Abstract
Adaptive laboratory evolution (ALE) has emerged as a valuable method by which to investigate microbial adaptation to a desired environment. Here, we performed ALE to 42 °C of ten parallel populations of Escherichia coli K-12 MG1655 grown in glucose minimal media. Tightly controlled experimental conditions allowed selection based on exponential-phase growth rate, yielding strains that uniformly converged toward a similar phenotype along distinct genetic paths. Adapted strains possessed as few as 6 and as many as 55 mutations, and of the 144 genes that mutated in total, 14 arose independently across two or more strains. This mutational recurrence pointed to the key genetic targets underlying the evolved fitness increase. Genome engineering was used to introduce the novel ALE-acquired alleles in random combinations into the ancestral strain, and competition between these engineered strains reaffirmed the impact of the key mutations on the growth rate at 42 °C. Interestingly, most of the identified key gene targets differed significantly from those found in similar temperature adaptation studies, highlighting the sensitivity of genetic evolution to experimental conditions and ancestral genotype. Additionally, transcriptomic analysis of the ancestral and evolved strains revealed a general trend for restoration of the global expression state back toward preheat stressed levels. This restorative effect was previously documented following evolution to metabolic perturbations, and thus may represent a general feature of ALE experiments. The widespread evolved expression shifts were enabled by a comparatively scant number of regulatory mutations, providing a net fitness benefit but causing suboptimal expression levels for certain genes, such as those governing flagellar formation, which then became targets for additional ameliorating mutations. Overall, the results of this study provide insight into the adaptation process and yield lessons important for the future implementation of ALE as a tool for scientific research and engineering.
Keywords: adaptive laboratory evolution, temperature stress, Escherichia coli, MAGE, transcriptomics
Introduction
Adaptive laboratory evolution, or ALE, has developed over the years into a potent tool for biological discovery and engineering (Dragosits and Mattanovich 2013). By exploiting the inherent competition at play between organisms and the natural accumulation of mutations within a microbial population, desired phenotypic traits can be selected for without requiring a priori knowledge on how the traits might arise. These adaptively evolved organisms can then be subjected to whole-genome resequencing, uncovering the genetic changes that enabled their phenotypic alteration. Additional data types, such as transcriptomics or metabolic uptake and secretion rates, serve to characterize the evolved strains and how they diverged from their ancestor, a divergence which must be enabled by their altered genotype. This analysis shines light on the functionality of particular genes (Cheng et al. 2014) and the dynamics of the evolution process (Wiser et al. 2013), increasing the biological knowledge base. While serving as a method to perform basic scientific inquiry such as this, ALE can be an equally useful tool for applied research, pairing with synthetic and systems biology to aid in the engineering of strains (Portnoy et al. 2011).
ALE experiments often examine adaptation following a perturbation, either metabolic (e.g., growth on alternate carbon sources [Ibarra et al. 2002] or following knock-out of metabolic genes [Charusanti et al. 2010]) or stressful (e.g., exposure to osmotic stress [Stoebel et al. 2009] or high ethanol concentrations [Goodarzi et al. 2010]). However, the selective pressure guiding the adaptation can also be influenced in large part by the environment in which the strain is evolved. Evolutionary environments typically involve either batch culturing, wherein populations (often several in parallel) are serially propagated to new flasks with fresh growth medium at regular intervals, or chemostats, in which growth in a bioreactor allows for tight control of nutrient levels and other factors such as pH and oxygenation. In either case, “fitness” ostensibly refers to a growth advantage, but this becomes more complicated by the existence of spatial or temporal inhomogeneities in the culturing environment that can lead to ecological niches. For example, in a chemostat bacteria can persist by adhering to the walls of the bioreactor (Rao and Rao 2004), whereas batch cultures that reach stationary phase before passage can spawn subpopulations optimized for different phases of growth (Rozen et al. 2009). If the target of investigation is the method by which a cell will evolve to a particular perturbation, it can be desirable to confine “fitness” to a single aspect, reducing the potentially confounding variables toward which a population might additionally be evolving. To this effect, batch culture serial propagation in midexponential phase ensures that selection occurs primarily for growth rate.
In this study we sought to examine the ALE process by adaptively evolving the wild-type mesophilic bacterium Escherichia coli K-12 MG1655, arguably the most highly studied and well-characterized microorganism, to constant exponential growth at a stressful elevated temperature in glucose minimal media. Ten parallel populations were evolved at 42 °C using an automated system allowing for passage of batch cultures in midexponential phase multiple times a day, enabling many generations of growth in a relatively short time. Although genes that mutate in parallel across independently evolved populations are often taken to be the likely causes for the fitness increase (Wood et al. 2005), true causal determination would require knocking-in each mutation to the starting strain in all possible combinations and comparing the resultant fitness, which would capture the individual effects of each mutation as well as their epistatic interactions. However, this quickly becomes prohibitively time consuming in strains with greater than three mutations, thus a different tool with which to probe mutational causality would be advantageous. For this reason, we examined multiplex automated genome engineering (MAGE) (Wang et al. 2009) as a technique to supplement ALE experiments. After identifying the mutations occurring in the endpoints of this evolution study, MAGE was used to introduce these ALE-acquired alleles in a random fashion into the starting strain, allowing the combinatorial knock-in method to be somewhat mimicked. By competing the heterogeneous populations of genetically engineered strains against one another and determining the mutants that frequently emerged victorious, causal mutations were identified and compared with those inferred by mutational parallelism across the ALE populations.
Elevated temperature was selected as the perturbation of interest for several reasons. First, to aid in the analysis of mutational causality it would be beneficial to have more than simply one or two frequent gene targets, and adaptation to a global stress provided a diverse set of genetic changes. Additionally, a previous study (Tenaillon et al. 2012) investigated evolution of a large number of replicates in a very similar environment, differing only moderately in ancestral strain and method by which the batch cultures were serially propagated. Comparison with this work allowed examination of the extent to which mutational parallelism persists across studies. Furthermore, examining temperature stress was desirable because two other studies, both involving metabolic perturbations (Fong et al. 2005; Carroll and Marx 2013), have provided strong evidence that a large feature of evolutionary adaptation involves acclimatizing changes in gene expression back toward preperturbed levels. By evaluating the transcriptome of our temperature-evolved strains we sought to determine whether this trend extended to stress perturbations, which would indicate that it may be a general feature of evolution, irrespective of the nature of the perturbation.
