Abstract
The major families of chromatin remodelers have been conserved throughout eukaryotic evolution. Because they play broad, pleiotropic roles in gene regulation, it was not known if their functions could change rapidly. Here, we show that major alterations in the use of chromatin remodelers are possible, because the nucleosome remodeling factor (NURF) complex has acquired a unique role in the sperm/oocyte decision of the nematode Caenorhabditis briggsae. First, lowering the activity of C. briggsae NURF-1 or ISW-1, the core components of the NURF complex, causes germ cells to become oocytes rather than sperm. This observation is based on the analysis of weak alleles and null mutations that were induced with TALENs and on RNA interference. Second, qRT–polymerase chain reaction data show that the C. briggsae NURF complex promotes the expression of Cbr-fog-1 and Cbr-fog-3, two genes that control the sperm/oocyte decision. This regulation occurs in the third larval stage and affects the expression of later spermatogenesis genes. Third, double mutants reveal that the NURF complex and the transcription factor TRA-1 act independently on Cbr-fog-1 and Cbr-fog-3. TRA-1 binds both promoters, and computer analyses predict that these binding sites are buried in nucleosomes, so we suggest that the NURF complex alters chromatin structure to allow TRA-1 access to Cbr-fog-1 and Cbr-fog-3. Finally, lowering NURF activity by mutation or RNA interference does not affect this trait in other nematodes, including the sister species C. nigoni, so it must have evolved recently. We conclude that altered chromatin remodeling could play an important role in evolutionary change.
Keywords: Caenorhabditis briggsae, chromatin remodelers, NURF complex, evolution
Introduction
To control development, gene expression must be regulated in time and space. The organization of DNA into chromatin plays a key role in this process, by restricting the accessibility of promoters and enhancers (Clapier and Cairns 2009). This restriction also increases precision, because chromatin-remodeling complexes can actively cooperate with transcription factors to control gene expression. However, the pleiotropic phenotypes of most chromatin remodelers have made it difficult to evaluate their roles in evolutionary change.
Nematodes provide an ideal way to address this problem. Most of the known chromatin-remodeling complexes exist in nematodes and control various aspects of development (Cui and Han 2007). Moreover, detailed molecular models have been established for two developmental processes in Caenorhabditis elegans. The first involves the specification of sensory rays in the male tail—the trithorax group of chromatin regulators promotes the expression of two Hox genes in the seam cells V5 and V6, whereas the Polycomb group blocks their expression (Chamberlin and Thomas 2000; Ross and Zarkower 2003; Zhang et al. 2003). The other involves the induction of vulval development by an EGF signal from the anchor cell. In the nearby hypodermis, the Nucleosome Remodeling Deacetylase (NURD) and Retinoblastoma (Rb) complexes from the SynMuvB group block the expression of the EGF gene lin-3 (Cui et al. 2006), preventing the inappropriate activation of the Ras pathway. Other chromatin remodeling complexes help regulate the development of the vulva, but their targets remain unknown (Fay and Yochem 2007). For example, the Tip60/NuA4 histone acetyl transferase (HAT) complex blocks vulval development (Ceol and Horvitz 2004), whereas the nucleosome remodeling factor (NURF) complex promotes vulval cell fates (Andersen et al. 2006).
Chromatin regulators also play an important role in the establishment of the nematode germline (Schaner et al. 2003), and three observations raise the possibility that some of them influence the sperm/oocyte decision. First, the C. elegans tra-4 gene encodes a PLZF-containing protein that works with histone chaperones and deacetylases to promote many female cell fates (Grote and Conradt 2006), although a role in oogenesis has not been detected. Second, natural variation in the C. elegans NATH-10 acetyltransferase controls the number of sperm produced by hermaphrodites (Duveau and Félix 2012), though how it does so remains unknown. Third, the Tip60 HAT complex regulates the sperm/oocyte decision in both C. elegans and C. briggsae (Guo et al. 2013), although it remains unclear whether Tip60 directly acetylates the transcription factor TRA-1 or works with TRA-1 to acetylate histones in the promoters of targets like fog-3. With these cases in mind, we began investigating the role of chromatin remodelers in the sperm/oocyte decision.
This regulatory decision is ideal for comparative evolutionary studies (Haag 2005), because it played a critical role in the origin of self-fertile hermaphrodites (Baldi et al. 2009). Both C. briggsae and C. elegans evolved hermaphroditic reproduction independently (Cho et al. 2004; Kiontke et al. 2004, 2011). In each species, the XX animals have female bodies but make sperm during the L4 larval stage and oocytes as adults, an arrangement that allows self-fertilization. Several studies have shown that this evolutionary step involved independent modifications to the sex-determination pathway, which controls the sperm/oocyte decision. In C. elegans, FOG-2 and GLD-1 cause spermatogenesis in hermaphrodites, by blocking the translation of tra-2 messages (Clifford et al. 2000). By contrast, there is no fog-2 gene in C. briggsae (Nayak et al. 2005), gld-1 has a different function (Nayak et al. 2005; Beadell et al. 2011) and tra-2 is regulated by the novel protein SHE-1 (Guo et al. 2009). The FEM complex is also required for male cell fates in C. elegans (Doniach and Hodgkin 1984; Kimble et al. 1984; Hodgkin 1986) but is dispensable for hermaphrodite spermatogenesis in C. briggsae (Hill et al. 2006). All of these regulatory genes act through the transcription factor TRA-1 to control fog-1 and fog-3, which promote spermatogenesis in both sexes (Chen and Ellis 2000; Chen et al. 2001; Jin et al. 2001). Because the Tip60 HAT complex is involved in this decision, we began studying the roles of other chromatin remodelers.
Here, we show that NURF-1A and ISW-1, the C. briggsae homologs of Drosophila NURF301 and ISWI, promote spermatogenesis in both sexes. These proteins are the principal components of the NURF complex (Tsukiyama and Wu 1995; Tsukiyama et al. 1995), which is a member of the imitation switch family of chromatin remodelers (Corona and Tamkun 2004). These complexes use the energy of ATP hydrolysis to slide nucleosomes along the DNA, which increases chromatin fluidity and alters the accessibility of target sites to transcription factors and other regulatory proteins. In Drosophila, NURF301 and ISWI are essential for the maintenance of germline stem cells (Cherry and Matunis 2010). In C. elegans, they also promote germ cell proliferation and antagonize the Tip60 HAT complex to allow normal vulval development (Andersen et al. 2006). Here, we show that C. briggsae NURF-1 and ISW-1 also initiate spermatogenesis, by promoting the expression of fog-1 and fog-3. Furthermore, this regulation is independent of the Gli protein TRA-1. Surprisingly, this role of the NURF complex in the sperm/oocyte decision is unique to C. briggsae, although its other functions in embryonic and vulval development have been conserved.
Results
The C. briggsae NURF Complex Is Required for Germ Cells to Adopt Male Fates
In Drosophila, the NURF complex regulates many aspects of development and is critical for the proliferation of germ cells (Alkhatib and Landry 2011). Although the Drosophila complex contains four components, ISWI and NURF301 are sufficient to reconstitute its activity (Xiao et al. 2001). In C. elegans, mutations in the corresponding NURF genes isw-1 and nurf-1 cause sterility and suppress the vulval defects of lin-15AB mutants (Andersen et al. 2006). Thus, we focused on orthologs of these genes in the related nematode C. briggsae. Using RT-PCR and RACE, we defined the Cbr-isw-1 and Cbr-nurf-1 genes (fig. 1A and D).
