Abstract
The zinc finger transcription factor CONSTANS has a well-established central role in the mechanism for photoperiod sensing in Arabidopsis, integrating light and circadian clock signals to upregulate the florigen gene FT under long-day but not short-day conditions. Although CONSTANS-LIKE (COL) genes in other species have also been shown to regulate flowering time, it is not clear how widely this central role in photoperiod sensing is conserved. Legumes are a major plant group and various legume species show significant natural variation for photoperiod responsive flowering. Orthologs of several Arabidopsis genes have been shown to participate in photoperiodic flowering in legumes, but the possible function of COL genes as integrators of the photoperiod response has not yet been examined in detail. Here we characterize the COL family in the temperate long-day legume Medicago truncatula, using expression analyses, reverse genetics, transient activation assays and Arabidopsis transformation. Our results provide several lines of evidence suggesting that COL genes are unlikely to have a central role in the photoperiod response mechanism in this species.
Keywords: legume, flowering, photoperiod, Medicago, CONSTANS
Introduction
The length of the daily photoperiod is an important environmental variable that influences plant development. The most widely-recognized response to photoperiod is the induction of flowering, but photoperiod also controls other vegetative and reproductive characteristics, including formation of storage organs, axillary branching, and vegetative bud dormancy (Thomas and Vince-Prue, 1997). Within individual species, genetic variation for photoperiod responsiveness can be a major feature of adaptation to different latitudes and is therefore significant both in the natural environment and for agriculture.
As a result, there is widespread interest in the mechanism by which plants measure and respond to photoperiod, and this has been extensively examined in both Arabidopsis and rice. The study of induced mutants and natural variants affecting photoperiod responsiveness in both species has identified genes in the FT florigen family as the major target of photoperiod regulation, and have highlighted the general importance of light signaling pathways and the circadian clock for photoperiod measurement (Andres and Coupland, 2012; Brambilla and Fornara, 2013; Song et al., 2013; Tsuji et al., 2013).
In Arabidopsis, one gene in particular, CONSTANS (CO), has a central role in the mechanism of photoperiod measurement, integrating clock and light signals to provide photoperiod-specific induction of FT expression (Andres and Coupland, 2012; Song et al., 2013). CO was originally defined on the basis of a long day (LD)-specific late-flowering mutant phenotype (Koornneef et al., 1991), and encodes a B-box zinc finger transcription factor (Putterill et al., 1995). Transgenic plants overexpressing CO are extremely early flowering, and epistatic and regulatory interactions position CO genetically between GI and FT (Onouchi et al., 2000; Suárez-López et al., 2001). It has subsequently been shown that FT is an early transcriptional target of CO (Samach et al., 2000), and that the CO protein binds to the FT promoter (Tiwari et al., 2010).
The LD-specificity for activation of FT by CO is achieved through regulation of CO protein abundance at both transcriptional and post-translational level. CO mRNA is rhythmically expressed under the control of the circadian clock, such that peak expression occurs at night under short days (SD) but in the afternoon under LD (Suárez-López et al., 2001). Afternoon CO expression in LD is reinforced by action of the FKF1 blue light photoreceptor, which interacts with GI to degrade CDF proteins, which are transcriptional repressors of CO (Fornara et al., 2009; Song et al., 2012). CO protein accumulation is prevented in darkness by the ubiquitin ligase COP1 (Jang et al., 2008) but permitted in the afternoon under LD where phyA suppresses COP1 activity (Valverde et al., 2004) and FKF1 directly stabilizes CO (Song et al., 2012).
In rice, a warm-season crop with a short-day requirement for flowering, the CO-like gene Hd1 also contributes to photoperiod measurement and photoperiod-specific regulation of FT family genes (Brambilla and Fornara, 2013). In contrast to Arabidopsis CO, Hd1 appears to be a bifunctional regulator, acting to promote FT expression in SD and to repress it in LD (Izawa et al., 2002; Kojima et al., 2002). These observations have suggested that CO function may be widely conserved across the angiosperms. This conclusion has been tested in expression and functional analyses in a number of other species. In some species such as potato and sugar beet, CO-like genes do seem to be involved in photoperiod responses (Chia et al., 2008; Gonzalez-Schain et al., 2012), whereas evidence from other species such as barley, and poplar is less clear or inconclusive (Campoli et al., 2012; Hsu et al., 2012).
In the legume species pea (Pisum sativum L.), cloning of several flowering loci has demonstrated conserved roles for Arabidopsis circadian clock genes GI, ELF4 and ELF3 in the regulation of FT genes and the control of photoperiod-responsive flowering (Hecht et al., 2007; Liew et al., 2009; Weller et al., 2012). A similar role has also been demonstrated for GI in soybean (Watanabe et al., 2011). However, the endogenous function of CO-like (COL) genes in legumes has not been directly tested, and the possibility that they may participate in photoperiod measurement is still unresolved. In this study we have examined the potential involvement of COL genes in photoperiodic flowering of the temperate long-day legume Medicago truncatula, using expression analyses, Arabidopsis complementation, and loss-of-function mutants.
Materials and methods
Plant material
The experiments shown in Figures 2, 4 used the Medicago truncatula line R108 and derived mutants obtained from reverse-screening the Tnt1 insertion population described by Tadege et al. (2008). The Medicago sequences used for the experiments in Figure 3 were obtained from cv Jester (MtFTa1 promoter, MtCOLa-d) or R108 (MtCOLe-h).
Growth conditions
Arabidopsis plants were grown under long day photoperiod (16 h light/8 h dark) in growth cabinets maintained at 21°C with 30% to 40% humidity, and an irradiance of approximately 115 μmol m−2 s−1. Medicago plants were grown in growth cabinets maintained at 22°C under either long (16-h) or short-day (8-h) photoperiods.
Expression analysis
Analysis of MtCOL expression followed procedures described by Hecht et al. (2011). Harvested material consisted of all expanded leaves from three-week-old plants, with each sample consisting of material pooled from two plants. Two technical replicates and three biological replicates were performed for each timepoint. Transcript levels for experimental genes were evaluated as previously described (Weller et al., 2009), relative to the reference gene MtTEF1α. Primer sequences are given in Supplemental Table 2.