Results
Evolution Process and the Endpoint Phenotypes
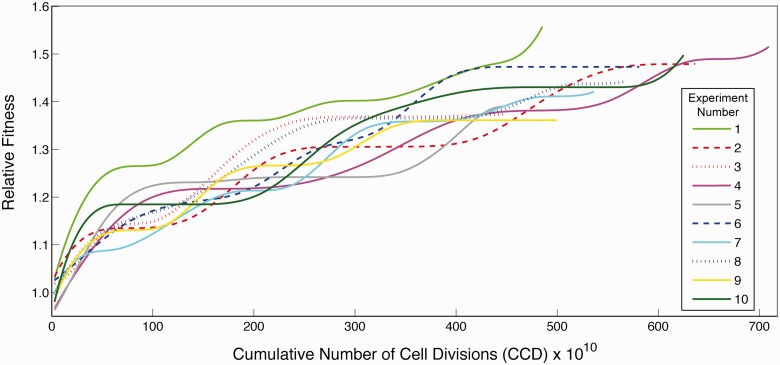
Ten independent populations, started from wild-type E. coli K-12 MG1655, were adaptively evolved in M9 minimal media supplemented with 4 g/l glucose at 42 °C for approximately 45 days. Cultures were serially passed (∼5 times per day) to flasks with fresh media once reaching a target optical density (OD) such that stationary phase was never reached and glucose concentrations were always in great excess (never dropping below 3 g/l). As mutations accrued and gained dominance within the separate populations, their fitness increased markedly relative to the ancestral strain. The populations followed different trajectories along the fitness landscape, arriving at final growth rates on average 1.45 (±0.06, standard deviation)-fold higher than the starting point (fig. 1; raw fitness data shown in supplementary fig. S1, Supplementary Material online). The populations underwent approximately 1,500 generations of growth (supplementary fig. S2A, Supplementary Material online), but because mutations occur predominantly due to DNA polymerase errors in genomic replication during cell division (Camps et al. 2003), the cumulative number of cell divisions (CCD) serves as a more meaningful scale for the time coordinate of an ALE than do generations (Lee et al. 2011). This metric accounts for the population subsampling inherent to serial passage of cultures (supplementary fig. S2B, Supplementary Material online). The CCD reached by the independent populations at the conclusion of the experiment ranged from 4.5 to 7.1 × 1012 with an average of 5.5 × 1012. With the exception of the outlier experiment #1, CCD and final population fitness were significantly correlated (Pearson’s r = 0.93 or 0.58 excluding or including experiment #1, respectively). This correlation is lost when using generations as the time coordinate (Pearson’s r = 0.35 or 0.06 excluding or including experiment #1, respectively).
Fig. 1.
Fitness trajectories of the evolving populations. Plotted is the population increase in fitness relative to the initial average growth rate of the common starting strain. At the approximate CCD of 200 × 1010, the even-numbered experiments’ serial passage volumes were increased 10-fold to examine the impact of CCD on overall fitness. The variability in CCD across experiments is due to this as well as fluctuations encountered in passage cell density over the course of the ALE. Dashed and dotted lines represent populations that became dominated by hypermutators, with identical colors indicating a similar hypermutator genotype.
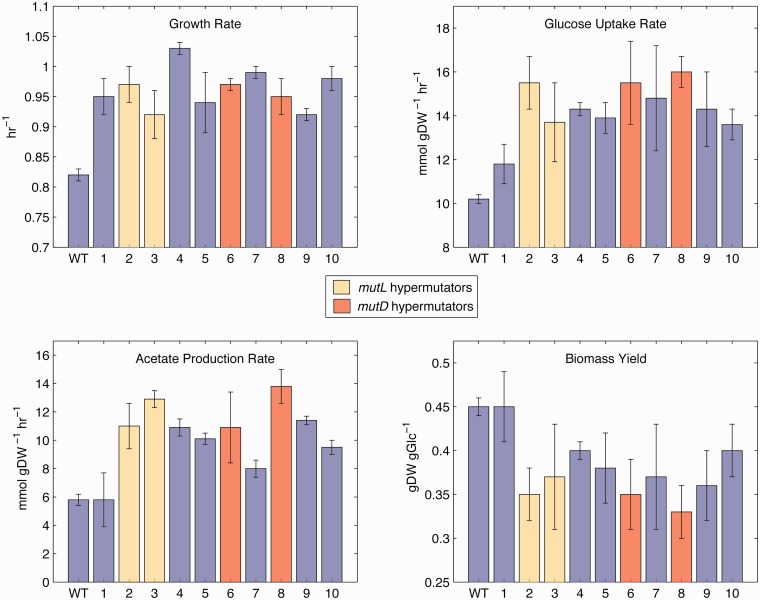
Clones were isolated from each of the evolved population endpoints and subjected to further analysis to determine the physiological differences at 42 °C between the wild-type and the evolved strains (table 1 and fig. 2). Interestingly, the fitness (i.e., growth rate) increase of the isolated clones relative to the ancestor is noticeably lower than that of the populations (paired t-test, P < 10−7). Although the discrepancy could be due to population-level altruistic interactions (Lee et al. 2010), it may result simply from the evolved populations being fully physiologically adapted to constant exponential growth after approximately 1,500 generations under static experimental conditions. This adapted intracellular state is in contrast with the evolved clones, for which growth curves were started up from stationary phase overnight cultures, potentially resulting in suboptimal performance due to insufficient time for reacclimation of their protein expression machinery to the exponential growth, an effect previously documented (Shachrai et al. 2010). Indeed, additional growth rate tests on the fastest and slowest growing endpoints clones (strains 4 and 9, respectively) and the populations from which they were isolated support this hypothesis, and reveal that the clone and population growth rates are in fact in excellent agreement (supplementary fig. S3, Supplementary Material online). Thus, the selected clones were assumed to be representative of the dominant phenotype and genotype within the population endpoints.
Table 1.
Physiological Characterization of Colonies Isolated from Evolved Population Endpoints.
| Strain No. | New Mutations Relative to WT | Growth Rate (h−1) | Relative Fitness Increase | GUR (mmol gDW−1 h−1) | APR (mmol gDW−1 h−1) | Biomass Yield (gDW gGlc−1) |
|---|---|---|---|---|---|---|
| WT | 0 | 0.82 ± 0.01 | 1 | 10.2 ± 0.2 | 5.8 ± 0.4 | 0.45 ± 0.02 |
| 1 | 6 | 0.95 ± 0.03 | 1.17 ± 0.04 | 11.8 ± 0.9 | 5.8 ± 1.9 | 0.45 ± 0.04 |
| 2a | 34 | 0.97 ± 0.03 | 1.19 ± 0.04 | 15.5 ± 1.2 | 11.0 ± 1.6 | 0.35 ± 0.03 |
| 3a | 30 | 0.92 ± 0.04 | 1.12 ± 0.06 | 13.7 ± 1.8 | 12.9 ± 0.6 | 0.37 ± 0.06 |
| 4 | 8 | 1.03 ± 0.01 | 1.26 ± 0.01 | 14.3 ± 0.3 | 10.9 ± 0.6 | 0.40 ± 0.01 |
| 5 | 8 | 0.94 ± 0.05 | 1.15 ± 0.06 | 13.9 ± 0.7 | 10.1 ± 0.4 | 0.38 ± 0.04 |
| 6b | 41 | 0.97 ± 0.01 | 1.19 ± 0.02 | 15.5 ± 1.9 | 10.9 ± 2.5 | 0.35 ± 0.04 |
| 7 | 8 | 0.99 ± 0.01 | 1.21 ± 0.03 | 14.8 ± 2.4 | 8.0 ± 0.6 | 0.37 ± 0.06 |
| 8b | 55 | 0.95 ± 0.03 | 1.17 ± 0.02 | 16.0 ± 0.7 | 13.8 ± 1.2 | 0.33 ± 0.03 |
| 9 | 6 | 0.92 ± 0.01 | 1.12 ± 0.02 | 14.3 ± 1.7 | 11.4 ± 0.3 | 0.36 ± 0.04 |
| 10 | 8 | 0.98 ± 0.02 | 1.19 ± 0.03 | 13.6 ± 0.7 | 9.5 ± 0.5 | 0.40 ± 0.03 |
Note.—For growth rate and relative fitness, the standard deviation based on triplicate experiments is given; for other values, the 95% confidence interval is given.
a and b are hypermutator strains of the same lineage.
Fig. 2.
Physiology of the temperature-evolved strains. Strain numbers are listed below their respective bars. The error bars for growth rate represent standard deviation of three biological replicates, whereas error bars for the other traits represent 95% confidence intervals.