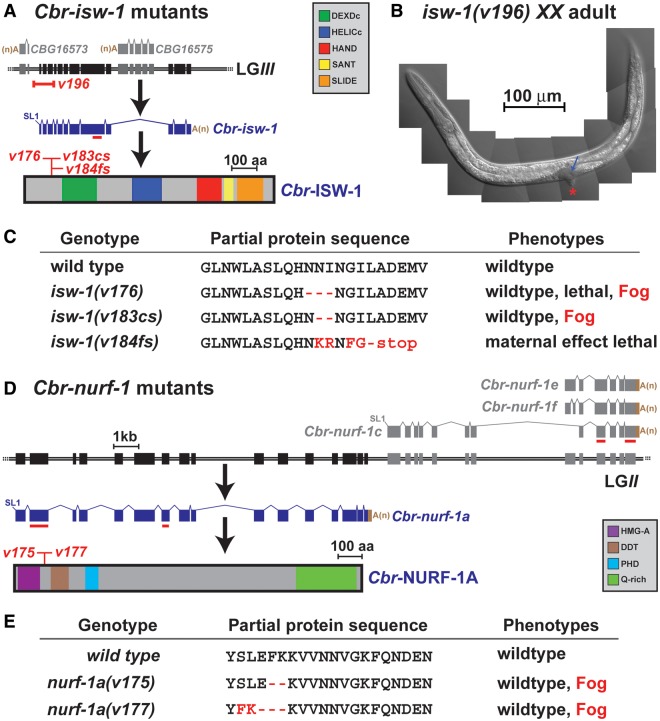
Fig. 1.
The Caenorhabditis briggsae NURF complex regulates the sperm/oocyte decision. (A) Location of mutations affecting Cbr-isw-1. Caenorhabditis briggsae genomic DNA is shown as a double line, with exons as boxes. Nearby transcripts are above the line in gray, and isw-1 is below the line, in blue. The position of each new mutation is marked in red, and critical domains of the protein are coded by color. Regions of the transcript targeted by RNAi are underlined in red. (B) Nomarski photomicrograph of a Cbr-isw-1(v196) XX adult, showing a protruding vulva (red *) and small germ line (blue arrow). (C) Molecular lesions and phenotypes for each isw-1 mutation (for details, see table 1). (D) Location of mutations affecting Cbr-nurf-1. Conventions are like those for isw-1. (E) Molecular lesions and phenotypes for each nurf-1 mutation (for details, see table 1).
To learn the function of the C. briggsae NURF complex, we used Transcription Activator-Like Effector Nucleases (TALENs) (Wood et al. 2011; Wei et al. 2014) to make deletions in each gene (fig. 1). We isolated four mutations in Cbr-isw-1–v184, a frameshifting deletion; v196, an 862-bp deletion that removes the first three and a half exons; and v176 and v183, small in-frame deletions. We also recovered two in-frame deletions in Cbr-nurf-1–v175 and v177.
Homozygous isw-1(v196) null mutants had a tiny germ line and a protruding vulva and did not produce offspring (fig. 1B). Although Cbr-isw-1(v184) frameshift mutants were viable if maternal transcripts were present, their progeny died as embryos (fig. 1C and table 1). Thus, C. briggsae ISW-1 is essential for the development of the vulva and the proliferation of germ cells, and maternal ISW-1 is needed for embryonic viability. Surprisingly, the in-frame mutants of isw-1 and nurf-1 were not only healthy but occasionally Fog (Feminization of the germline; fig. 1 and table 1). Thus, the C. briggsae NURF complex not only has pleiotropic functions, as in other species, but appears to regulate the sperm/oocyte decision. The Fog phenotype of C. briggsae NURF mutants was surprising, because nothing similar had been seen in C. elegans (Andersen et al. 2006). Hence, we began a detailed exploration of this trait and then used the results for comparison with other nematode species.
Table 1.
Caenorhabditis briggsae NURF-1 and ISW-1 Promote Spermatogenesis, Viability, and Fertility.
| Germline Phenotypes |
||||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | Oocytes Only (%) | Sperm and Oocytes (%) | Sperm Only (%) | Immature Germ Cells (%) | Lethal (%) | Other (%) | n | |
| nurf-1(v175) | XX | 0.8 | 94.2 | 0 | 0 | 2 | 3a | 408 |
| nurf-1(v177) | XX | 0.5 | 93.5 | 0 | 0 | 2 | 4a | 336 |
| isw-1(v176) | XX | 1 | 89 | 0 | 0 | 4 | 6a | 815 |
| isw-1(v183cs) | XX | 11 | 88 | 0 | 0 | 0 | 1b | 67 |
| isw-1(v183cs) | XO | 0 | 0 | 100 | 0 | 0 | 0 | 20 |
| isw-1(v184fs)M+ | XX | 0 | 100 | 0 | 0 | 0 | 0 | 24 |
| isw-1(v184fs)M− | XX | 0 | 0 | 0 | 0 | 100 | 0 | 52 |
| isw-1(v184fs)/+ | XOc | 0 | 0 | 99d | 0 | 0 | 1e | 101 |
| isw-1(v196df) | XX | 0 | 0 | 0 | 100f | 0 | 0 | 16 |
| isw-1(v196df)/+ | XOg | 0 | 4 | 96 | 0 | 0 | 0 | 80 |
| isw-1(v196df) | XO | 0 | 0 | 0 | 100h | 0 | 0 | 21 |
Note.—Animals were scored at 20 °C, except isw-1(v183cs), which was scored at the restrictive temperature of 15 °C.
aEgg-laying defective.
bDisorganized germ line.
cMales were progeny from crosses of dpy-18 hermaphrodites with v184/dpy-18 or v184 males.
dOne male had sperm at both ends of the gonad. Five males made very few sperm.
eThe male had a ruptured gonad, produced sperm, and appeared to be starting oogenesis.
fAll animals were smaller in soma and germ line than AF16, and had a protruding vulva.
gMales were progeny from crosses of dpy-18 hermaphrodites with v196/dpy-18 or v196 males.
hAll animals were smaller in soma and germ line, and had a blunt, defective tail.
The C. briggsae NURF Complex Is Required to Initiate Spermatogenesis in Both Sexes
Because weak alleles of nurf-1 and isw-1 caused a Fog phenotype in some animals, we used RNA interference to lower gene activity further, without eliminating it altogether (Montgomery et al. 1998). Knocking down either Cbr-nurf-1 or Cbr-isw-1 caused animals of both sexes to make oocytes instead of sperm (fig. 2 and supplementary fig. S1, Supplementary Material online). This effect was complete in XX animals and highly penetrant in XO males (tables 2 and 3). As in C. elegans, RNAi against the remaining NURF components was lethal (tables 2 and 3). Thus, the C. briggsae NURF complex is required in both sexes for germ cells to initiate spermatogenesis rather than oogenesis.
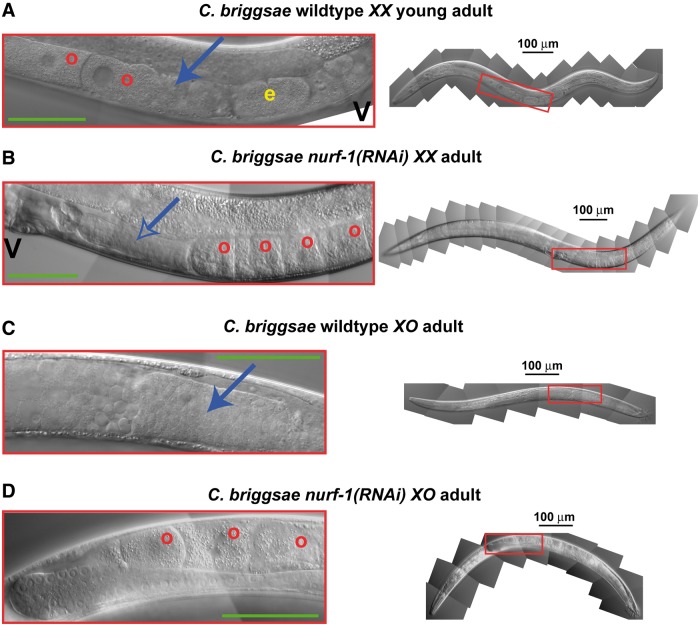
Fig. 2.
Caenorhabditis briggsae NURF-1 controls the sperm/oocyte decision in both sexes. (A) Caenorhabditis briggsae wild-type XX young adult. (B) Caenorhabditis briggsae nurf-1(RNAi) XX adult. This animal is older than the control in panel A, because the nurf-1 germ line develops more slowly. (C) Caenorhabditis briggsae wild-type male. (D) Caenorhabditis briggsae nurf-1(RNAi) Fog male. In all panels, anterior is left and ventral is down. The size of each inset is shown by a box on the adjacent animal; the green bar in the inset is 50 µm. Finally, “o” indicates oocytes, “e” embryos, “V” the vulva, a solid blue arrow marks a spermatheca filled with sperm, and a hollow blue arrow marks an empty spermatheca.