Arabidopsis transformation
DNA fragments containing full-length coding sequences of MtCOLa-COLh were amplified by PCR from cDNA and cloned into the pCR8/GW/TOPO TA vector (Invitrogen). The resulting entry vector was then recombined into plant transformation vector, pB2GW7 (Karimi et al., 2002) to generate the 35S:MtCOLa-h constructs. Transgenic plants were produced by applying Agrobacterium tumefaciens strain LBA4404 containing the pB2GW7 vectors to Arabidopsis co-2 mutant flowers using the protocol described by Martinez-Trujillo et al. (2004). Seeds from these plants were collected and sown directly onto soil and selected using Basta herbicide. Putative transformants were confirmed by qRT-PCR analysis.
Transient assays
The transient expression assays were performed by infiltrating Nicotiana benthamiana leaves, as described by Hellens et al. (2005). Agrobacterium strains containing either the FT promoter–reporter construct or a 35S:COL construct were co-infiltrated into leaves using a mixture of the two strains at a ratio of 7:1, respectively. Firefly luciferase and Renilla luciferase were assayed 4 d after infiltration using the Dual-Luciferase Reporter Assay System (Promega) as described by Hellens et al. (2005).
Results
Defining the CONSTANS-LIKE (COL) gene family in legumes
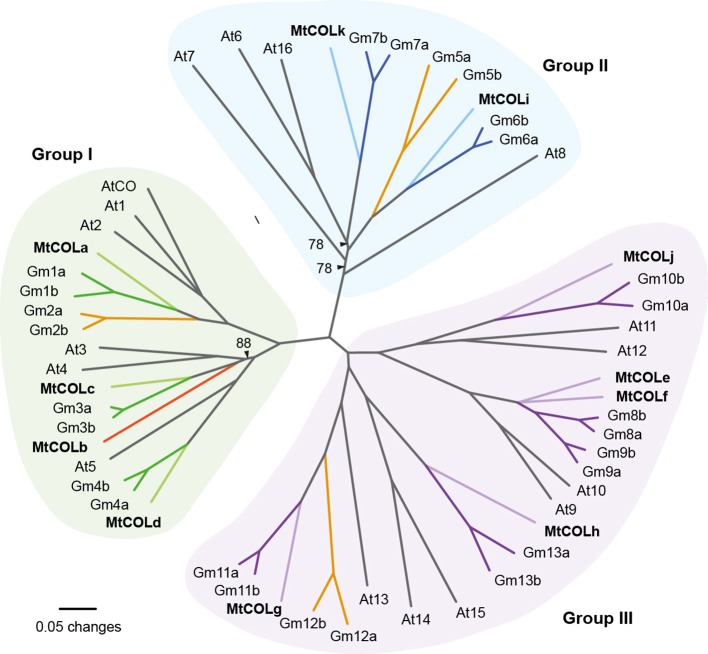
We previously reported a partial characterization of the COL gene family in legumes (Hecht et al., 2005) focusing on the so-called Group I COL genes (Griffiths et al., 2003). This group of genes includes Arabidopsis CO and is characterized by two B-box domains within an N-terminal Zn finger region, and a conserved C-terminal (CCT) domain that is also found in the circadian clock-related pseudo-response regulator gene TOC1 and related PRR (Strayer et al., 2000; Griffiths et al., 2003). To extend our understanding of legume COL genes, we used a combination of database searches and PCR-based approaches to isolate additional COL genes in Medicago truncatula. We identified a total of 11 expressed and apparently full-length COL coding sequences (Figure 1, Supplemental Figure 1) that included four Group I genes (COLa-COLd), two group II genes (COLi, COLk) and four Group III genes (COLe-COLh, COLj). It thus appears that all major groups within the COL family are represented in legumes, but some degree of independent expansion has occurred within Groups II and III.
Figure 1.
The CONSTANS-LIKE (COL) gene family in Medicago. Phylogram of legume and Arabidopsis COL protein sequences. The analysis is based on the sequence alignment shown in Supplemental Figure 1 online. Sequence details are available in Supplemental Table 1 online. Groups I (green shading), II (blue shading) and III (purple shading) correspond to the classification of Griffiths et al. (2003). Branches representing legume proteins are shaded in color consistent with each group, with soybean proteins shown in dark and Medicago genes in light shading. Branches shown in orange indicate soybean homeolog pairs for which no corresponding Medicago ortholog was found, and the single Medicago gene without a soybean counterpart is shown in red. Branches with bootstrap values <50% have been collapsed, and black arrowheads indicate branches with support >50% but <90%. All other branches have support >90%. At, Arabidopsis thaliana; Gm, Glycine max; Mt, Medicago truncatula.
Consistent with a previous report (Hecht et al., 2005) we identified only a single group Ia gene in Medicago (MtCOLa) and found that the three Arabidopsis group Ia genes AtCO, AtCOL1 and AtCOL2 were more similar to each other than to MtCOLa.
A recent report from soybean has identified 26 COL genes, representing 13 pairs of homeologs (Wu et al., 2014). For nine of these pairs, we identified a single Medicago ortholog (Figure 1), and the clade containing GmCOL8a/b and COL9a/b also included two Medicago genes; MtCOLe and COLf. The MtCOLb gene had no corresponding pair of genes in soybean, and three soybean homeolog pairs were not represented by Medicago genes. This latter situation could imply the existence of additional Medicago COL genes not represented in the current genome build (Mt4.0), and we were particularly interested in a comparison of the Group Ia genes as this clade contains most of the genes known to have CO-like function in other species. In soybean, there are four Group Ia COL genes; GmCOL1a/b and COL2a/b. The single Group Ia gene MtCOLa is clearly orthologous to the GmCOL1a/b pair, implying that Medicago might possess a second Group Ia COL gene orthologous to GmCOL2a/b. To address the possibility, we examined the genomic regions containing GmCOL2a and COL2b for evidence of microsynteny with the Medicago genome. Supplemental Figure 2 shows that genes in the GmCOL2a/COL2b regions showed highest similarity to genes on Medicago chromosome 6, with clear evidence of microsynteny, but there was no MtCOL gene in this location, suggesting that this gene may have been lost from the Medicago lineage. Similarly, microsynteny between regions containing GmCOL5a/b and another part of Medicago chromosome 6, and between regions containing GmCOL12a/b and Medicago chromosome 2 (Supplemental Figure 2) also suggests that orthologs of these genes are also absent from the Medicago genome. We therefore tentatively conclude that the 11 Medicago COL genes we have identified represent the entire gene family.