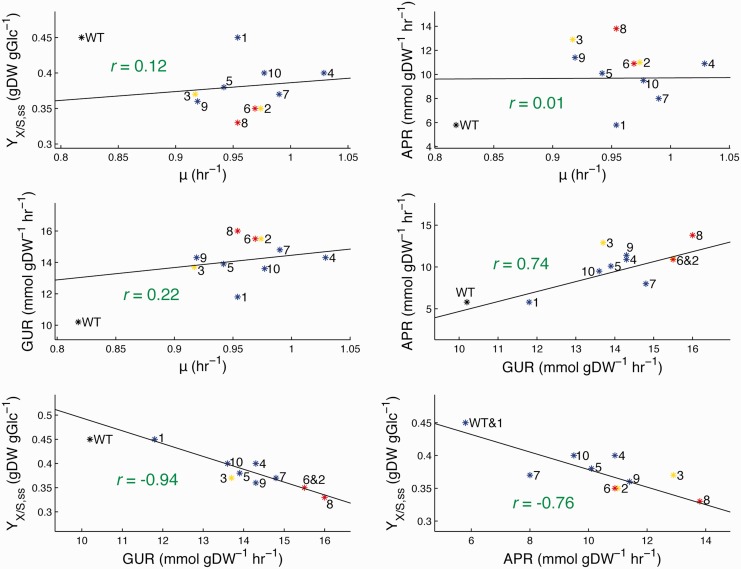
The evolved strains all displayed similar physiological changes. However, genome examination revealed that strains 2 and 3 and strains 6 and 8 likely shared a lineage at some point (see Mutational Analysis), thus strains 3 and 8 were omitted from the following statistics to ensure that only fully independently evolved phenotypes were considered (changing which two strains to omit did not significantly alter any values). On average the independently evolved strains increased their growth rate (μ) by 0.15 h−1 (equivalent to a decrease of 7.7 min in doubling time), increased their glucose uptake rate (GUR) by 4.0 mmol gDW−1 h−1, increased their acetate production rate (APR) by 3.9 mmol gDW−1 h−1, and decreased in biomass yield by 0.07 gDW gGlc−1 (YX/S,ss; calculated at steady state by dividing growth rate by GUR). Correlation plots between the pairwise combinations of these characteristics highlight the physiological divergence of the evolved strains from the wild-type, as well as the relation or lack thereof between specific traits (fig. 3). There is a strong negative correlation (Pearson’s r = −0.94) between biomass yield and GUR among the evolved strains and a notable positive correlation (r = 0.74) between APR and GUR, both of which contribute to the negative correlation between biomass yield and APR (r = −0.76). These results imply that the strains adopted a similar phenotypic change, but to a varying extent, of increased glucose uptake at the cost of increased acetate overflow metabolism (Vemuri et al. 2006), utilizing this greater metabolic flux not to create biomass but rather to generate more energy. Notably, growth rate itself was not correlated with any of the other traits (no Pearson’s r with a magnitude above 0.22). These physiological trends replicate what is observed upon evolution to glucose minimal media at 37 °C (LaCroix RA, unpublished data).
Fig. 3.
Pairwise correlation plots between the physiological traits of the evolved strains. Strain numbers are listed next to their respective points, with the mutL hypermutators in yellow and the mutD hypermutators in red. The least squares linear best-fit line to the evolved strains (excluding the wild-type starting strain from the fit as well as strains 3 and 8, so that only fully independently evolved phenotypes are considered) is overlaid along with its Pearson correlation coefficient (YX/S,ss = biomass yield at steady state, μ = growth rate).
Mutational Analysis
Whole-genome resequencing was performed on isolated clones from each experiment in order to investigate the genetic basis underlying their phenotypic changes. A total of 161 unique de novo mutations relative to the starting strain were found across all ten endpoints (supplementary data set S1, Supplementary Material online), with a number of these being shared among two or more of the strains, or occurring within the same genes. The emergence of several “hypermutators,” a recurring feature of many ALE experiments (Sniegowski et al. 1997), accounted for the majority of these unique mutations. Four of the ten strains were hypermutators, possessing on average 40 mutations, whereas the remaining six “nonmutators” had an average of seven mutations. The resequencing results indicate that two of the four hypermutators likely resulted from unanticipated cross-mixing between the evolving populations, thus only two hypermutators can be said to have occurred independently: One likely due to a single nucleotide polymorphism (SNP) in mutL (strains 2 and 3) (Ban and Yang 1998) and the other due to an SNP in mutD, also known as dnaQ (strains 6 and 8) (Echols et al. 1983). Analysis of the nonmutator genotypes does not indicate that they suffered from similar occurrences of cross-mixing (see Materials and Methods).
The observed mutations in the clones isolated from each population were compared. Fourteen genes or intergenic regions were found to mutate in parallel across two or more of the evolved strains (discounting those mutations shared due to cross-mixing), which fell into three general functional categories: Mutations affecting metabolism, regulation, or the cell envelope (table 2). Intergenic mutations were categorized based on their position relative to the genes (e.g., SNPs downstream of one gene and upstream of another likely change expression of the latter) and transcriptomic data obtained from RNA-seq analysis. The key mutations within each of the three functional categories are now described:
Metabolic mutations: Only three metabolic genes were found to mutate in more than one strain, but two of these occurred in half or more of the strains. Foremost among all mutations, regardless of category, is an 82-bp deletion between pyrE and rph that occurred in every strain except the two mutD hypermutators. This mutation does not appear to be specifically temperature-related, having been observed in several other ALE studies on adaptation to lactate- or glucose-supplemented minimal media at 30 or 37 °C, respectively (Conrad et al. 2009; Blank et al. 2014), and is hypothesized to relieve a defect in pyrimidine biosynthesis present in the starting strain (Jensen 1993). The second most predominant metabolic mutation was in pykF, or pyruvate kinase I, which experienced one intergenic SNP (accompanied by a 2.7-fold downregulation in gene expression) and two different nonsynonymous mutations that may cause PykF inactivation through premature truncation of the enzyme (K286*) or alteration of a putative substrate binding residue (T278S) (UniProt 2014). As with pyrE/rph this is likely not a temperature-specific beneficial mutation, given that PykF inactivation is a recurring feature of E. coli adaptation to glucose minimal media, hypothesized to allow for increased glucose uptake by decreasing the metabolism of phosphoenolpyruvate to pyruvate (Woods et al. 2006; Kishimoto et al. 2010; Blank et al. 2014).