Table 2.
Caenorhabditis briggsae NURF-1A Controls Germ Cell Fates in Both Sexes.
| Targets of RNAi | Oocytes Only (%) | Sperm and Oocytes (%) | Sperm Only (%) | Other (%) | n | |
|---|---|---|---|---|---|---|
| Cbr-nurf-1a | XX | 99.6 | 0.4 | 0 | 0 | 421 |
| Cbr-nurf-1a | XO | 62 | 9 | 20 | 9a | 44 |
| Cbr-nurf-1cefb | XX | 0 | 100 | 0 | 0 | 207 |
| Cbr-nurf-1a + nurf-1cef | XX | 86 | 12 | 0 | 2c | 253 |
aUndifferentiated germ cells, some with vacuoles in the germ line.
bRNAi was used to simultaneously target the nurf-1.2c, e, and f transcripts.
cDead eggs.
Table 3.
Caenorhabditis briggsae ISW-1 Acts with NURF-1A to Control Germ Cell Fates.
| Drosophila Homolog | Target of RNAi | Oocytes Only (%) | Sperm and Oocytes (%) | Sperm Only (%) | Dead Eggs (%) | Dead Larvae (%) | n | |
|---|---|---|---|---|---|---|---|---|
| ISWI | Cbr-isw-1 | XX | 100 | 0 | 0 | 0 | 0 | 398 |
| ISWI | Cbr-isw-1 | XO | 52 | 18 | 30 | NA | NA | 33 |
| NURF55 | Cbr-lin-53 | XX | 0 | 1a | 0 | 91 | 8 | 274 |
| NURF55 | Cbr-rba-1 | XX | 0 | 3 | 0 | 96 | 1 | 125 |
| NURF38 | Cbr-pyp-1 | XX | 0 | 0 | 0 | 23 | 77 | 150 |
aTwo animals produced both sperm and oocytes but were sterile.
To see whether the oocytes made by these mutants were functional, we crossed individual Fog animals with wild-type males and studied the eggs they laid over the following 20 h. We found that 92% of the progeny from Cbr-nurf-1(RNAi) mothers hatched and grew into healthy adults, and only 8% died as embryos (n = 751). Similarly, 86% of the progeny from Cbr-isw-1(RNAi) mothers became healthy adults, and 14% died as embryos (n = 280). As a control, we found that 7% of the progeny from she-1(v35) mothers died as embryos (n = 242). This level of lethality is similar to that observed for the first oocytes fertilized in C. elegans Fog mothers (Andux and Ellis 2008). Thus, the oocytes of Cbr-nurf-1(RNAi) and Cbr-isw-1(RNAi) Fog animals function normally.
A Single NURF-1 Product Regulates the Sperm/Oocyte Decision in C. briggsae
Alternative splicing produces several NURF301 isoforms in Drosophila (Kwon et al. 2009). When we used RT-PCR to characterize the C. briggsae messages, we detected four nurf-1 transcripts (fig. 2D), which correspond to prominent nurf-1 transcripts identified in C. elegans (Andersen et al. 2006). One message is produced from the left half of the nurf-1 locus, which we name nurf-1A. The remaining transcripts are produced from the right half of the locus, which we call nurf-1B. Neither C. briggsae nor C. elegans makes a product like full-length Drosophila NURF301, which would span the entire region (Xiao et al. 2001; Andersen et al. 2006), so the range of NURF-1 isoforms differs from that seen in fruit flies.
Both C. elegans and C. briggsae share nurf-1a, c, and e transcripts (Andersen et al. 2006). Although knocking down nurf-1A caused a Fog phenotype, knocking down the remaining transcripts did not (tables 2 and 3). Furthermore, we could detect no other defects in the nurf-1B(RNAi) animals, and mutations that affect the corresponding C. elegans transcripts also had no phenotype (Andersen et al. 2006). Thus, nurf-1a might encode the only NURF-1 isoform that regulates developmental decisions in nematodes. NURF-1A is also the only isoform to contain an HMG-A domain, which is known to bind DNA (Travers 2000).
As in C. elegans, Cbr-isw-1 is trans-spliced to SL1 and produces a single transcript. When we used RT-PCR to study nurf-1a, isw-1, and lin-53 transcript levels, we found that each was predominantly expressed in germ cells, as expected from the phenotypes we observed (supplementary fig. S2, Supplementary Material online).
NURF-1A and ISW-1 Promote the Expression of the Male Genes fog-1 and fog-3
In C. elegans, fog-1 and fog-3 are required for germ cells to become sperm rather than oocytes (Barton and Kimble 1990; Ellis and Kimble 1995), and the function of each gene is conserved in C. briggsae (Chen et al. 2001). Because the NURF complex is predicted to regulate transcription by altering chromatin (Tsukiyama and Wu 1995), we used real-time RT-PCR to study these transcripts in wild-type, Cbr-nurf-1a(RNAi) and Cbr-isw-1(RNAi) animals.
In C. elegans hermaphrodites, fog-1 and fog-3 expression is high during larval development, when germ cells are undergoing spermatogenesis, and low in adults, during oogenesis (Chen and Ellis 2000; Lamont and Kimble 2007). Thus, we focused on three developmental stages of C. briggsae XX animals: larvae at the L3/L4 molt, L4 larvae, and adults. The levels of Cbr-fog-1 and Cbr-fog-3 transcripts were highest at the L3/L4 molt (fig. 3A), when germ cells were beginning to select male fates (Lee et al. 2011; Morgan et al. 2013). In L4 larvae, the levels of Cbr-fog-1 and Cbr-fog-3 transcripts had begun to decline, but genes like Cbr-spe-4, which are active in primary spermatocytes (Arduengo et al. 1998; Gosney et al. 2008), were expressed at high levels. In adults, Cbr-fog-1, Cbr-fog-3, and Cbr-spe-4 transcripts were all found at low or undetectable levels.
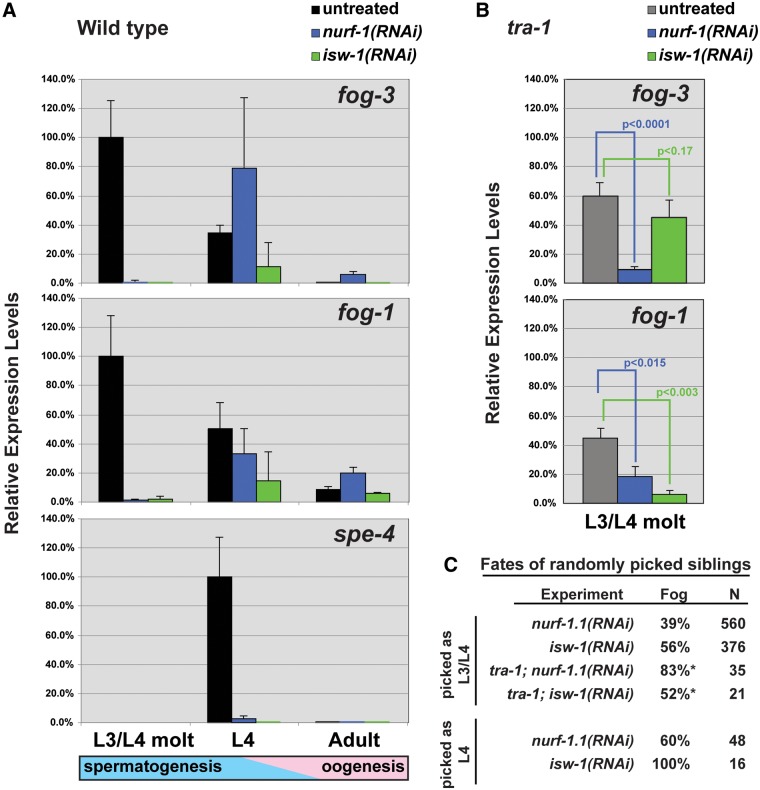
Fig. 3.