Diurnal rhythms of COL gene expression
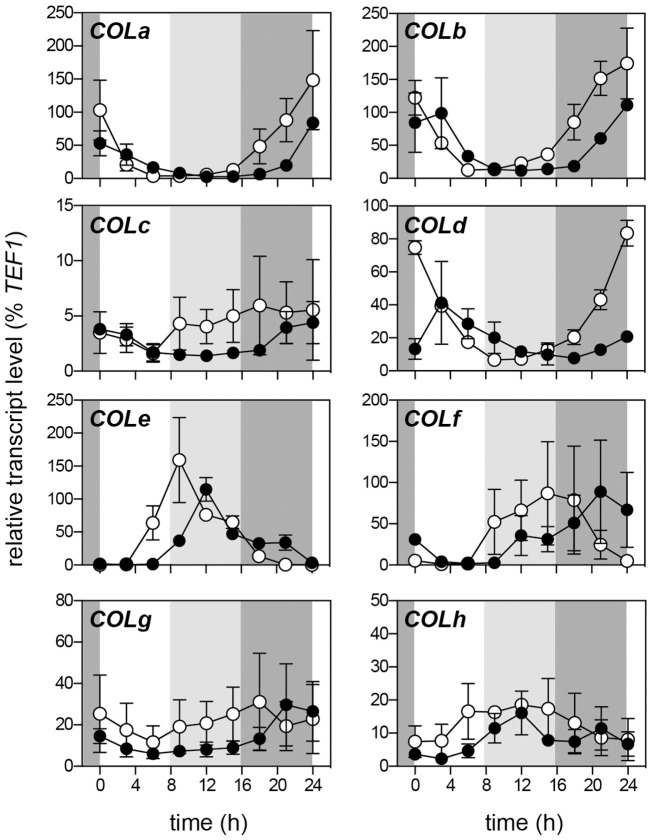
In Arabidopsis, the characteristic diurnal mRNA expression rhythm of CO is linked to its function in photoperiod measurement. Under SD, CO expression peaks in the night and is low throughout the day. Under LD, CO expression increases during the afternoon, and this increase is reinforced by an additional relief from repression through the action of the blue-light photoreceptor FKF1 (Imaizumi et al., 2003). We reasoned that if transcriptional regulation of COL genes was similarly important for photoperiod responses in temperate legumes, one or more COL genes might show distinctly different expression rhythms in long and short days. We therefore examined the diurnal expression rhythms for eight of the 11 MtCOL genes (COLa-COLh).
We previously reported that the single group Ia COL gene in pea, PsCOLa, shows a morning-phased expression rhythm in LD (Hecht et al., 2007) that is similar to the Arabidopsis Group Ia genes COL1 and COL2 (Ledger et al., 2001). Figure 2 shows that MtCOLa expression also follows a similar LD rhythm with a peak at dawn. The level of expression under SD was not significantly different than under LD throughout the daily timecourse, and under both conditions, COLa showed significant morning expression, which declined to basal level by ZT9. Under LD specifically, COLa expression remained very low during the afternoon, with no evidence of the afternoon “shoulder” to the LD rhythm that is characteristic of Arabidopsis CO (Imaizumi et al., 2003). More generally, there was no evidence for any difference in COLa expression during the light phase in LD compared to SD.
Figure 2.
Rhythmic regulation of MtCOL expression under SD and LD. Transcript levels were determined in fully-expanded leaves taken from 3-week-old R108 seedlings grown under 8-h (short-day; open symbols) or 16-h long-day photoperiods (filled symbols) in growth cabinets at 22°C. The night period common to both treatments is represented by dark gray shading, with the period in which plants are in the light in long days but not short days is represented by light gray shading. Data represent mean ± SE for n = 2 biological replicates.
Like COLa, the Group Ic genes COLb and COLd also showed a morning-phased rhythm. For both genes, the phase of the expression rhythm was earlier in SD than in LD, typical of the response of many rhythmically-regulated genes to photoperiod. However, as for COLa, there was no evidence of a qualitative difference in expression during the light phase between LD and SD conditions for either gene. COLc was only expressed at a very low level and showed minimal diurnal variation. In contrast to the Group I genes, the Group III genes generally showed an evening-phased rhythm under LD, which in most cases, was shifted earlier in SD. COLe showed the most strongly rhythmic expression with an afternoon peak in LD at around ZT12, and COLf expression was also clearly rhythmic, with peak expression under LD during the night. COLg and COLh were at most weakly rhythmic. The closest similarity to the Arabidopsis CO rhythm was seen for COLf, which was not expressed at dawn or at either of the two timepoints during the light phase under SD, but showed significant expression at dawn and during the afternoon in LD.
Activity of legume COL genes
Arabidopsis CO is a potent inducer of flowering, and Arabidopsis plants overexpressing CO flower very early under both LD and SD (Onouchi et al., 2000). To test whether any of the MtCOL genes might be similarly effective in flowering regulation, we assessed their ability to complement the late flowering phenotype of the Arabidopsis co-2 mutant. Figure 3A shows that none of the eight MtCOL genes that we tested caused early flowering when overexpressed from the cauliflower mosaic virus 35S promoter in the late-flowering Arabidopsis co-2 mutant plants.
Figure 3.
