stable complex (binding energy -84.71 Kcal/mol) with it as compared to human M1- aminopeptidases (docking score -3.95 to -8.98). Amongst human M1-aminopeptidases APA, THRDE, APB, APB-L, APO and LTA4 showed weak binding affinity for Compound 3.
Discussion
Conserved motifs between PfA-M1 and its human homologues, especially at active site are not only involved in interaction with metal ion but also with the other ligands (inhibitors). Hence, most of the compounds that show high binding affinity for PfA-M1 also had high binding affinity for human M1- aminopeptidases as well, limiting their applications as potential anti-malarial drugs. However, phylogenetic shows that PfA-M1 is evolutionary diverged from all of the human aminopeptidases (Supplementary data Figure 2) that provides an opportunity to design PfA-M1 specific inhibitors. In the present study, three compounds showed preference toward PfA-M1 in comparison to human M1-aminopeptidases. Although these compounds will require further validation through enzyme inhibition assays. Nevertheless a good correlation observed between molecular docking with in vitro enzyme inhibition assay provides us confidence to believe that the three PfA-M1 inhibitors selected in the present study could be PfA-M1 specific inhibitors. In cases where a PfA-M1 inhibitor shows good docking score against some human aminopeptidase as well, it does not completely preclude its possibility of using as anti malaria drug. Because some enzymes like PLAP and TRHDE that are target of selected compounds are required for certain physiological functions only viz PLAP in degradation of oxytocin and vasopressin etc and TRHDE in specific inactivation of Thyrotropin Releasing Hormone after its release.
Moreover, aminopeptidases are being targeted for drug development against other human parasitic infections also viz Trypanosoma cruze, Fasciola gigantic and Paragonimus westermani [35, 37, 38]. The availability of human M1-aminopeptidases structural models will be useful for studying the specificity of inhibitors designed against these pathogens also. Besides, these three potential lead compounds can act as scaffold for developing inhibitors with increased specificity towards PfAM1.
Conflict of interest
Authors declare no conflict of interest.
Supplementary material
Acknowledgments
The financial assistance of the Department of Biotechnology (DBT): Ministry of Science and Technology, Government of India towards this research is hereby duly acknowledged
Footnotes
Citation:Sahi et al, Bioinformation 10(8): 518-525 (2014)
References
- 1.Enserink M, et al. Science. 2008;321:1620. doi: 10.1126/science.321.5896.1620b. [DOI] [PubMed] [Google Scholar]
- 2.McGowan S, et al. Proc Natl Acad Sci U S A. 2010;107:2449. doi: 10.1073/pnas.0911813107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olotu A, et al. N Engl J Med. 2013;368:1111. doi: 10.1056/NEJMoa1207564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sa JM, et al. Essays Biochem. 2011;51:137. doi: 10.1042/bse0510137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koenderink JB, et al. Trends Parasitol. 2010;26:440. doi: 10.1016/j.pt.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Padmanaban G, Rangarajan PN. Biochem Biophys Res Commun. 2000;268:665. doi: 10.1006/bbrc.1999.1892. [DOI] [PubMed] [Google Scholar]
- 7.Skinner-Adams TS, et al. Trends Biochem Sci. 2010;35:53. doi: 10.1016/j.tibs.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Ragheb D, et al. Journal of Biological Chem. 2011;286:27255. doi: 10.1074/jbc.M111.225318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin RE, Kirk K. Blood. 2007;109:2217. doi: 10.1182/blood-2005-11-026963. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, et al. Proc Natl Acad of Sci USA. 2006;103:8840. [Google Scholar]
- 11.Stack CM, et al. J Biol Chem. 2007;282:2069. doi: 10.1074/jbc.M609251200. [DOI] [PubMed] [Google Scholar]
- 12.Rawlings ND, et al. Nucleic acids research. 2012;40:D343. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harbut MB, et al. Proc Natl Acad Sci U S A. 2011;108:E526. doi: 10.1073/pnas.1105601108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puente XS, et al. Nat Rev Genet. 2003;4:544. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- 15.Chen L, et al. Curr Med Chem. 2011;18:964. doi: 10.2174/092986711794940879. [DOI] [PubMed] [Google Scholar]
- 16.Davies DR, et al. Journal of Medicinal Chemistry. 2009;52:4694. doi: 10.1021/jm900259h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen TT, et al. Nat Struct Mol Biol. 2011;18:604. doi: 10.1038/nsmb.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birtley JR, et al. Biochemistry. 2012;51:286. doi: 10.1021/bi201230p. [DOI] [PubMed] [Google Scholar]
- 19.Wong AHM, et al. Journal of Biological Chemistry. 2012;287:36804. doi: 10.1074/jbc.M112.398842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, et al. J Biol Chem. 2013;288:25638. doi: 10.1074/jbc.M113.494955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DÃaz-Perales A, et al. Journal of Biological Chemistry. 2005;280:14310. doi: 10.1074/jbc.M413222200. [DOI] [PubMed] [Google Scholar]
- 22.Kelley LA, Sternberg MJE. Nat Protocols. 2009;4:363. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 23.Sali A, Blundell TL. J Mol Biol. 1993;234:779. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 24.Kim DE, et al. Nucleic Acids Research. 2004;32:W526. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. http://www.schrodinger.com/Maestro.
- 26.Luthy R, et al. Nature. 1992;356:83. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski RA, et al. J Appl Cryst. 1993;26:283. [Google Scholar]
- 28.Benkert P, et al. Proteins: Structure, Function, and Bioinformatics. 2008;71:261. doi: 10.1002/prot.21715. [DOI] [PubMed] [Google Scholar]
- 29.Benkert P, et al. Bioinformatics. 2011;27:343. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friesner RA, et al. J Med Chem. 2004;47:1739. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 31.Maruyama M, et al. J Biol Chem. 2007;282:20088. doi: 10.1074/jbc.M702650200. [DOI] [PubMed] [Google Scholar]
- 32.Yamada M, et al. FEBS Lett. 1994;342:53. doi: 10.1016/0014-5793(94)80583-0. [DOI] [PubMed] [Google Scholar]
- 33.Bauer K. Eur J Biochem. 1994;224:387. doi: 10.1111/j.1432-1033.1994.00387.x. [DOI] [PubMed] [Google Scholar]
- 34.Nakanishi Y, et al. Placenta. 2000;21:628. doi: 10.1053/plac.2000.0564. [DOI] [PubMed] [Google Scholar]
- 35.Song SM, et al. Parasitol Int. 2008;57:334. doi: 10.1016/j.parint.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 36.McGowan S, et al. Proc Natl Acad Sci U S A. 2009;106:2537. doi: 10.1073/pnas.0807398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Changklungmoa N, et al. Exp Parasitol. 2012;131:283. doi: 10.1016/j.exppara.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Cadavid-Restrepo G, et al. BMC Biochem. 2011;12:46. doi: 10.1186/1471-2091-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