Caenorhabditis briggsae NURF-1A and ISW-1 control the expression of fog-1 and fog-3. Transcript levels were measured by real-time RT-PCR and calculated using 2exp(−ΔΔCT), with rpb-1 transcripts as a reference; fog-1 and fog-3 were normalized to wild-type animals at the XX L3/L4 molt and spe-4 to the XX L4. Error bars represent standard error of the mean. (A) Wild-type animals subjected to RNAi. (B) tra-1(nm2) animals subjected to RNAi. P values were determined with a Student’s t-test, one-tailed, with unequal variance. (C) The phenotypes of siblings of animals that were collected for transcript analysis.
However, in both Cbr-nurf-1a(RNAi) and Cbr-isw-1(RNAi) animals, the levels of fog-1 and fog-3 transcripts were dramatically lower at the L3/L4 molt, a period that is critical for the initiation of spermatogenesis (fig. 3A). By the L4 stage, the levels of fog-1 and fog-3 transcripts were nearly wild type, but this recovery was too late to alter development, since about half of the animals only produced oocytes (fig. 3C). This Fog phenotype was confirmed by the absence of spe-4 transcripts in the Cbr-nurf-1a(RNAi) and Cbr-isw-1(RNAi) L4 larvae (fig. 3A). Finally, fog-1 and fog-3 transcripts were found at higher levels in Cbr-nurf-1a(RNAi) adults than in the wild type (P < 0.003, fig. 3A). Thus, NURF-1A and ISW-1 promote high levels of fog-1 and fog-3 expression in larvae, but NURF-1A might help repress these transcripts in adults.
The NURF Complex and TRA-1 Act Independently on fog-1 and fog-3
TRA-1 is a Gli transcription factor that acts at the end of the sex-determination pathway (Hodgkin and Brenner 1977; Zarkower and Hodgkin 1992; Kelleher et al. 2008). Furthermore, there are conserved TRA-1 binding sites in the fog-1 and fog-3 promoters, and C. elegans TRA-1 can bind these sites in vitro (Chen and Ellis 2000; Jin et al. 2001). To see if the NURF complex acts upstream of TRA-1 to promote the expression of fog-1 and fog-3, we examined double mutants with Cbr-tra-1(nm2), a nonsense mutation (Kelleher et al. 2008), and Cbr-tra-1(v181), which causes a frameshift prior to the zinc finger domain (see Materials and Methods). These null mutations transform XX animals into males that make sperm as larvae and oocytes as adults. Surprisingly, Cbr-tra-1; Cbr-nurf-1a(RNAi) and Cbr-tra-1; Cbr-isw-1(RNAi) XX young adults only made oocytes (table 4). Thus, NURF-1A and ISW-1 can promote spermatogenesis even in the absence of TRA-1 and must act near the end of the sex-determination pathway in germ cells.
Table 4.
NURF-1A and ISW-1 Are Epistatic to TRA-1.
| Genetic Background | Target Gene | Oocytes Only (%) | Sperm and Oocytes (%) | Sperm Only (%) | n |
|---|---|---|---|---|---|
| tra-1(nm2)a | 0 | 96 | 4 | 25 | |
| tra-1(nm2)a | nurf-1a | 86 | 8 | 4 | 48 |
| tra-1(nm2)a | isw-1 | 56 | 44 | 0 | 50 |
| tra-1(v181fs)a | 0 | 88 | 12 | 60 | |
| tra-1(v181fs)a | nurf-1a | 100 | 0 | 0 | 122 |
| tra-1(v181fs)a | isw-1b | 87 | 13 | 0 | 160 |
aXX tra-1 animals were self-progeny of tra-1 +/ +dpy-18 mothers. They were picked as L4s and scored with DIC 1 day later.
bFour micrograms per microliter isw-1 dsRNA was injected.
We also studied double mutants with Cbr-tra-2. The weak allele tra-2(nm9ts) and NURF complex RNAi mutually suppressed each other (table 5), which is consistent with models in which TRA-1 and the NURF complex act independently on the fog promoters. NURF RNAi also caused oogenesis in tra-2(nm1) null mutants, which normally make only sperm. Over 50% of the Cbr-tra-2(nm1); Cbr-nurf-1a(RNAi) animals and some Cbr-tra-2(nm1); Cbr-isw-1(RNAi) animals made oocytes, but we never saw oogenesis in the controls (table 5).
Table 5.
Mutations in the NURF Complex and Cbr-tra-2 Are Mutually Antagonistic.
| Genetic Background | Target Gene | Fog (%) | Sperm Only (%) | Othera (%) | n |
|---|---|---|---|---|---|
| tra-2(nm9ts) | 0 | 100 | 0 | 21 | |
| tra-2(nm9ts) | nurf-1a | 55 | 45 | 0 | 22 |
| tra-2(nm9ts) | isw-1b | 32 | 42 | 26 | 31 |
| tra-2(nm1) cby-15c | 0 | 100 | 0 | 36 | |
| tra-2(nm1) cby-15 | nurf-1a | 50 | 38 | 12 | 24 |
| tra-2(nm1) cby-15 | isw-1 | 0 | 100 | 0 | 195 |
| tra-2(nm1) | 0 | 96 | 4 | 25 | |
| tra-2(nm1) | nurf-1a | 62 | 32 | 6 | 72 |
| tra-2(nm1) | isw-1 | 3 | 55 | 42 | 77 |
| tra-2(nm1) | isw-1b | 3 | 76 | 21 | 38 |
aAnimals had defective germ lines with no differentiated cells. Some had vacuoles.
bFour micrograms per microliter isw-1 dsRNA was injected.
ccby-15 was used as a linked marker for tra-2 in this set of crosses.
To confirm these results, we studied Cbr-tra-1(nm2); nurf-1a(RNAi) and Cbr-tra-1(nm2); isw-1(RNAi) mutants at the L3/L4 molt. Both strains had lower levels of fog-1 and fog-3 transcripts than tra-1 controls (fig. 3B). Because NURF-1A and ISW-1 do not require TRA-1 to regulate these genes, the NURF complex is likely to regulate their promoters independently.
The Role of the NURF Complex Changed during Recent Nematode Evolution
In C. elegans, mutations that affect the NURF complex did not block spermatogenesis (Andersen et al. 2006), whereas in C. briggsae, lowering NURF activity even slightly feminized the germ line. To see whether the C. elegans NURF complex plays any role in the sperm/oocyte decision, we performed more detailed studies of one isw-1 mutant. The Cel-isw-1(n3297) mutation alters an amino acid located just after the DEXDc domain (Andersen et al. 2006), whereas the Cbr-isw-1(v176) and v183 mutations delete a few amino acids just before this domain. All of these mutants are viable and none show the complete sterility of null alleles, so they each cause a partial loss of function. We found that 38% of Cel-isw-1(n3297) XX mutants were sterile, but none of them appeared Fog (n = 213). Furthermore, we examined some of the sterile animals with differential interference contrast (DIC) optics, and 67% had sperm and defective oocytes, whereas the rest had small germ lines without differentiated gametes (n = 33). Finally, 90% of Cel-isw-1(n3297) XO males made sperm, 4% had tumorous germ lines, and 3% had small germ lines but none made oocytes (n = 49). Since these C. briggsae and C. elegans mutations all decrease ISW-1 activity, it seems likely that lowering the activity of the NURF complex alters the sperm/oocyte decision in C. briggsae but not in C. elegans.
Second, we compared null alleles. The Cbr-isw-1(v196) deletion is a null allele (supplementary fig. S3, Supplementary Material online). Although homozygotes are sterile because of severe germ line defects, some Cbr-isw-1(v196)/+ males made oocytes (supplementary fig. S4, Supplementary Material online, and table 1). By contrast, no Cel-isw-1(ok1951)/+ males made oocytes (n = 131). This difference is significant at P = 1%, in a z-test comparing the two proportions.