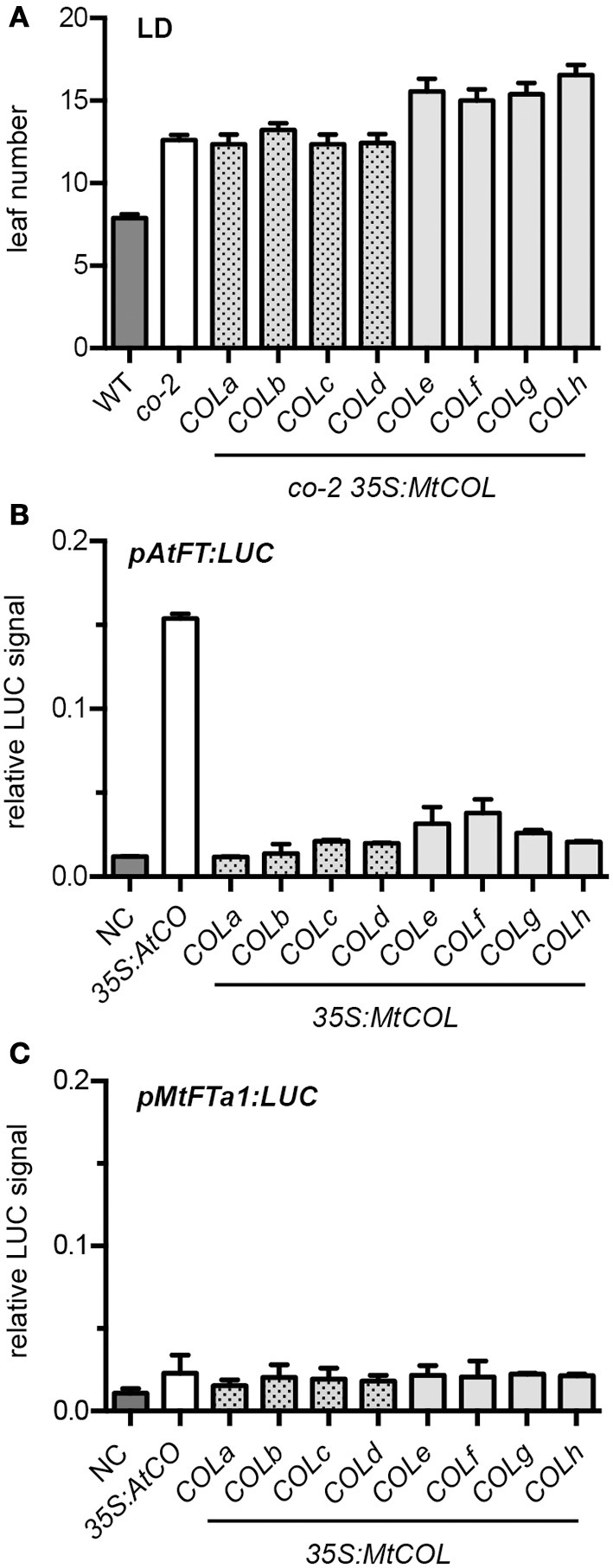
Functional analyses of MtCOL genes. (A). Overexpression of MtCOLa-COLh genes does not promote flowering in the Arabidopsis co-2 mutant. Flowering time is indicated by leaf number at flowering. Data represent a minimum of 10 plants for each line ± SE. (B). MtCOL genes are unable to induce expression from the Arabidopsis FT promoter in transient expression assays. The 35S:AtCO construct and 35S:MtCOL constructs were co-infiltrated with AtFT promoter fused to the luciferase (LUC) reporter gene into N. benthamiana leaves. Only 35S:AtCO and 35S:MtCOLf resulted in statistically significant upregulation of the AtCO promoter compared with the NC (no construct) control, P < 0.0001 and P = 0.045, respectively (C). MtCOL genes are unable to induce expression from the Medicago FTa1 promoter in transient expression assays. The 35S:AtCO construct and 35S:MtCOLa-h constructs were co-infiltrated with MtFTa1 promoter:LUC into N. benthamiana leaves. No statistically-significant difference in relative LUC signal between the NC control and 35S:AtCO or the 35S:MtCOLs was observed. NC (no construct) refers to leaves infiltrated with untransformed Agrobacterium along with the AtFT or MtFTa1 promoter:LUC constructs. Relative LUC signal is a ratio of LUC activity versus Renilla luciferase activity to correct for variation in transformation efficiencies between infiltrated N. benthamiana leaves and data represent the mean ± SE of three biological replicates. Statistical analysis was performed using Student's t-test. In all panels, Group I and Group III MtCOL genes are represented by dark gray and light gray shading, respectively.
Next, we examined the ability of the MtCOL genes to directly activate the Arabidopsis FT promoter using a transient assay system. In this system, the Arabidopsis FT promoter was fused to the luciferase reporter gene and infiltrated into Nicotiana benthamiana leaves together with different transcription factors. Figure 3B shows that expression of AtCO resulted in substantial upregulation of luciferase expression from the Arabidopsis FT promoter (P < 0.0001). In contrast, the majority of MtCOL genes had no clear statistically significant effect consistent with their inability to complement the co-2 mutant. The one possible exception was MtCOLf, which showed a small increase in LUC signal with marginal statistical significance (P = 0.045).
The transient assay system was also used to investigate if any of the MtCOL genes are able to activate the Medicago FTa1 promoter. The Medicago FTa1 gene plays a key role in promoting flowering in response to both vernalization and LD (Laurie et al., 2011). When Medicago plants are shifted from SD to LD, FTa1 is upregulated by exposure to a single long day (Laurie et al., 2011). An MtFTa1 promoter sequence comprising 2017 bp upstream of the start codon was fused to the luciferase reporter gene and infiltrated into Nicotiana benthamiana leaves together with different MtCOL genes. Neither Arabidopsis CO nor any of the MtCOL genes was able to induce LUC expression from this promoter sequence (P > 0.05 in all cases) (Figure 3C).
Overall, these results provide further evidence that that none of the MtCOL genes are functionally equivalent to AtCO, with respect to their ability to induce expression of AtFT. In addition they also suggest that neither AtCO nor any of the tested MtCOL genes are able to induce MtFTa1 expression. Although the specific reason for the inactivity of MtCOL genes on AtFT, and AtCO on MtFTa1 is not yet clear, it could partially reflect divergence in FT promoter sequences and/or DNA binding characteristics of CO and COL proteins. An alignment of the AtFT proximal promoter with regions upstream of the transcriptional start site in the Medicago and chickpea FTa1 genes (Supplemental Figure 3) shows that neither of the two CO-responsive (CORE) elements defined in the AtFT promoter are significantly conserved in the legume promoters, which may provide an explanation for the inactivity of AtCO on the MtFTa1 promoter.