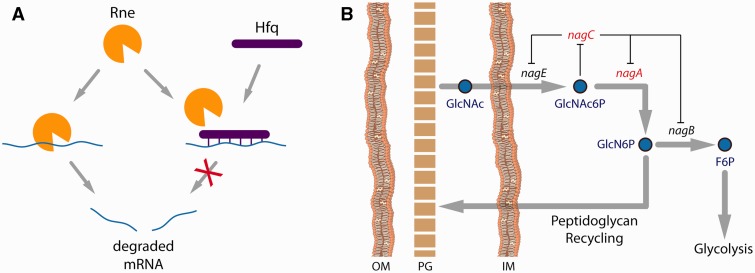
Regulatory mutations: Five of the fourteen recurring mutations were found in regulatory genes, with three of these occurring in half of the strains: rpoC, rne, and ygaH/mprA. RpoC is a subunit of the RNA polymerase complex, which, given its ability to function as a global regulator, is a frequent target of mutations in bacterial adaptation (Murphy and Cashel 2003; Klein-Marcuschamer et al. 2009). Similar ALE-identified rpoC mutations have been found to increase the general metabolic efficiency of E. coli grown in minimal media (Conrad et al. 2010; Cheng et al. 2014) or are inferred to adapt it to higher temperatures (Tenaillon et al. 2012). The ribonuclease rne had four different SNPs, the most diverse set of mutations observed in any one gene in this study. Rne is an essential enzyme involved in rRNA and tRNA processing and, as a key component of the RNA degradosome, is the rate-limiting or sole degrader of many transcripts (Jain et al. 2002). A previously discovered rne mutant was nonviable at 44 °C and significantly defective in rRNA/tRNA processing as well as mRNA degradation, but could be rescued at the elevated temperature by SNPs in the N-terminal catalytic domain (in which all four ALE-acquired SNPs occur) that restored the rRNA/tRNA processing to wild-type levels but did not undo the 2- to 3-fold decrease in mRNA decay rate (Perwez et al. 2008). Relevantly, in prokaryotes the stability of mRNA is directly correlated with the optimal growth temperature of the organism (Gu et al. 2010), suggesting that adaptation strategies to increased temperature might include increasing the stability of mRNA transcripts. Taken together, this implies that the rne mutations found in this study may function to improve the enzyme’s rRNA/tRNA processing capabilities at 42 °C without likewise improving its endonuclease efficacy. It is also of note that an SNP in hfq was observed in two strains, and Hfq binding can prevent mRNA degradation by Rne (fig. 4A) (Folichon et al. 2003). The three different intergenic ygaH/mprA SNPs all occurred between 10 and 14 nt upstream of the mprA start codon, likely influencing translation efficiency by modulation of the ribosomal binding site (Ringquist et al. 1992). MprA is a transcriptional repressor for a number of genes that code for multidrug resistance pumps (Rodionov et al. 2001). Although drug resistance is not a factor in this evolution, increased expression of the pumps leads to altered membrane flux for a variety of compounds (Piddock 2006), thus this regulatory mutation could also perhaps be classified as cell envelope-related.
Cell envelope mutations: Despite no single cell envelope-related gene being mutated in more than three of the ten endpoints, there was nevertheless a clear selective pressure on the cell envelope in general, with 6 of the 14 recurring mutations falling into this category. These range from phospholipid transporters to membrane proteins and channels to genes involved in the levels of cell envelope components. Note that although nagC and nagA could be classified as regulatory and metabolic, respectively, their role in the recycling of cell wall peptidoglycan (fig. 4B) is responsible for their classification as cell envelope-related mutations (Plumbridge 2009). This category of mutations is a feature of ALEs in general (Vijayendran et al. 2008; Conrad et al. 2009) and in this particular study may help the envelope maintain its essential physical properties at the elevated temperatures of the experiment (Sinensky 1974).
Table 2.
Recurring Mutations Identified across the ALE Endpoint Strains.
| Gene | Specific Function | Category | Mutation | Protein Change | Strain Number(s) |
|---|---|---|---|---|---|
| pyrE/rph | Orotate phosphoribosyltransferase/RNase PH | M | 82-bp deletion | Frameshift | 1, 2, 3, 4, 5, 7, 9, 10 |
| rpoC | RNA polymerase subunit | R | SNP | A734V (GCG→GTG) | 1, 2 |
| SNP | Q1367* (CAG→TAG) | 5, 7, 10 | |||
| pykF | Pyruvate kinase I | M | SNP | ydhZ/pykF intergenic (−498/−59) | 3 |
| SNP | T278S (ACC→TCC) | 10 | |||
| SNP | K286* (AAA→TAA) | 4, 5, 7 | |||
| rne | Ribonuclease E | R | SNP | D415N (GAC→AAC) | 6/8 |
| SNP | H243Y (CAT→TAT) | 3 | |||
| SNP | G124S (GGT→AGT) | 9 | |||
| SNP | V19A (GTA→GCA) | 2 | |||
| ygaH/mprA | L-Valine efflux transporter/MprA repressor | R | SNP | Intergenic (+77/−14) | 6/8 |
| SNP | Intergenic (+80/−11) | 10 | |||
| SNP | Intergenic (+81/−10) | 4, 5 | |||
| mlaE | Phospholipid ABC transporter | C | SNP | L107F (TTG→TTT) | 4, 5, 7 |
| yfdI | Predicted inner membrane protein | C | 1-bp insertion | Frameshift (519/1,332 nt) | 8 |
| SNP | Q186* (CAA→TAA) | 6 | |||
| 1-bp deletion | Frameshift (1,274/1,332 nt) | 3 | |||
| nagC | PTS regulator | C | SNP | C264S (TGC→AGC) | 6/8 |
| 1-bp deletion | Frameshift (218/1,221 nt) | 2 | |||
| hns/tdk | H-NS regulator/thymidine kinase | R | SNP | Intergenic (−22/−583) | 6/8 |
| Insertion sequence | Intergenic (−111/−486) | 9 | |||
| hfq | RNA binding protein | R | SNP | D9A (GAT→GCT) | 1, 7 |
| nagA | N-acetylglucosamine-6-phosphate deacetylase | C | SNP | G265D (GGC→GAC) | 2 |
| 21-bp deletion | In-frame (381 − 401/1,149 nt) | 1 | |||
| secD | Membrane protein channel component | C | SNP | R181L (CGC→CTC) | 8 |
| SNP | G499G (GGC→GGT) | 2 | |||
| dinQ/arsR | Toxic membrane peptide/metal-responsive regulator | C | SNP | Intergenic (−209/−486) | 8 |
| 1-bp deletion | Intergenic (−305/−390) | 3 | |||
| ilvL/ilvX | Leader peptide regulating isoleucine and valine biosynthesis operons | M | Double SNP | Intergenic (+47/−39) | 6 |
| SNP | Intergenic (+48/−39) | 8 |
M, metabolic; R, regulatory; C, cell envelope.
Fig. 4.
Repeatedly mutated genes with related functionality. (A) Both rne and hfq, mutually involved in mRNA degradation, mutated in multiple of the temperature-evolved ALE strains. (B) Genes in the nag operon facilitate uptake of GlcNAc from the periplasmic space, which can be channeled away into glycolysis or reincorporated into the peptidoglycan of the cell wall. Genes in red (nagC, nagA) were recurring mutational targets (OM, outer membrane; PG, peptidoglycan; IM, inner membrane; GlcNAc, N-acetylglucosamine; GlcNAc6p, N-acetylglucosamine-6-phosphate; GlcN6P, glucosamine-6-phosphate; F6P, fructose-6-phosphate).
Although a number of ALE studies examining adaptation to increased temperature have been performed to date (Riehle et al. 2001; Kishimoto et al. 2010; Blaby et al. 2012; Tenaillon et al. 2012), the one by Tenaillon et al. serves as an ideal point of mutational comparison given the significant similarity in culture environment between our two studies and the large number of independent lines they evolved, providing a statistically significant basis against which to compare the mutations observed herein. However, despite the similarity in evolution environment, the difference in mutational frequency is extremely pronounced. Of the 14 genes in this study that mutated in two or more strains, only four were found to mutate in any of the 114 evolved lines sequenced by Tenaillon. Both rne (four different SNPs across five strains) and ygaH/mprA (three different SNPs across five strains) had a mutation in only 1 of the 114 lines. Mutations in rpoC and those in or around ilvL were the only noticeably recurring feature: 5 of 10 and 2 of 10 rpoC and ilvL mutants, respectively, in the evolved strains of this study, versus 21 of 114 and 29 of 114 identified in the Tenaillon strains. Even given the relatively small sample size in this study, to have greater than 20% mutational infiltration of 12 genes in one instance and less than 1% in another implies a substantial difference in evolutionary trajectory. Though the recurring mutations in this study (table 2) overlap poorly with those identified by Tenaillon, there is slightly more overlap between the functional units determined to be significant in their study (possessing five or more mutations across the 114 strains) and the mutations observed in this work: 9 of the 26 functional units share one or more mutated genes, despite most mutational overlap occurring in only a single one of our evolved strains (supplementary table S1, Supplementary Material online).