Third, we studied the function of the NURF complex in mutants that were predisposed to producing oocytes. Specifically, we used RNAi to knock down Cel-nurf-1a or Cel-isw-1 in either Cel-fog-1(q253ts) or Cel-fem-1(hc17ts) mutants, at the intermediate temperature of 20 °C. Although NURF RNAi did not cause synthetic feminization (fig. 4 and supplementary table S1, Supplementary Material online), similar experiments using trr-1 caused strong synthetic feminization (Guo et al. 2013). Thus, none of these studies identified any role for the NURF complex in the C. elegans sperm/oocyte decision.
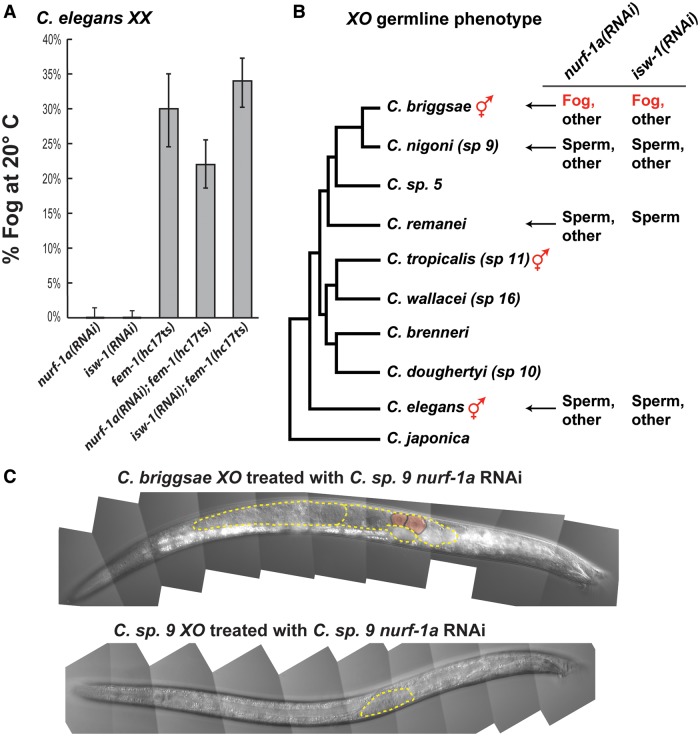
Fig. 4.
The role of NURF-1 and ISW-1 in germ cells changed during evolution. (A) At 20 °C, Caenorhabditis elegans NURF-1A or ISW-1 were knocked down by RNAi in fem-1(hc17ts) mutants. The double mutants were not more feminized than fem-1. Error bars represent 95% confidence intervals for a proportion. (B) Phylogeny of Caenorahbditis (Kiontke et al. 2011; Felix et al. 2014) showing the results of RNAi experiments (supplementary table S2, Supplementary Material online). (C) Caenorhabditis nigoni dsRNA causes a Fog phenotype in C. briggsae males (top) but not in C. nigoni (bottom). Gonads are outlined with dotted yellow lines, and two oocytes are shaded pink.
Finally, we studied the NURF complex in male/female species related to C. briggsae. Knocking down either nurf-1a or isw-1 in C. nigoni or C. remanei did not cause males to produce oocytes but did cause occasional sterility (fig. 4 and supplementary table S2, Supplementary Material online). In a control experiment, fog-3(RNAi) caused 42% of C. nigoni males to become Fog (n = 38), so this cell-fate decision is sensitive to RNA interference. Finally, when we injected C. nigoni nurf-1a or isw-1 dsRNA into C. briggsae wild-type strains, each one caused a Fog phenotype in males (fig. 4C and supplementary table S2, Supplementary Material online). Thus, the NURF complex has either acquired a new function in C. briggsae, or its role in the sperm/oocyte decision has changed so much that it can only be detected in C. briggsae.
Discussion
Caenorhabditis briggsae NURF-1A and ISW-1 Are Required for Germ Cells to Adopt Male Fates
In C. briggsae, knocking down either NURF-1A or ISW-1 by RNA interference causes animals of both sexes to make oocytes instead of sperm. Furthermore, partial loss-of-function mutations in either gene cause some animals to become Fog, and loss of just one copy of isw-1 causes rare males to make oocytes. Thus, NURF-1A and ISW-1 promote spermatogenesis in C. briggsae, and small decreases in their activities cause germ cells to differentiate as oocytes. These two proteins are likely to work together as part of a NURF chromatin remodeling complex.
The Role of the NURF Complex in Germ Cells Changed during Recent Evolution
This role of the NURF complex in the sperm/oocyte decision appears to be unique to C. briggsae. In C. elegans, nurf-1 and isw-1 mutants make both sperm and oocytes (Andersen et al. 2006). Furthermore, lowering C. elegans NURF activity does not change the sperm/oocyte decision in mutants that are predisposed to a transformation in germ cell fates, whereas lowering Tip60 HAT activity causes synthetic feminization in these genetic backgrounds (Guo et al. 2013). Finally, C. elegans isw-1(null)/+ males do not make oocytes, unlike some of their C. briggsae counterparts.
We also found no role for the NURF complex in the sperm oocyte decision of male/female nematodes. Caenorhabditis nigoni is so closely related to C. briggsae that they can mate and produce viable offspring (Woodruff et al. 2010), and these species appear to have diverged on the order of 107 generations ago (Cutter et al. 2010). However, knocking down NURF activity in C. nigoni did not cause males to make oocytes, and similar experiments with the more distant relative C. remanei had the same result. We know the dsRNA we used was of high quality, because C. briggsae germ cells became oocytes in response to both C. nigoni nurf-1a and C. nigoni isw-1 dsRNA, even though C. nigoni germ cells did not. Furthermore, these differences were not due to variable sensitivity to RNA interference, because knocking down NURF activity in C. nigoni and C. remanei strongly affected germline traits like proliferation. We conclude that the role of the NURF complex changed during recent evolution.
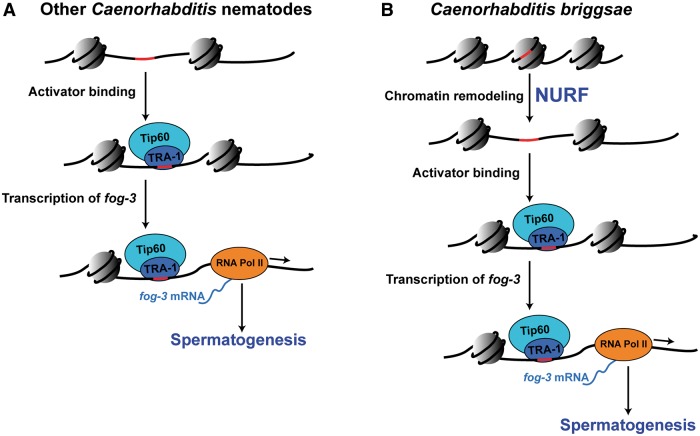
Two types of models could explain this change. In the first, the C. briggsae NURF complex was recruited to regulate the sperm/oocyte decision; NURF-1A might be critical for this function because it has an HMG-A domain, which can bind DNA. In the most parsimonious version, the configuration of chromatin in this region of the genome has be opened in C. briggsae larvae but is already accessible in C. elegans larvae (fig. 5). In the figure, we propose that the NURF complex allows TRA-1 access to the fog promoters, because it is the only transcription factor known to interact with them; however, it remains possible that an unknown activator regulates these genes. This model is consistent with data from Berkseth et al. (2013), who showed that TRA-1 binds the fog-3 promoter at higher levels in C. elegans than in C. briggsae. However, the underlying differences responsible for altered chromatin structure remain unknown, because the C. briggsae and C. nigoni fog-3 promoters are similar in sequence, and the C. briggsae fog-3 promoter is regulated normally in C. elegans (Chen et al. 2001).
Fig. 5.
Model for the initiation of spermatogenesis in Caenorhabditis. (A) In most species, the NURF complex is not required. For spermatogenesis, an unknown activator promotes the expression of fog-3. Currently, the only candidate for this activator is full-length TRA-1, perhaps in association with the Tip60 HAT complex (Guo et al. 2013). (B) In C. briggsae, the NURF complex is required for fog-3 to promote spermatogenesis. Because NURF is known to remodel chromatin, the simplest possibility is that it opens up the fog-3 promoter, so that TRA-1 binding sites (shown in red) are accessible. TRA-1 then works with the Tip60 HAT complex to promote expression of fog-3, which works with fog-1 to cause germ cells to initiate spermatogenesis.