Genetic analysis of COL function
Finally, in order to directly examine COL gene function, we made use of the Medicago Tnt1 insertion platform (Tadege et al., 2008) to identify putative insertion mutants for three of the four Group I MtCOL genes. Insertions in COLa, COLb and COLc were verified by sequencing and mutant lines shown to specifically lack the corresponding transcript (Figure 4A). For phenotypic comparisons we vernalized seeds for 2 weeks at 4°C and grew seedlings under an 18 h photoperiod. In addition to a pure line of the progenitor line R108, we also included WT lines selected from individual segregating progenies for each mutant as controls. Figure 4B shows that neither colb nor colc mutants flowered significantly differently from their corresponding control lines in terms of either flowering time (P > 0.5 and P > 0.2 for colb and colc, respectively) or for node of first flower (P = 0.088 and P > 0.5, respectively). The genetic background carrying the cola mutation was slightly later flowering than the R108 control line, in terms of days (18.9 vs. 16.2 days, P < 0.001), but slightly earlier in terms of nodes (5.2 vs. 5.9 nodes, P < 0.01). The cola mutant line was marginally later than its WT control line for both time (19.9 vs. 18.9 days, P = 0.044) but not for node number (5.7 vs. 5.2 nodes, P = 0.088). However, importantly, the variation in flowering time or node observed within and between these lines was negligible relative to the strong delay of flowering in vernalized R108 plants under SD, indicating that none of these three COL genes contributes significantly to the promotion of flowering by LD.
Figure 4.
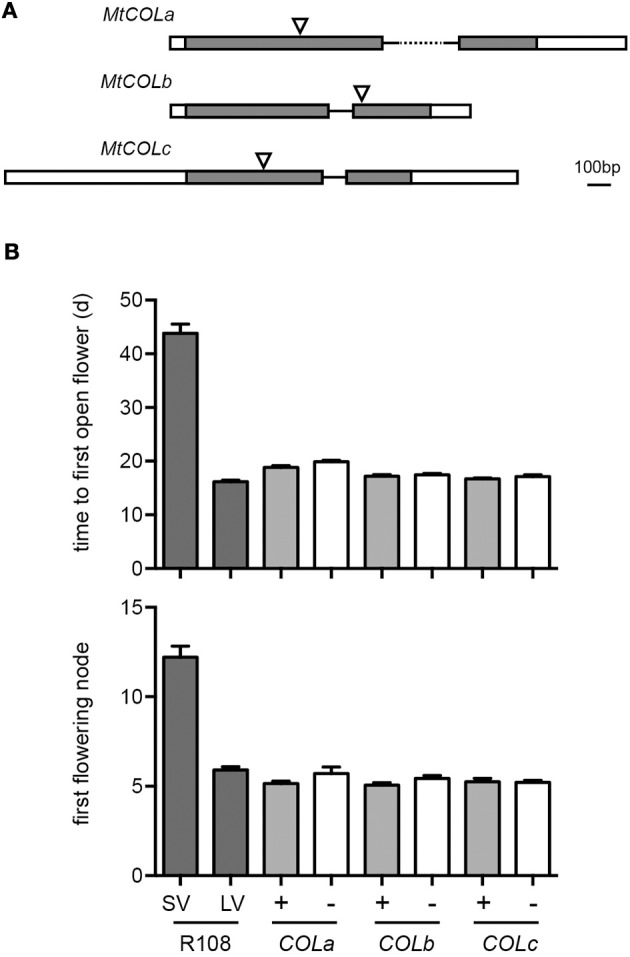
Characterisation of mutants for MtCOLa, COLb and COLc. (A) Diagram of MtCOLa, COLb and COLc genes showing gene structure and site of the Tnt1 insertion. Exons are shown as boxes with the coding sequence in dark gray, and the 5′ and 3′ untranslated regions in white. (B) Flowering time and node of first flower for WT and COL mutant plants vernalized for 14 days at 4°C before transfer to LD at 22°C. Homozygous WT (+) and mutant (−) genotypes were selected from segregating progenies and are represented by by shaded and empty bars, respectively. Vernalized R108 plants grown under LD (LV) or SD (SV) were included as a control. Values represent mean ± SE for n = 12–20.
Discussion
The CONSTANS protein has been a central feature of models explaining the molecular basis for plant responses to photoperiod, and the potential conservation of CO function across flowering plants has been a topic of considerable interest. In legumes, several studies have identified conserved elements of the photoperiod response pathway, including homologs of GIGANTEA (Hecht et al., 2007; Watanabe et al., 2011), PHYA (Weller et al., 2004; Liu et al., 2008a; Watanabe et al., 2009), FT (Kong et al., 2010; Hecht et al., 2011; Laurie et al., 2011; Sun et al., 2011) and circadian clock genes (Liew et al., 2009, 2014; Weller et al., 2012), but the potential role of CO-like genes has received less attention. In this study we have identified 11 COL genes in the model long-day legume Medicago truncatula, and investigated the regulation and function of eight of these. Collectively our results provide several strong lines of evidence that the three genes most similar to Arabidopsis CO, MtCOLa, MtCOLb and MtCOLc, genes do not participate in the induction of flowering by photoperiod. Our results also indicate that the five other genes we examined (MtCOLd-COLh) are also unlikely to function in a manner similar to AtCO.
The first line of evidence comes from regulation of MtCOL expression. Rhythmic expression of group I and group III MtCOL genes showed broad similarity to reported results from other species, with group I genes showing peak expression around dawn and other genes generally more strongly expressed late in the day or in the early part of the night (Figure 2). However, with the possible exception of MtCOLf, we found no evidence for photoperiod-specific coincidence of MtCOL expression with the light phase in LD, or for the afternoon peak that is characteristic of the AtCO transcriptional rhythm under LD (Imaizumi et al., 2003) (Figure 2). Nevertheless, it should be noted that the absence of these regulatory features does not in itself exclude the possibility that these genes have CO-like function. First, because Arabidopsis CO is known to undergo significant post-transcriptional regulation, and it is conceivable that photoperiod-specific activity could be conferred on one or more of the MtCOL genes predominantly through regulation at the protein level. Second, because the link between CO expression dynamics and photoperiod responsiveness has only been elaborated in detail for Arabidopsis and it is not yet clear how widely this may be conserved (Ballerini and Kramer, 2011).
We obtained more direct evidence on MtCOL functions from Arabidopsis complementation and experiments using a tobacco transient assay system. None of the MtCOL genes was able to promote flowering when overexpressed in the Arabidopsis co-2 mutant (Figure 3A), in contrast to the strong flower promoting activity of Arabidopsis CO (Onouchi et al., 2000). This contrast was also seen in transient assays, where none of the eight tested MtCOL genes was able to activate transcription from the Arabidopsis FT promoter, even though AtCO clearly possessed this ability (Figure 3B). Finally, transcript-null mutants for the Group Ia gene MtCOLa and two other Group I genes all flowered normally under LD after vernalization (Figure 4), clearly indicating that these genes are not needed for, and likely do not participate in, the promotion of flowering by LD.