Analysis of Mutational Causality Using MAGE
MAGE (Wang et al. 2009) was utilized to examine causality of the various mutations identified in the evolved strains. To limit the scale of the experiments, only those mutations found in the six nonmutator strains were selected as targets to introduce through recombineering. This yielded 31 distinct mutations between the strains, resulting in 29 unique oligos with which to perform recombineering (two of the mutations were intergenic insertion sequences, infeasible for use in MAGE due to their size, whereas insertion sequences occurring within genes were assumed to function equivalently to knock-outs). These oligos were organized into seven distinct pools—six pools which contained only those oligos corresponding to the mutations found within each of the nonmutator-evolved strains (strains 1, 4, 5, 7, 9, and 10) and one pool which contained all 29 oligos. Nine rounds of recombineering were performed on the starting strain for each of these seven pools, after which serial growth and passage of the cultures occurred to enrich for those strains existing within the highly heterogeneous starting populations that were most fit for growth at 42 °C. Multiple colonies were isolated from each of these enriched populations and subjected to whole-genome resequencing (supplementary data set S2, Supplementary Material online).
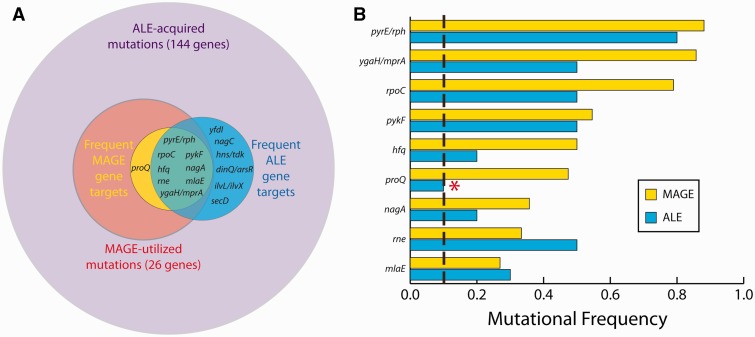
The most frequently observed MAGE mutations (occurring in at least three of the enriched strains, with a frequency >25%) are given in table 3. Frequency for a gene is defined as the number of resequenced strains which possessed a mutation divided by the number of resequenced strains which potentially could have possessed it given the pool of oligos used in recombineering. This recapitulates what was observed in the evolved ALE strains—there is excellent agreement between the most frequent MAGE and ALE mutations, keeping in mind that the use of only nonmutator strain mutations in MAGE explains the absence of yfdI, nagC, secD, dinQ/arsR, ilvL/ilvX, and hns/tdk (fig. 5). This leaves proQ, a regulator of the membrane transporter ProP, as the sole disagreement between tables 2 and 3, which mutated in only ALE strain 7 (a missense SNP, Q216P) and yet was found in 9 of 19 relevant MAGE strains. These results reinforce the conclusions drawn from examination of mutational parallelism across the independently evolved ALE strains—that the identified genes are likely key targets for adaptation to growth at 42 °C in glucose minimal media.
Table 3.
The Most Significantly Recurring Genes Following Enrichment of MAGE Strains.
| Gene | MAGE Frequency | ALE Frequency | New Off-Target Mutationsa |
|---|---|---|---|
| pyrE/rph | 0.88 | 0.80 | 26 (including nine frameshifts) |
| ygaH/mprA | 0.86 | 0.50 | 1 (intergenic 1-bp deletion) |
| rpoC | 0.79 | 0.50 | 2 (I1357I, N762S) |
| pykF | 0.55 | 0.50 | 0 |
| hfq | 0.50 | 0.20 | 0 |
| proQ | 0.47 | 0.10 | 1 (G212G) |
| nagA | 0.36 | 0.20 | 2 (G127G, H129N) |
| rne | 0.33 | 0.50 | 0 |
| mlaE | 0.27 | 0.30 | 1 (L99L) |
aProtein alterations given in parentheses.
Fig. 5.
Mutational agreement between the MAGE strain enrichment and ALE experiment results. (A) A subset of the novel alleles acquired through evolution were randomly introduced into the ancestral strain with MAGE, and these MAGE strains were competed against one another in the enrichment process. “Frequent ALE gene targets” refer to genes that mutated in at least two of the ten ALE endpoint strains, whereas “Frequent MAGE gene targets” refer to those that mutated in at least 25% of the relevant postenrichment MAGE strains. Circle areas are proportional to the number of genes contained within each category. (B) Frequency comparison between MAGE and ALE strains for the nine most frequent MAGE gene targets. The only disagreement occurs for proQ (marked by *), which was not a frequent ALE gene target. The vertical dashed line corresponds with 10%, the frequency for a gene mutating in only a single one of the ten ALE strains.
On average, enriched strains possessed four mutated genes targeted by recombineering and three mutations in secondary locations throughout the genome. However, several of the targeted genes experienced mutations that differed from what would be expected given the design of the recombineering oligos. With the exception of one, all of these novel, “off-target” mutations occurred within the 70-bp region introduced into the genome by allelic replacement and thus likely resulted from incorporation of missynthesized oligos containing erroneous bases, as has been observed with MAGE previously (Wang et al. 2009). Nevertheless, examining these unintentional mutations that were able to fix in the postenrichment populations provides insight into the nature of the causal genetic changes.
In the case of pyrE/rph, 26 new SNPs and indels were found, and in no instance did a strain possess some combination of rph or pyrE/rph mutations that did not include the introduction of a shifted rph stop codon. Rph is naturally defective in the E. coli strain used in this study (Jensen 1993), so alterations to its coding sequence should be phenotypically neutral. These mutations support the mechanism put forth previously (Conrad et al. 2009), whereby a frameshift that moves the rph stop codon closer to the pyrE attenuator loop allows for improved regulation of pyrE expression. If these rph frameshifts and nonsense SNPs all yield roughly equivalent outcomes, then this begs the question of why only the same 82-bp deletion was observed in the ALE strains. This disparity in variation can be explained by the different acquisition method of ALE versus MAGE mutations. The 82-bp pyrE/rph deletion is flanked by two 10-bp repeats, thus its prevalence in the adaptively evolved strains may be due to the relative frequency of DNA polymerase slippage during DNA replication (Michel 2000; Conrad et al. 2009), and once this fixes within the populations there is no longer a selective force for continued genetic alteration in the region. This mutational bias is decreased under MAGE conditions—even with only a small fraction of oligos with synthesis errors, a variety of mutations can be acquired across each round of recombineering before the populations are subjected to growth-based selection in the form of enrichment, negating the ease-of-acquisition benefit the 82-bp deletion has in gaining dominance.
Unlike with pyrE/rph, where most frameshifts will be beneficial and gene inactivation is not a concern, one would expect that in the other MAGE gene targets the introduction of missynthesized oligos would in most instances result in a fitness decrease by altering the amino acid sequence of a functioning protein. This fitness decrease would render the mutants unable to survive the enrichment process, yielding few off-target mutations within the relevant genes of the enriched MAGE strains. The data support this conclusion; other than the 26 new pyrE/rph mutations, only eight off-target mutations were found across all other MAGE-targeted genes in the resequenced strains, with more than half of these resulting in no protein sequence alteration (one intergenic 1-bp deletion, four synonymous SNPs, and three missense SNPs). There is no reason that the oligos used for pyrE/rph should be more erroneous than those for any other gene, thus the difference in number of off-target mutations that survived enrichment highlights the relative specificity of the ALE-identified mutations for effecting a fitness increase. However, pykF experienced no off-targets despite its two separate oligo-introduced mutations both potentially leading to gene inactivation, either by altering a potential binding residue or truncating the protein. It may be that these mutations only decreased the enzymatic activity of PykF rather than completely eliminating it, and reduced PykF functionality happens to be more beneficial than the complete inactivation that would likely result from the introduction of random, missynthesized oligos. It should also be noted that one off-target mutation, an SNP in rpoC (N762S), fell outside of an oligo-targeted region and thus may have arisen independently.