In the second type of model, C. briggsae changed, so that it now depends on NURF activity, which is not needed for spermatogenesis in other worms. For example, C. briggsae might have lost a redundant chromatin regulator that controls the fog-3 locus, which made the NURF complex essential in that species. In either scenario, broad changes in the regulation of chromatin structure have occurred during short evolutionary time spans, and these changes played important roles in shaping the expression of genes such as fog-1 and fog-3.
Although chromatin states can be assayed directly in single cells like yeast (Boeger et al. 2008; Yen et al. 2012), such experiments are more difficult in multicellular organisms. In this case, only 75 germ cells in young L4 hermaphrodites will become spermatocytes, out of a total of about 1,500, and these cells cannot be isolated from the rest of the animal. Thus, distinguishing among these models will require a new array of tools. We expect genome editing might help create these tools (Wood et al. 2011; Lo et al. 2013; Wei et al. 2014) and will become a standard part of evolutionary studies in the future.
NURF-1A Is a Critical Component of the Nematode NURF Complex
In Drosophila, NURF301 and ISWI are core components of the NURF complex, which also contains NURF38 and NURF55. In C. briggsae, mutants in NURF-1A and ISW-1 have similar phenotypes, so they probably form a nematode NURF complex. Because the homologs of NURF38 and NURF55 are essential, they could not be tested for a role in the sperm/oocyte decision but probably also form part of this complex. Because null mutations in isw-1 block germ cell proliferation, at least one function of the NURF complex is broadly conserved in animals.
However, the structure of the NURF complex remains unclear. In Drosophila, nurf301 produces multiple transcripts, whose differing roles are poorly understood (Kwon et al. 2009). In C. elegans, only NURF-1A regulates vulval cell fates (Andersen et al. 2006), and in C. briggsae, only NURF-1A regulates germ cell fates. Inactivating other nurf-1 products causes no phenotype either species. Because Cbr-NURF-1A, Cel-NURF-1A, and Drosophila NURF301C are similar in structure to each other and to the human protein FAC1 (Bowser et al. 1995), they might share a conserved chromatin regulatory function.
Caenorhabditis briggsae fog-3 Is Ideal for Studying Interactions of Gli Proteins with Chromatin Regulators
Developing germ cells make two major decisions: Whether to proliferate or enter meiosis, and whether to become spermatocytes or oocytes (Kimble and Crittenden 2007; Morgan et al. 2013). In nematodes, FOG-1 and FOG-3 influence the mitosis–meiosis decision and are absolutely required for germ cells to adopt male fates (Barton and Kimble 1990; Ellis and Kimble 1995; Thompson et al. 2005; Snow et al. 2013). Three sets of proteins are known to regulate the expression of these genes. First, TRA-1 is the sole nematode Gli protein. Cleaved TRA-1 favors oogenesis by repressing fog-1 and fog-3, whereas full-length TRA-1 appears to favor spermatogenesis by promoting their expression (Chen and Ellis 2000; Schvarzstein and Spence 2006). Second, the Tip60 HAT complex is required for TRA-1 to promote fog-3 expression (Guo et al. 2013). It might act on TRA-1 or work with TRA-1 to modify chromatin. Third, NURF activity is needed in C. briggsae larvae for the expression of fog-1 and fog-3, when germ cells are beginning to commit to spermatogenesis.
Although TRA-1 acts directly on the fog-3 promoter, knocking down NURF activity lowers the expression of fog-1 and fog-3 in tra-1(null) mutants. Thus, the NURF complex must act downstream of TRA-1, and we infer that TRA-1 and the NURF complex act independently on these two promoters. This idea is supported by one other observation—Cbr-tra-2(null) mutants partially suppress the Cbr-nurf-1a(RNAi) and Cbr-isw-1(RNAi) phenotypes. Because these tra-2 mutations act through TRA-1 to promote spermatogenesis, they might increase its activator function enough to promote fog-3 expression without chromatin remodeling. Thus, we favor models in which the NURF complex remodels chromatin to provide TRA-1 and Tip60 access to the fog-3 promoter (fig. 5). This possibility is consistent with predictions that the TRA-1 binding sites in the Cbr-fog-1 and Cbr-fog-3 promoters are normally buried in nucleosomes (supplementary fig. S5, Supplementary Material online; Xi et al. 2010) and might require NURF activity to become accessible.
In yeast and humans, HAT complexes and chromatin remodelers can work as coactivators to regulate gene expression (Featherstone 2002). Furthermore, studies in Drosophila showed that the Gcn5 HAT complex and the NURF complex regulate a common set of target genes, and that the NURF complex is required for Gcn5 to access these targets (Carre et al. 2008). Because the Gli protein TRA-1, the NURF chromatin-remodeling complex, and Tip60 HAT complex all regulate fog-3 expression in C. briggsae, this system provides an ideal model for exploring how chromatin-remodelers interact with Gli transcription factors to control target genes.
Materials and Methods
Strains and Genetics
Caenorhabditis briggsae mutants were derived from the wild isolate AF16 (Fodor et al. 1983). They include LGII: dpy(nm4) (Hill et al. 2006), tra-2(nm1) and tra-2(nm9ts) (Kelleher et al. 2008), and cby-15(sy5418) (Sternberg P, personal communication); LGIII: tra-1(nm2) (Kelleher et al. 2008) and dpy-18(mf104) (Felix M, personal communication); and LGIV: she-1(v49) (Guo et al. 2009). We also used the C. briggsae wild isolate HK104 (Kagawa H, personal communication; Stein et al. 2003).
Caenorhabditis elegans mutants were derived from the wild isolate N2 (Brenner 1974) and include LGI: fog-1(q253ts) (Barton and Kimble 1990), LGIII: isw-1(ok1951) (Andersen et al. 2006), and LGIV: fem-1(hc17ts) (Nelson et al. 1978). All C. nigoni RNAi experiments were done using JU1421 (Felix M, personal communication), and C. remanei RNAi experiments were done using PB4641 (Baird S, personal communication).
RNA Interference
Each template was amplified from mixed-stage cDNA by the PCR, using primers that contained a T7 promoter (supplementary table S3, Supplementary Material online). Templates were purified with a PCR Purification kit (Qiagen) and transcribed using MegaScript (Ambion). After annealing, double-stranded RNA was purified with MegaClear (Ambion). RNAi was performed by injection, using 1 µg/µl solutions of dsRNA (Fire et al. 1998).
Semiquantitative RT-PCR
For each genotype, groups of five adult worms of the desired age and phenotype were collected and processed as described (Chen and Ellis 2000); three independent samples were prepared to confirm reproducibility. RT-PCR was performed as described, using HotMaster Taq DNA polymerase (5PRIME) and MMLV Reverse Transcriptase (Invitrogen). The PCR reactions were run for 35 cycles using primers from supplementary table S4A, Supplementary Material online.
Real-Time Quantitative RT-PCR
Groups of five worms of the desired age and phenotype were picked and prepared as described above, and three or four independent biological replicates and two technical replicates were assayed for each data point. Siblings of the collected animals were allowed to mature and scored for phenotype, to determine the efficiency of the RNAi. For each reaction, 1/20 of the total cDNA sample was used in a final volume of 25 µl, which included 12.5 µl of FastStart Universal SYBR Green Master (Rox) (Roche) and 6 µM primers (supplementary table S4B, Supplementary Material online). Amplification was 40 cycles, using Applied Biosystems 7500 Real-Time PCR Systems. Samples that did not show detectable amplification by the final cycle were arbitrarily assigned a Ct value of 40.
Microscopy
Worms were observed with DIC microscopy. Images were captured with a Zeiss Axiocam digital camera and Zeiss AxioVision software, and assembled using Adobe Photoshop.