Overall, the lack of any clear effect of MtCOL genes on flowering is somewhat surprising, particularly in the case of COLa, in view of the fact that Group Ia COL genes across a range of species have been shown to some degree of function in flowering regulation. Outside of Arabidopsis, the involvement of Group Ia COL genes in photoperiod responsiveness is most conclusive in rice, where the single group Ia COL gene Hd1 underlies a major-effect QTL for flowering, and has a bidirectional role in regulation of FT homologs (Yano et al., 2000). The potato CO gene also has a clear endogenous role in photoperiod responsiveness, although this is less pronounced for flowering induction than for tuberization (Navarro et al., 2011). Group Ia COL genes in a number of other species have shown flower-promoting activity in Arabidopsis. Genes from potato, tomato, poplar and sugar beet are able to at least partially complement Arabidopsis co mutants (Ben-Naim et al., 2006; Chia et al., 2008; Gonzalez-Schain et al., 2012; Hsu et al., 2012). The barley Hd1 ortholog CO1 has no activity in transgenic Arabidopsis but does promote flowering when overexpressed in barley itself (Campoli et al., 2012).
In Arabidopsis, activation of the Arabidopsis FT promoter by CO requires several regulatory motifs located within 500 bp of the FT transcriptional start site (Adrian et al., 2010; Tiwari et al., 2010). Full FT activation in planta requires the additional association of CO with proteins bound to distal promoter regions and the formation of chromatin loops (Ben-Naim et al., 2006; Adrian et al., 2010; Cao et al., 2014). However, a 1 kb proximal fragment of the AtFT promoter is sufficient for maximal induction by AtCO in a transient assay system (Adrian et al., 2010) and our results show that MtCOL genes lack this activity, consistent with their lack of flower-promoting activity when overexpressed in Arabidopsis (Figure 4A). While the reason for this is not clear, the simplest explanation may be that MtCOL proteins either do not bind to the CO-responsive elements in this region, or simply do not function as transcriptional regulators. Our results also show that an equivalent region of the MtFTa1 promoter is activated neither by AtCO nor MtCOL genes. The lack of AtCO activity may reflect the fact that none of the functionally validated proximal elements in the Arabidopsis FT promoter are significantly conserved in the corresponding regions of the Medicago or chickpea FTa1 genes (Supplemental Figure 3).
The argument may also be made that CO function may be preserved in the MtCOL family but is comprised of small individual contributions from multiple members. Clearly the present data also do not exclude this possibility, with some features of MtCOLf (Figures 2, 3B) consistent with a weak CO-like effect. However, it is worth noting that loss-of-function variants for Arabidopsis CO and rice Hd1 both have large phenotypic effects and were first identified through relatively direct forward genetic analysis. This is not proof that deep redundancy within the COL family is not an explanation for our results, but does make it seem less likely. It also remains possible that CO function could be carried out one or more of the other MtCOL genes that we did not examine, which include both group II (COLi, COLk) and Group III (COLj) genes. Although no gene outside the group Ia COL clade has been implicated in photoperiodic flowering, a more general effect on flowering has been demonstrated for certain other COL genes. In Arabidopsis, the group Ic gene COL3 (Datta et al., 2006) and the group III gene COL9 (Cheng and Wang, 2005) both inhibit flowering, whereas a second group Ic gene, COL5, may have a promotive function (Hassidim et al., 2009). In rice, the Group Ic gene COL4 gene inhibits flowering under SD and LD through repression of FT homologs RFT and Hd3a (Lee et al., 2010).
Recently, new information has emerged on COL genes in the short-day legume soybean. Soybean has multiple group Ia COL genes, which comprise two pairs of homeologs; COL1a/b and COL2a/b. Two recent studies show that one of these, GmCOL2a, is able to complement the Arabidopsis co-2 mutant (Fan et al., 2014; Wu et al., 2014), and there is evidence that the remaining genes COL1a/b and COL2b may also have some activity in Arabidopsis (Wu et al., 2014). However, our phylogenetic comparisons indicate that Medicago has only a single Group Ia COL gene orthologous to Arabidopsis CO/COL1/COL2, and does not have an ortholog of the GmCOL2 genes (Supplemental Figure 2). Sequence searches in other temperate legumes (including pea, and chickpea and Lotus japonicus) also identify only a single group Ia COL gene in these species, indicating that loss of COL2 orthologs may have occurred relatively early in this temperate legume lineage.
Overall, it seems on balance likely that COL genes do not function as central integrators of photoperiod responsive flowering in Medicago. This may also be more generally true across the temperate legumes, at least for COLa, as COLa orthologs in pea and Lotus japonicus also do not show characteristic regulatory features of Arabidopsis CO (Hecht et al., 2007; Yamashino et al., 2013). Instead, CO-independent pathways may have a more prominent role in this plant group. In Arabidopsis, a number of other factors contribute to direct regulation of FT expression. These include the positive factors PIF4 and CIB1, bHLH proteins involved in light signaling, and SPL3, which is a target of the miRNA156 pathway controlling juvenility (Liu et al., 2008b; Kim et al., 2012; Kumar et al., 2012). Factors repressing FT include the CDF family of Dof transcription factors, and a number of AP2 domain proteins that are targets of miR172 (Jung et al., 2007; Fornara et al., 2009). In particular, it is intriguing that despite the apparent absence of CO-like function in Medicago, evidence from the related legume pea shows a major role for the GI ortholog LATE1 (Hecht et al., 2007, 2011). In Arabidopsis GI has been shown to promote FT transcription independently of CO, both by contributing to degradation of CDF proteins (Song et al., 2012), and also by positive effects on miR172 biogenesis (Jung et al., 2007). In addition, the recently-identified B3-transcription factor-like gene E1 in soybean is also important for photoperiod-dependent regulation of FT expression, and an ortholog is present in Medicago (Xia et al., 2012; Zhai et al., 2014). Whether one or more of these mechanisms contribute to the photoperiod response in temperate legumes will be an important question for future investigation.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jacqueline Vander Schoor and Robyn Lee for technical assistance, Ian Cummings, Tracey Winterbottom and Michelle Lang for help with care of plants, and George Coupland for providing the co-2 mutant. Funding for this work was provided by the Australian Research Council (James L. Weller) and the New Zealand Foundation for Research, Science and Technology (Richard C. Macknight).