The resequenced MAGE strains were subjected to growth rate tests to ensure that they were in fact adapted to growth at 42 °C. The tests revealed two strains that, despite possessing mutations and being picked from enriched populations, did not grow faster than the starting wild-type strain. These two strains were precluded from the MAGE mutational frequency analysis, but nevertheless highlighted an interesting feature: All adapted strains possessed mutations in pyrE/rph and/or rpoC, and the two unadapted strains were the only ones to have neither.
Transcriptomic Profiling through RNA-Seq
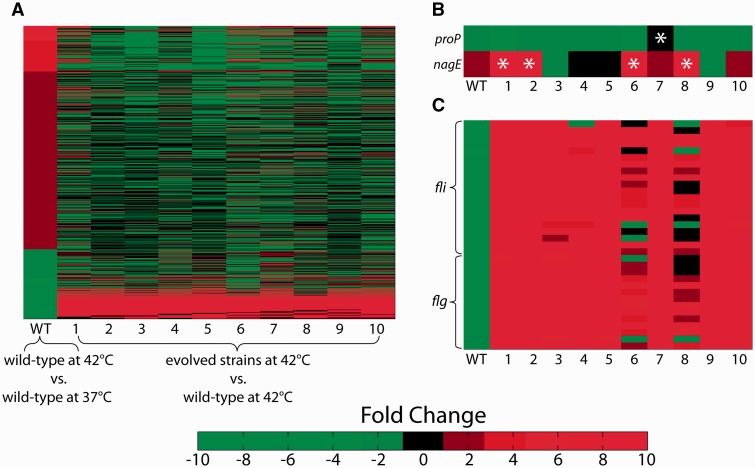
RNA-seq was performed to examine the global shifts in gene expression resulting from the altered genotypes of the evolved strains as compared with the wild-type ancestral strain at 42 °C. To complement this analysis, the expression shift of the wild-type strain when grown at 42 versus 37 °C was also determined (supplementary data set S3, Supplementary Material online). Figure 6A shows a heat map of fold changes for the 1,208 genes deemed to be significantly differentially expressed (q value < 0.05; Storey and Tibshirani 2003) in the wild-type strain when grown at the higher temperature. The differentially expressed wild-type genes and their pattern of up- and downregulation are in good agreement with a previous study examining differential expression in E. coli after growth at 43 °C (Gunasekera et al. 2008), including such features as upregulation of heat shock proteins (e.g., ClpB, DnaK, GrpE, and GroL, among others) and sulfur metabolism genes (cys genes), and downregulation of genes involved in flagellar synthesis (fli and flg genes) and putrescine catabolism (puu genes).
Fig. 6.
Heat maps of differential expression between the ancestral and evolved strains. (A) The 1,208 genes which are significantly differentially expressed (q value < 0.05) in the wild-type after growth at 42 °C, rank-ordered. (B) Mutations acquired by the evolved strains (relevant strains marked with *) could either serve to resist (proP; strain 7) or aid in (nagE; strains 1, 2, 6, and 8) the reinforcement of the wild-type expression shift. (C) Widespread upregulation of flagellar genes after evolution, partially resisted by two strains (6 and 8) possessing similar genotypes.
In most of these cases, and indeed as a general trend across many of the genes (fig. 6A), the mutations of the evolved strains served to reverse the heat-induced transcriptional shift, restoring the expression state back toward the levels of the wild-type at 37 °C. On average, each evolved strain had restorative shifts for 73% of the 1,208 genes, and principal component analysis provides an additional means of visualizing these evolved shifts in gene expression (supplementary fig. S4, Supplementary Material online). Such restoration has been documented in two other instances; in one study examining evolution of E. coli onto lactate or glycerol as the sole carbon source as opposed to glucose (Fong et al. 2005), and in another examining evolution of Methylobacterium extorquens following replacement of a native central metabolism reaction with a functionally analogous, heterologous pathway (Carroll and Marx 2013). In both cases there was widespread restoration of expression back to the wild-type levels, highlighted by the less common “reinforcement” of a change (i.e., down- or upregulated genes at the start of evolution becoming increasingly down- or upregulated following evolution, respectively). This reinforcement, when occurring across multiple of the independently evolved strains, was taken as evidence for the reinforcing shift being an important factor in their increased fitness. In this study, highly parallel (occurring in at least eight of the ten evolved strains) transcriptional reinforcement occurs in 101 genes, compared with 703 genes that experience highly parallel restorative shifts. It should be noted that expression levels can be influenced by growth rate-dependent effects (Gyaneshwar et al. 2005), but this cannot explain the observed transcriptional restoration given that the growth rate of the wild-type strain at 42 °C is higher than at 37 °C (0.82 vs. 0.7 h−1), both of which are lower than the evolved strains.
The reinforcing shifts in gene expression across the temperature-evolved strains were examined in more detail. Evidence was found both supporting and refuting the assumption that these reinforcements point to key mechanisms of adaptation. Supporting data include the expression shifts observed in nagE, which increased 1.6-fold in the wild-type at 42 °C followed by a further increase between 15- and 71-fold in evolved strains 1, 2, 6, and 8, in stark contrast with the remaining strains which changed in expression by no more than 1.6-fold (fig. 6B). The strains exhibiting the strongly reinforcing increase in nagE expression are those possessing mutations in nagC and/or nagA, both determined to be significant gene targets in the adaptation to increased temperature (table 2). These mutations are likely responsible for the observed nagE shifts, given the genes’ mutual involvement in peptidoglycan recycling (Plumbridge 2009). This finding is similar to the strongly reinforced upregulation of pntAB observed by Carroll and Marx in the M. extorquens-evolved strains, which additional analysis showed to be a major contributor to their fitness increase, and the discovery of mutations within this operon.
In contrast with nagE, expression shifts in proP serve as an example of a reinforcing shift seemingly acting against the best interest of the cells. Upon initial growth at 42 °C the wild-type strain decreases in proP expression by 1.5-fold, a decrease which is reinforced uniformly across the ten evolved strains. However, strain 7 exhibits the smallest expression decrease (an insignificant −1.2 fold, compared with an average of −2.7 fold across the other nine strains) and is also the strain that acquired an SNP in proQ. Though not appearing in multiple of the ALE endpoint strains, the proQ mutation was found to be significant in the MAGE enrichment analysis and is likely responsible for the greater comparative proP expression, given that ProQ’s only known function is as a regulator of ProP levels (Chaulk et al. 2011). This implies that the reinforced change in expression of proP is actually detrimental to growth at increased temperatures, enough to allow for fixation of this mutation that combats the change and provides a fitness increase of suitable magnitude to repeatedly survive MAGE enrichment. However, although no epistatic interactions could be inferred from the MAGE results (the proQ mutation did not appear solely in the presence of any other single mutation), the possibility cannot be discounted that different expression shifts in proP, or indeed any gene, could have different effects in different strains.