Determination of Gene Structures
For isw-1, nurf-1a, and nurf-1c, e, and f, 3′-RACE, 5′-RACE (Frohman 1994), and RT-PCR with an SL1 primer were used to identify the ends of each transcript. The internal splice sites were identified by RT-PCR. Because of the duplication and specialization of two exons, the genomic sequences that encode the C. briggsae nurf-1a transcript do not overlap those that encode the remaining nurf-1 transcripts. We are naming this entire complex locus “nurf-1” to be consistent with the literature from other species. The portion that encodes the nurf-1a transcript is officially the nurf-1A gene. The portion that encodes the remaining nurf-1 transcripts is nurf-1B.
TALEN Knockout Mutants
TALENs were designed and produced as described by Wei et al. (2014). To create mutants, mRNA was injected into the gonad of adult hermaphrodites (Wood et al. 2011). The injection solution contained a pair of TALEN mRNAs, each at 3 µg/µl. At 20 °C, the F1 progeny from a 6- to 32-h time window were singled to new plates at the L4 stage, and F2 animals that were heterozygous or homozygous for new mutations were identified by phenotype or by PCR analysis of the target site.
The v181 mutation deletes nucleotides 276–280 from the coding region of tra-1a, creating a frameshift and early stop codon. As a consequence, it also removes nt 150–154 of tra-1b. Because v181 eliminates the products of both tra-1 transcripts, it must be a null allele.
Fertility and Oocyte Quality Assays
To assess fertility and oocyte quality for nurf-1a(RNAi) and isw-1(RNAi) Fog mutants, ten individuals were singled and each crossed with six wild-type males for 24 h at 20 °C. Afterward, they were scored for fertility, and their progeny for lethality.
Statistical Analyses
To determine the significance of the difference between two proportions, we used www.vassarstats.net/propdiff_ind.html (last accessed June 30, 2014) to calculate the z-ratio and P values.
Supplementary Material
Supplementary figures S1–S5 and tables S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank E. Moss, E. Andersen, M. Sundaram, and Y. Guo for comments, and the Caenorhabditis Genetics Center for strains. This work was supported by the National Institutes of Health (R01 GM085282).
References
- Alkhatib SG, Landry JW. The nucleosome remodeling factor. FEBS Lett. 2011;585:3197–3207. doi: 10.1016/j.febslet.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen EC, Lu X, Horvitz HR. C. elegans ISWI and NURF301 antagonize an Rb-like pathway in the determination of multiple cell fates. Development. 2006;133:2695–2704. doi: 10.1242/dev.02444. [DOI] [PubMed] [Google Scholar]
- Andux S, Ellis RE. Apoptosis maintains oocyte quality in aging Caenorhabditis elegans females. PLoS Genet. 2008;4:e1000295. doi: 10.1371/journal.pgen.1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arduengo PM, Appleberry OK, Chuang P, L’Hernault SW. The presenilin protein family member SPE-4 localizes to an ER/Golgi derived organelle and is required for proper cytoplasmic partitioning during Caenorhabditis elegans spermatogenesis. J Cell Sci. 1998;111:3645–3654. doi: 10.1242/jcs.111.24.3645. [DOI] [PubMed] [Google Scholar]
- Baldi C, Cho S, Ellis RE. Mutations in two independent pathways are sufficient to create hermaphroditic nematodes. Science. 2009;326:1002–1005. doi: 10.1126/science.1176013. [DOI] [PubMed] [Google Scholar]
- Barton MK, Kimble J. fog-1, a regulatory gene required for specification of spermatogenesis in the germ line of Caenorhabditis elegans. Genetics. 1990;125:29–39. doi: 10.1093/genetics/125.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadell AV, Liu Q, Johnson DM, Haag ES. Independent recruitments of a translational regulator in the evolution of self-fertile nematodes. Proc Natl Acad Sci U S A. 2011;108:19672–19677. doi: 10.1073/pnas.1108068108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkseth M, Ikegami K, Arur S, Lieb JD, Zarkower D. TRA-1 ChIP-seq reveals regulators of sexual differentiation and multilevel feedback in nematode sex determination. Proc Natl Acad Sci U S A. 2013;110:16033–16038. doi: 10.1073/pnas.1312087110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Kornberg RD. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell. 2008;133:716–726. doi: 10.1016/j.cell.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowser R, Giambrone A, Davies P. FAC1, a novel gene identified with the monoclonal antibody Alz50, is developmentally regulated in human brain. Dev Neurosci. 1995;17:20–37. doi: 10.1159/000111270. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carre C, Ciurciu A, Komonyi O, Jacquier C, Fagegaltier D, Pidoux J, Tricoire H, Tora L, Boros IM, Antoniewski C. The Drosophila NURF remodelling and the ATAC histone acetylase complexes functionally interact and are required for global chromosome organization. EMBO Rep. 2008;9:187–192. doi: 10.1038/sj.embor.7401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol CJ, Horvitz HR. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev Cell. 2004;6:563–576. doi: 10.1016/s1534-5807(04)00065-6. [DOI] [PubMed] [Google Scholar]
- Chamberlin HM, Thomas JH. The bromodomain protein LIN-49 and trithorax-related protein LIN-59 affect development and gene expression in Caenorhabditis elegans. Development. 2000;127:713–723. doi: 10.1242/dev.127.4.713. [DOI] [PubMed] [Google Scholar]
- Chen PJ, Cho S, Jin SW, Ellis RE. Specification of germ cell fates by FOG-3 has been conserved during nematode evolution. Genetics. 2001;158:1513–1525. doi: 10.1093/genetics/158.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PJ, Ellis RE. TRA-1A regulates transcription of fog-3, which controls germ cell fate in C. elegans. Development. 2000;127:3119–3129. doi: 10.1242/dev.127.14.3119. [DOI] [PubMed] [Google Scholar]
- Cherry CM, Matunis EL. Epigenetic regulation of stem cell maintenance in the Drosophila testis via the nucleosome-remodeling factor NURF. Cell Stem Cell. 2010;6:557–567. doi: 10.1016/j.stem.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Jin SW, Cohen A, Ellis RE. A phylogeny of Caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res. 2004;14:1207–1220. doi: 10.1101/gr.2639304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Clifford R, Lee MH, Nayak S, Ohmachi M, Giorgini F, Schedl T. FOG-2, a novel F-box containing protein, associates with the GLD-1 RNA binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development. 2000;127:5265–5276. doi: 10.1242/dev.127.24.5265. [DOI] [PubMed] [Google Scholar]
- Corona DF, Tamkun JW. Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim Biophys Acta. 2004;1677:113–119. doi: 10.1016/j.bbaexp.2003.09.018. [DOI] [PubMed] [Google Scholar]
- Cui M, Chen J, Myers TR, Hwang BJ, Sternberg PW, Greenwald I, Han M. SynMuv genes redundantly inhibit lin-3/EGF expression to prevent inappropriate vulval induction in C. elegans. Dev Cell. 2006;10:667–672. doi: 10.1016/j.devcel.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Cui M, Han M. 2007. Roles of chromatin factors in C. elegans development. In The CeRC editor. WormBook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Yan W, Tsvetkov N, Sunil S, Felix MA. Molecular population genetics and phenotypic sensitivity to ethanol for a globally diverse sample of the nematode Caenorhabditis briggsae. Mol Ecol. 2010;19:798–809. doi: 10.1111/j.1365-294X.2009.04491.x. [DOI] [PubMed] [Google Scholar]
- Doniach T, Hodgkin J. A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev Biol. 1984;106:223–235. doi: 10.1016/0012-1606(84)90077-0. [DOI] [PubMed] [Google Scholar]
- Duveau F, Félix MA. Role of pleiotropy in the evolution of a cryptic developmental variation in Caenorhabditis elegans. PLoS Biol. 2012;10:e1001230. doi: 10.1371/journal.pbio.1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RE, Kimble J. The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics. 1995;139:561–577. doi: 10.1093/genetics/139.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS, Yochem J. The SynMuv genes of Caenorhabditis elegans in vulval development and beyond. Dev Biol. 2007;306:1–9. doi: 10.1016/j.ydbio.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone M. Coactivators in transcription initiation: here are your orders. Curr Opin Genet Dev. 2002;12:149–155. doi: 10.1016/s0959-437x(02)00280-0. [DOI] [PubMed] [Google Scholar]