Supplementary material
The supplementary material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00486/abstract
References
- Adrian J., Farrona S., Reimer J. J., Albani M. C., Coupland G., Turck F. (2010). cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22, 1425–1440 10.1105/tpc.110.074682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres F., Coupland G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639 10.1038/nrg3291 [DOI] [PubMed] [Google Scholar]
- Ballerini E.S., Kramer E.M. (2011). In the light of evolution: a reevaluation of conservation in the CO-FT regulon and its role in photoperiodic regulation of flowering time. Front. Plant Sci. 2:81 10.3389/fpls.2011.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Naim O., Eshed R., Parnis A., Teper-Bamnolker P., Shalit A., Coupland G., et al. (2006). The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 46, 462–476 10.1111/j.1365-313X.2006.02706.x [DOI] [PubMed] [Google Scholar]
- Brambilla V., Fornara F. (2013). Molecular control of flowering in response to day length in rice. J. Integr. Plant Biol. 55, 410–418 10.1111/jipb.12033 [DOI] [PubMed] [Google Scholar]
- Campoli C., Drosse B., Searle I., Coupland G., Von Korff M. (2012). Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. Plant J. 69, 868–880 10.1111/j.1365-313X.2011.04839.x [DOI] [PubMed] [Google Scholar]
- Cao S., Kumimoto R.W., Gnesutta N., Calogero A.M., Mantovani R., Holt B.F., 3rd (2014). A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 26, 1009–1017 10.1105/tpc.113.120352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X.-F., Wang Z.-Y. (2005). Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 43, 758–768 10.1111/j.1365-313X.2005.02491.x [DOI] [PubMed] [Google Scholar]
- Chia T. Y., Muller A., Jung C., Mutasa-Gottgens E. S. (2008). Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus. J. Exp. Bot. 59, 2735–2748 10.1093/jxb/ern129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Hettiarachchi G.H., Deng X.W., Holm M. (2006). Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18, 70–84 10.1105/tpc.105.038182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C., Hu R., Zhang X., Wang X., Zhang W., Zhang Q., et al. (2014). Conserved CO-FT regulons contribute to the photoperiod flowering control in soybean. BMC Plant Biol. 14:9 10.1186/1471-2229-14-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F., Panigrahi K. C., Gissot L., Sauerbrunn N., Ruhl M., Jarillo J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17, 75–86 10.1016/j.devcel.2009.06.015 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Schain N.D., Diaz-Mendoza M., Zurczak M., Suarez-Lopez P. (2012). Potato CONSTANS is involved in photoperiodic tuberization in a graft-transmissible manner. Plant J. 70, 678–690 10.1111/j.1365-313X.2012.04909.x [DOI] [PubMed] [Google Scholar]
- Griffiths S., Dunford R. P., Coupland G., Laurie D. A. (2003). The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 131, 1855–1867 10.1104/pp.102.016188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassidim M., Harir Y., Yakir E., Kron I., Green R.M. (2009). Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta 230, 481–491 10.1007/s00425-009-0958-7 [DOI] [PubMed] [Google Scholar]
- Hecht V., Foucher F., Ferrandiz C., Macknight R., Navarro C., Morin J., et al. (2005). Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 137, 1420–1434 10.1104/pp.104.057018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V., Knowles C. L., Vander Schoor J. K., Liew L. C., Jones S. E., Lambert M. J. M., et al. (2007). Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 144, 648–661 10.1104/pp.107.096818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V., Laurie R. E., Vander Schoor J. K., Ridge S., Knowles C. L., Liew L. C., et al. (2011). The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 23, 147–161 10.1105/tpc.110.081042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R. P., Allan A. C., Friel E. N., Bolitho K., Grafton K., Templeton M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1:13 10.1186/1746-4811-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C. Y., Adams J. P., No K., Liang H., Meilan R., Pechanova O., et al. (2012). Overexpression of CONSTANS homologs CO1 and CO2 fails to alter normal reproductive onset and fall bud set in woody perennial poplar. PLoS ONE 7:e45448 10.1371/journal.pone.0045448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T., Tran H. G., Swartz T. E., Briggs W. R., Kay S. A. (2003). FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426, 302–306 10.1038/nature02090 [DOI] [PubMed] [Google Scholar]
- Izawa T., Oikawa T., Sugiyama N., Tanisaka T., Yano M., Shimamoto K. (2002). Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering in rice. Genes Dev. 16, 2006–2020 10.1101/gad.999202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K. C., Wenkel S., Soppe W., Deng X. W., et al. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27, 1277–1288 10.1038/emboj.2008.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.-H., Seo Y.-H., Seo P. J., Reyes J. L., Yun J., Chua N.-H., et al. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19, 2736–2748 10.1105/tpc.107.054528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inze D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 10.1016/S1360-1385(02)02251-3 [DOI] [PubMed] [Google Scholar]
- Kim J.J., Lee J.H., Kim W., Jung H.S., Huijser P., Ahn J.H. (2012). The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol. 159, 461–478 10.1104/pp.111.192369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S., Takahashi Y., Kobayashi Y., Monna L., Sasaki T., Araki T., et al. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1105 10.1093/pcp/pcf156 [DOI] [PubMed] [Google Scholar]
- Kong F., Liu B., Xia Z., Sato S., Kim B.M., Watanabe S., et al. (2010). Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 154, 1220–1231 10.1104/pp.110.160796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C. J., Van Der Veen J. H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66 10.1007/BF00264213 [DOI] [PubMed] [Google Scholar]
- Kumar S. V., Lucyshyn D., Jaeger K. E., Alos E., Alvey E., Harberd N. P., et al. (2012). Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484, 242–245 10.1038/nature10928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie R.E., Diwadkar P., Jaudal M., Zhang L., Hecht V., Wen J., et al. (2011). The Medicago FLOWERING LOCUS T homolog, MtFTa1, is a key regulator of flowering time. Plant Physiol. 156, 2207–2224 10.1104/pp.111.180182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledger S., Strayer C., Ashton F., Kay S.A., Putterill J. (2001). Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 26, 15–22 10.1046/j.1365-313x.2001.01003.x [DOI] [PubMed] [Google Scholar]