In the same way that the reinforced expression shifts may not all necessarily aid in the fitness of the cells, so too is this the case for the restorative shifts. The most noticeably counterintuitive restorative shift is the massive upregulation of flagellar genes in the evolved strains following their initial downregulation in the wild-type upon growth at 42 °C (fig. 6C). In the well-mixed environment of the ALE experiment there is no evident need for motility, so increased expression of these genes should incur a significant energetic cost (Soutourina and Bertin 2003) while providing no apparent benefit. Indeed, previous ALE studies, regardless of culturing temperature or carbon source in the growth medium, have yielded strains with mutations that led to the downregulation of flagellar genes (Cooper et al. 2003; Fong et al. 2005; LaCroix RA, unpublished data). Interestingly, strains 6 and 8 (the two mutD hypermutators) appear to have somewhat mitigated this flagellar upregulation in the same way that strain 7 did for proP downregulation. The SNP 22 bp upstream of the hns start codon that strains 6 and 8 share is a potential candidate for the genetic change behind their outlying behavior, given hns’s part in the regulation of flagellar genes (Landini and Zehnder 2002). Taken together, the data suggest that the evolved expression shifts, whether restorative or reinforcing, can sometimes be actively detrimental instead of beneficial.
Other than the influence of the nagA/nagC and proQ mutations on transcription of nagE and proP, respectively, the recurring ALE mutations (table 2) have only subtle, if at all discernible, effects on expression levels relative to the other endpoint strains lacking these mutations. This indistinct mutational influence on transcription includes both the genes that mutated, as well as separate genes that might logically experience an expression shift as a result (e.g., a proQ mutation causing proP shifts). Even the intergenic mutations, which are more likely to cause expression shifts than are protein sequence alterations (Wang et al. 2011), fail to noticeably distinguish themselves from the strains without the mutations, and can be inconsistent in their influence. For example, although the strain possessing an SNP 59 bp upstream of the pykF start codon has the greatest decrease in pykF expression (−2.7 fold; the second largest decrease being −2.2 fold), it fails to stand distinctly apart from the remaining strains (t-test, P > 0.16). Similarly, although eight of the evolved strains share the identical 82-bp pyrE/rph deletion that should influence pyrE expression (Conrad et al. 2009), their fold changes for the gene ranged between −3 and +5. Isolating the transcriptional effects of any individual mutation is clearly complicated by the presence of other genetic changes within the evolved strains. Elucidation of the expression shifts enabled by specific mutations would likely require the creation and transcriptomic characterization of single knock-in mutants.
Discussion
In this study, ALE of ten replicate cultures was performed under carefully controlled conditions to yield E. coli strains selected solely for faster exponential-phase growth on glucose minimal media at 42 °C. These strains were physiologically characterized, transcriptomically profiled, and subjected to whole-genome resequencing to determine the genetic basis of their adaptation. Selected mutations were knocked-in to the ancestral strain in random combinations through MAGE to gain further insight into the causal genetic changes behind the acquired fitness increase. It was found that 1) frequency-based assignment of causality to mutations is largely consistent across the naturally evolved and genetically manipulated strains, 2) the path of genetic adaptation is greatly influenced by both the genotype of the starting strain and the conditions under which the evolution is performed, and 3) a variety of mutations lead to the same general trend of phenotypic and transcriptional convergence among the evolved strains, highlighted by the occasional counterintuitive shift in gene expression.
Although the number and variety of mutations capable of conferring a growth advantage in any particular environment may be nearly limitless, those which strongly contribute to an improved phenotype have a much greater probability of fixing within the evolving populations than do detrimental, neutral, or slightly beneficial mutations. Thus, genes mutated in parallel across independently evolved cultures stand out as likely causes for the observed fitness increase, rather than simply being “hitchhiker” mutations (Wood et al. 2005). This frequency-based analysis pointed to 14 different genes, fewer than 10% of the 144 genes mutated across all strains, as the foremost genetic targets for adaptation to growth on glucose at 42 °C. A subset of these 144 altered genes, encompassing the mutations acquired by the six strains with few (≤8) genetic changes relative to the common ancestor, were genetically engineered into the starting strain in random combinations with the techniques of MAGE. Repeated culturing of the highly heterogeneous MAGE populations enriched for the most fit strains existing within them, and genome resequencing revealed the genetic changes possessed by these strains. It was found that frequency-based analysis of the mutated genes within the MAGE strains perfectly recapitulated the likewise-determined key genes of the ALE strains (at least, for those genes which fell within the subset used in the genetic engineering) and pointed to only one additional key gene target that did not happen to mutate in parallel in the ALE. Additionally, the unintentional inclusion of oligos with synthesis errors in the MAGE allelic replacement (Wang et al. 2009) highlighted the effects the evolved mutations had on the genes; many unintentional off-target mutations accumulated if fitness was positively or neutrally affected by inactivation of the gene in question, but off-targets were generally lacking or synonymous if the mutated gene likely retained some measure of functionality. While not apparent in this study, MAGE analysis would also serve to highlight the existence of positive epistasis between acquired mutations if they predominantly showed up together in the postenrichment strains, implying that their combined fitness benefit was greater than either individually. MAGE thus presents itself as a useful tool for reaffirmation of mutational causality following an ALE experiment.
Though the excellent agreement in mutational frequency between ALE and MAGE strains implies that these recurring mutations would similarly appear in other E. coli temperature evolution experiments, comparison with the work of Tenaillon et al. revealed that for the most part this is untrue. Of the 14 recurring ALE mutations identified herein, ten experienced zero mutations across all 114 lines evolved by Tenaillon, whereas two mutated in only 1 of the 114 lines. To understand where this discrepancy arises, it is important to classify the ways in which the two studies differ. First, in this study K-12 MG1655 is used as the ancestral strain, differing from the B REL1206 strain used by Tenaillon (a descendant of B REL606 evolved for 2,000 generations on glucose minimal media at 37 °C; Barrick et al. 2009). This strain difference provides a clear explanation for several of the mutational discrepancies: The 82-bp pyrE/rph deletion widespread among the strains generated here is specific to MG1655, thought to relieve an inherent defect in pyrimidine biosynthesis (Conrad et al. 2009), and Tenaillon does not observe pykF mutations because B REL1206 already contains an inactivating mutation in pykF, precluding it from further selective forces. The use of different starting strains also provides a possible explanation for the lack of rne mutants found by Tenaillon. Rph is naturally defective in MG1655 and is known to interact in vivo with Rne (Duran-Figueroa et al. 2006), so it may be that certain detrimental effects resulting from elevated temperature do not manifest themselves when Rph and Rne are able to form functional assemblies, but the absence of Rph places pressure upon Rne to develop compensatory mutations for improvement of its functionality. On top of these mutational discrepancies that can be intuited, there are doubtless those for which explanations are not obvious; for example, several beneficial rpoB mutations identified in the Tenaillon study were actually detrimental to fitness, or potentially even lethal, when introduced into MG1655 (Rodriguez-Verdugo et al. 2013).
In addition to the different starting strains, differences in experimental methodology are likely to have a significant effect. Although evolved at essentially identical temperatures (42 vs. 42.2 °C) and on the same carbon source, the environments under which evolution occurred were nevertheless quite different. Whereas in this study constant exponential phase growth was maintained and all nutrients were always in great excess, thus allowing for selection based solely on growth rate, the methodology of Tenaillon mimics that of the long-term evolution experiment (LTEE) (Lenski et al. 1991)—the media contained only 25 mg/l of glucose (160 times less than the 4 g/l used here) and cultures were passed once per day, as a result spending the majority of their time in stationary phase. Significant differences in bacterial behavior arise during stationary phase (Navarro Llorens et al. 2010), and the process of repeated feast and famine causes the selective force to be nonconstant. This can give rise to unusual features, such as coexisting subpopulations that thrive in different phases of the daily cycle (Rozen et al. 2009). Furthermore, the Tenaillon strains evolved under poorly oxygenated conditions, given that 10-ml cultures were kept in 15-ml tubes shaken at only 100 rpm. The large difference in hypermutator prevalence between our two studies is also indicative of the methodological influence on genomic changes; at least 20% of our strains became hypermutators (discounting those likely caused by population cross-mixing), whereas Tenaillon evolved 115 lines of which only one became a hypermutator, and was precluded from further analysis.