- Felix MA, Braendle C, Cutter AD. A streamlined system for species diagnosis in Caenorhabditis (nematoda: rhabditidae) with name designations for 15 distinct biological species. PLoS One. 2014;9:e94723. doi: 10.1371/journal.pone.0094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fodor A, Riddle DL, Nelson FK, Golden JW. Comparison of a new wild-type Caenorhabditis briggsae with laboratory strains of C. briggsae and C. elegans. Nematologica. 1983;29:203–217. [Google Scholar]
- Frohman MA. On beyond classic RACE (rapid amplification of cDNA ends) PCR Methods Appl. 1994;4:S40–S58. doi: 10.1101/gr.4.1.s40. [DOI] [PubMed] [Google Scholar]
- Gosney R, Liau WS, LaMunyon CW. A novel function for the presenilin family member spe-4: inhibition of spermatid activation in Caenorhabditis elegans. BMC Dev Biol. 2008;8:44. doi: 10.1186/1471-213X-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote P, Conradt B. The PLZF-like protein TRA-4 cooperates with the Gli-like transcription factor TRA-1 to promote female development in C. elegans. Dev Cell. 2006;11:561–573. doi: 10.1016/j.devcel.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Guo Y, Chen X, Ellis RE. Evolutionary change within a bipotential switch shaped the sperm/oocyte decision in hermaphroditic nematodes. PLoS Genet. 2013;9:e1003850. doi: 10.1371/journal.pgen.1003850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Lang S, Ellis RE. Independent recruitment of F box genes to regulate hermaphrodite development during nematode evolution. Curr Biol. 2009;19:1853–1860. doi: 10.1016/j.cub.2009.09.042. [DOI] [PubMed] [Google Scholar]
- Haag ES. 2005. The evolution of nematode sex determination: C. elegans as a reference point for comparative biology. In: The CeRC editor. WormBook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RC, de Carvalho CE, Salogiannis J, Schlager B, Pilgrim D, Haag ES. Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev Cell. 2006;10:531–538. doi: 10.1016/j.devcel.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics. 1986;114:15–52. doi: 10.1093/genetics/114.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin JA, Brenner S. Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics. 1977;86:275–287. [PMC free article] [PubMed] [Google Scholar]
- Jin SW, Kimble J, Ellis RE. Regulation of cell fate in Caenorhabditis elegans by a novel cytoplasmic polyadenylation element binding protein. Dev Biol. 2001;229:537–553. doi: 10.1006/dbio.2000.9993. [DOI] [PubMed] [Google Scholar]
- Kelleher DF, de Carvalho CE, Doty AV, Layton M, Cheng AT, Mathies LD, Pilgrim D, Haag ES. Comparative genetics of sex determination: masculinizing mutations in Caenorhabditis briggsae. Genetics. 2008;178:1415–1429. doi: 10.1534/genetics.107.073668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL. Control of germline stem cells, entry into meiosis, and the sperm/oocyte decision in C. elegans. Annu Rev Cell Dev Biol. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- Kimble J, Edgar L, Hirsh D. Specification of male development in Caenorhabditis elegans: the fem genes. Dev Biol. 1984;105:234–239. doi: 10.1016/0012-1606(84)90279-3. [DOI] [PubMed] [Google Scholar]
- Kiontke K, Gavin NP, Raynes Y, Roehrig C, Piano F, Fitch DH. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc Natl Acad Sci U S A. 2004;101:9003–9008. doi: 10.1073/pnas.0403094101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke KC, Félix MA, Ailion M, Rockman MV, Braendle C, Penigault JB, Fitch DH. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol. 2011;11:339. doi: 10.1186/1471-2148-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SY, Xiao H, Wu C, Badenhorst P. Alternative splicing of NURF301 generates distinct NURF chromatin remodeling complexes with altered modified histone binding specificities. PLoS Genet. 2009;5:e1000574. doi: 10.1371/journal.pgen.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont LB, Kimble J. Developmental expression of FOG-1/CPEB protein and its control in the Caenorhabditis elegans hermaphrodite germ line. Dev Dyn. 2007;236:871–879. doi: 10.1002/dvdy.21081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Kim KW, Morgan CT, Morgan DE, Kimble J. Phosphorylation state of a Tob/BTG protein, FOG-3, regulates initiation and maintenance of the Caenorhabditis elegans sperm fate program. Proc Natl Acad Sci U S A. 2011;108:9125–9130. doi: 10.1073/pnas.1106027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo TW, Pickle CS, Lin S, Ralston EJ, Gurling M, Schartner CM, Bian Q, Doudna JA, Meyer BJ. Using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics. 2013;195:331–348. doi: 10.1534/genetics.113.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery MK, Xu S, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan CT, Noble D, Kimble J. Mitosis-meiosis and sperm-oocyte fate decisions are separable regulatory events. Proc Natl Acad Sci U S A. 2013;110:3411–3416. doi: 10.1073/pnas.1300928110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S, Goree J, Schedl T. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 2005;3:e6. doi: 10.1371/journal.pbio.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson GA, Lew KK, Ward S. Intersex, a temperature-sensitive mutant of the nematode Caenorhabditis elegans. Dev Biol. 1978;66:386–409. doi: 10.1016/0012-1606(78)90247-6. [DOI] [PubMed] [Google Scholar]
- Ross JM, Zarkower D. Polycomb group regulation of Hox gene expression in C. elegans. Dev Cell. 2003;4:891–901. doi: 10.1016/s1534-5807(03)00135-7. [DOI] [PubMed] [Google Scholar]
- Schaner CE, Deshpande G, Schedl PD, Kelly WG. A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev Cell. 2003;5:747–757. doi: 10.1016/s1534-5807(03)00327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvarzstein M, Spence AM. The C. elegans sex-determining GLI protein TRA-1A is regulated by sex-specific proteolysis. Dev Cell. 2006;11:733–740. doi: 10.1016/j.devcel.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Snow JJ, Lee MH, Verheyden J, Kroll-Conner PL, Kimble J. C. elegans FOG-3/Tob can either promote or inhibit germline proliferation, depending on gene dosage and genetic context. Oncogene. 2013;32:2614–2621. doi: 10.1038/onc.2012.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:166–192. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BE, Bernstein DS, Bachorik JL, Petcherski AG, Wickens M, Kimble J. Dose-dependent control of proliferation and sperm specification by FOG-1/CPEB. Development. 2005;132:3471–3481. doi: 10.1242/dev.01921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. Recognition of distorted DNA structures by HMG domains. Curr Opin Struct Biol. 2000;10:102–109. doi: 10.1016/s0959-440x(99)00056-1. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- Wei Q, Shen Y, Chen X, Shifman Y, Ellis RE. Rapid creation of forward-genetics tools for C. briggsae using TALENs: lessons for nonmodel organisms. Mol Biol Evol. 2014;31:468–473. doi: 10.1093/molbev/mst213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff GC, Eke O, Baird SE, Félix MA, Haag ES. Insights into species divergence and the evolution of hermaphroditism from fertile interspecies hybrids of Caenorhabditis nematodes. Genetics. 2010;186:997–1012. doi: 10.1534/genetics.110.120550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi L, Fondufe-Mittendorf Y, Xia L, Flatow J, Widom J, Wang JP. Predicting nucleosome positioning using a duration Hidden Markov Model. BMC Bioinformatics. 2010;11:346. doi: 10.1186/1471-2105-11-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu D, Wu C. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell. 2001;8:531–543. doi: 10.1016/s1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D, Hodgkin J. Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell. 1992;70:237–249. doi: 10.1016/0092-8674(92)90099-x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Azevedo RB, Lints R, Doyle C, Teng Y, Haber D, Emmons SW. Global regulation of Hox gene expression in C. elegans by a SAM domain protein. Dev Cell. 2003;4:903–915. doi: 10.1016/s1534-5807(03)00136-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.