- Lee Y. S., Jeong D. H., Lee D. Y., Yi J., Ryu C. H., Kim S. L., et al. (2010). OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J. 63, 18–30 10.1111/j.1365-313X.2010.04226.x [DOI] [PubMed] [Google Scholar]
- Liew L. C., Hecht V., Laurie R. E., Knowles C. L., Vander Schoor J. K., Macknight R. C., et al. (2009). DIE NEUTRALIS and LATE BLOOMER 1 contribute to regulation of the pea circadian clock. Plant Cell 21, 3198–3211 10.1105/tpc.109.067223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew L. C., Hecht V., Sussmilch F. C., Weller J. L. (2014). The pea photoperiod response gene STERILE NODES is an ortholog of LUX ARRHYTHMO. Plant Physiol. 165, 648–657 10.1104/pp.114.237008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Kanazawa A., Matsumura H., Takahashi R., Harada K., Abe J. (2008a). Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 180, 995–1007 10.1534/genetics.108.092742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yu X., Li K., Klejnot J., Yang H., Lisiero D., et al. (2008b). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539 10.1126/science.1163927 [DOI] [PubMed] [Google Scholar]
- Martinez-Trujillo M., Limones-Briones V., Cabrera-Ponce J.L., Herrera-Estrella L. (2004). Improving transformation efficiency of Arabidopsis thaliana by modifying the floral dip method. Plant Mol. Biol. Rep. 22, 22–30 10.1007/BF02773350 [DOI] [Google Scholar]
- Navarro C., Abelenda J. A., Cruz-Oro E., Cuellar C. A., Tamaki S., Silva J., et al. (2011). Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478, 119–122 10.1038/nature10431 [DOI] [PubMed] [Google Scholar]
- Onouchi H., Igeno M.I., Perilleux C., Graves K., Coupland G. (2000). Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12, 885–900 10.1105/tpc.12.6.885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857 10.1016/0092-8674(95)90288-0 [DOI] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S. E., Ditta G. S., Schwarz-Sommer Z., Yanofsky M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616 10.1126/science.288.5471.1613 [DOI] [PubMed] [Google Scholar]
- Song Y.H., Ito S., Imaizumi T. (2013). Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 18, 575–583 10.1016/j.tplants.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. H., Smith R. W., To B. J., Millar A. J., Imaizumi T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336, 1045–1049 10.1126/science.1219644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C., Oyama T., Schultz T.F., Raman R., Al E. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771 10.1126/science.289.5480.768 [DOI] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120 10.1038/35074138 [DOI] [PubMed] [Google Scholar]
- Sun H., Jia Z., Cao D., Jiang B., Wu C., Hou W., et al. (2011). GmFT2a, a soybean homolog of FLOWERING LOCUS T, is involved in flowering transition and maintenance. PLoS ONE 6:e29238 10.1371/journal.pone.0029238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M., Wen J., He J., Tu H., Kwak Y., Eschstruth A., et al. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54, 335–347 10.1111/j.1365-313X.2008.03418.x [DOI] [PubMed] [Google Scholar]
- Thomas B., Vince-Prue D. (1997). Photoperiodism in Plants. London: Academic Press [Google Scholar]
- Tiwari S. B., Shen Y., Chang H. C., Hou Y., Harris A., Ma S. F., et al. (2010). The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187, 57–66 10.1111/j.1469-8137.2010.03251.x [DOI] [PubMed] [Google Scholar]
- Tsuji H., Taoka K., Shimamoto K. (2013). Florigen in rice: complex gene network for florigen transcription, florigen activation complex, and multiple functions. Curr. Opin. Plant Biol. 16, 228–235 10.1016/j.pbi.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006 10.1126/science.1091761 [DOI] [PubMed] [Google Scholar]
- Watanabe S., Hideshima R., Xia Z., Tsubokura Y., Sato S., Nakamoto Y., et al. (2009). Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 182, 1251–1262 10.1534/genetics.108.098772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Xia Z., Hideshima R., Tsubokura Y., Sato S., Yamanaka N., et al. (2011). A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188, 395–407 10.1534/genetics.110.125062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J. L., Batge S. L., Smith J. J., Kerckhoffs L. H. J., Sineshchekov V. A., Murfet I. C., et al. (2004). A dominant mutation in the pea PHYA gene confers enhanced responses to light and impairs the light-dependent degradation of phytochrome A. Plant Physiol. 135, 2186–2195 10.1104/pp.103.036103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J. L., Hecht V., Vander Schoor J. K., Davidson S. E., Ross J. J. (2009). Light regulation of gibberellin biosynthesis in pea is mediated through the COP1/HY5 Pathway. Plant Cell 21, 800–813 10.1105/tpc.108.063628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J. L., Liew L. C., Hecht V. F., Rajandran V., Laurie R. E., Ridge S., et al. (2012). A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc. Natl. Acad. Sci. U.S.A. 109, 21158–21163 10.1073/pnas.1207943110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Price B.W., Haider W., Seufferheld G., Nelson R., Hanzawa Y. (2014). Functional and evolutionary characterization of the CONSTANS gene family in short-day photoperiodic flowering in soybean. PLoS ONE 9:e85754 10.1371/journal.pone.0085754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z., Watanabe S., Yamada T., Tsubokura Y., Nakashima H., Zhai H., et al. (2012). Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. U.S.A. 109, E2155–E2164 10.1073/pnas.1117982109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T., Yamawaki S., Hagui E., Ueoka-Nakanishi H., Nakamichi N., Ito S., et al. (2013). Clock-controlled and FLOWERING LOCUS T (FT)-dependent photoperiodic pathway in Lotus japonicus I: verification of the flowering-associated function of an FT homolog. Biosci. Biotechnol. Biochem. 77, 747–753 10.1271/bbb.120871 [DOI] [PubMed] [Google Scholar]
- Yano M., Katayose Y., Ashikari M., Yamanouchi U., Monna L., Fuse T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2484 10.1105/tpc.12.12.2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai H., Lu S., Liang S., Wu H., Zhang X., Liu B., et al. (2014). GmFT4, a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. PLoS ONE 9:e89030 10.1371/journal.pone.0089030 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.