As for the carbon source availability, in this study mutations facilitating faster glucose uptake are selected for only to the extent that they enable faster growth, whereas in LTEE conditions faster uptake of the scarce glucose in and of itself provides a significant advantage. This may be why under LTEE conditions pykF is known to acquire an internal insertion sequence (Barrick et al. 2009), almost certainly leading to full gene inactivation, but the pykF mutations identified herein potentially preserve some of the enzyme’s functionality given the failure of off-target pykF mutations to accrue in the MAGE strains. PykF is one of two enzymes catalyzing conversion of phosphoenolpyruvate to pyruvate in glycolysis, and it is hypothesized that PykF inactivation enables faster glucose uptake by increasing intracellular concentrations of phosphoenolpyruvate, which can be used by the phosphotransferase system to drive glucose uptake (Woods et al. 2006). In the glucose-rich conditions of this study, PykF impairment as opposed to inactivation could serve as a more beneficial alternative, balancing the pros of increased glucose uptake against the cons of a decrease in glycolytic flux capabilities. Another relevant comparison exists between this study and one with identical experimental conditions and starting strain, but with the culturing temperature set to 37 °C instead of 42 °C (LaCroix RA, unpublished data). The most notable genetic feature of the lower-temperature ALE was the various rpoB SNPs acquired by every evolved strain, but rpoB did not mutate in any of our higher-temperature ALE strains. Recurring mutational agreements exist only for pyrE/rph, hns/tdk, and pykF (which similarly experienced no alterations causing clear-cut inactivation). Together these mutational comparisons with other studies highlight the significant influence of experimental conditions and starting genotype on the resulting genetic adaptation. Thus, mutational reproducibility largely cannot be expected if even slightly different strains are used, if experimental methodology varies, or if evolution to multiple factors is simultaneous instead of sequential (e.g., evolving on glucose at 42 °C vs. evolving first on glucose at 37 °C, and subsequently at 42 °C).
Despite notable recurrence of multiple mutations across the evolved strains of this study, other than pyrE/rph no gene was mutated in more than half of the endpoints. In spite of these mostly distinct genotypes, convergence occurred toward the same physiological and transcriptional state. Growth, glucose uptake, and acetate production rates increased uniformly across all strains, whereas biomass yield decreased. Many genes were significantly differentially expressed in the wild-type upon an upshift from 37 to 42 °C, but following evolution at the elevated temperature more than 70% of these genes reverted their expression state back toward that of the “unperturbed” wild-type at 37 °C. Mimicking the uniform nature of the physiological changes, these expression shifts were generally highly parallel—over 700 genes shifted back toward the unperturbed state in eight or more of the ten evolved strains. Though a minority, highly parallel shifts that moved expression further away from the unperturbed state occurred for over 100 genes. These trends in frequency and direction of shifts are in good agreement with two previous studies, demonstrating a consistent method of adaptation whether for growth on new carbon sources (Fong et al. 2005), acclimation after perturbation of a metabolic pathway (Carroll and Marx 2013), or adjustment to a global stress such as elevated temperature. Any changes in physiology and gene expression observed between the wild-type and evolved strains must arise from the genetic differences that developed, thus the lack of uniformly occurring mutations means that the different genotypes acquired by each strain result in roughly the same overall state.
That large-scale alterations to the expression state can result from relatively few genetic changes points to the influence of regulatory mutations, a category into which many of the most frequently mutated genes fell, most notably rpoC, rne, and ygaH/mprA. At least one of these genes was mutated in every evolved strain, a prevalence matched or exceeded only by mutations in pyrE/rph and pykF. Interestingly, these latter two genes were the only metabolically related mutations to occur in more than two of the ten evolved strains, and neither is thought to result specifically from the increased temperature of the evolution, as discussed above; temperature-specific adaptation for the most part seems to involve changes in gene regulation or the cell envelope. The regulatory mutations clearly have a net positive benefit on the cell given their frequent occurrence, but this does not mean that every one of the widespread expression levels altered as a result is likewise beneficial. It could be that some expression shifts caused by the regulatory mutations are actively detrimental to the cell, just less so than the cumulative benefit of the global change. These deleterious changes would then be targets for further adaptation that could alleviate their effects. Here, as in previous works (Hindre et al. 2012), we find several pieces of evidence supporting this hypothesis, most strikingly in the massive and counterintuitive upregulation of flagellar genes following evolution, a trend that is resisted by two strains that succeeded in acquiring a mutation to mitigate the likely detrimental upregulation. Although overall fitness is always selected for, it seems that over the course of an evolution certain subsystems of the cell may experience transient fitness decreases before additional mutations can develop to undo them.
Overall, the results of this study yield lessons important for the future implementation of ALE as a tool for both biological discovery and engineering. First, we have demonstrated the utility of MAGE as a method by which to probe mutational causality, diminishing the need for the often-laborious creation of single and combinatorial knock-ins. Such a method would prove particularly useful in ALE studies lacking in replicates with which to examine mutational parallelism, or when hypermutability has produced an overabundance of genetic changes that obscure the strongly causal mutations hidden among them. Second, by comparing the mutations identified herein with those found in related studies, we have highlighted the great extent to which mutational development is predicated upon the ancestral genotype and influenced by all aspects of the evolutionary environment. Rarely will two studies examine evolution of identical strains under identical experimental conditions, thus authors should be wary of basing their expectations on comparisons with similar works. For example, if a more heat-tolerant production strain for a particular biochemical were desired, knocking-in the mutations identified in this study would not necessarily provide a benefit in the dissimilar genetic background and culturing environment. Finally, the previously documented restoration of expression state toward wild-type levels following metabolic perturbations and subsequent evolution has been shown to extend to global stresses, namely elevated temperature, as the perturbation. Transcriptomic analysis of our evolved strains demonstrated this, as well as revealing the apparent occurrence of likely transient, localized, detrimental changes in gene expression. In much the same way that overall entropy maximization can be achieved despite entropic decreases in certain components, as with a protein folding in solution (Harano and Kinoshita 2005), genetic subsystems can experience fitness decreases in the cellular pursuit of overall fitness maximization. Although given enough evolutionary time such suboptimal components would be ameliorated by compensatory mutations, this nevertheless provides an avenue for rational design to improve on the evolution process. Genome-scale metabolic models are capable of making a priori predictions on genes necessary for optimal cellular growth in a particular environment (Bordbar et al. 2014), so those genes predicted to be useless could be knocked out ahead of time, saving the cell from potentially energetically wasteful expression. Starting with a “preadapted” strain such as this would expedite the evolution process and eliminate the need for mutations solely to counteract superfluous expression.
Materials and Methods
Adaptive Evolution
An automated system was used to propagate the evolving populations over the course of the ALE and monitor their growth rates. Flasks filled with 25 ml of 4 g/l M9 minimal media were kept at 42 °C through placement in a heat block and aerated by magnetic tumble stirrers at 1,800 rpm. At the start of the experiment, a flask of the wild-type strain E. coli K-12 MG1655 (ATCC47076) was grown up to stationary phase in the same conditions and used to inoculate ten independent flasks with 900 µl of culture. As the bacteria grew, the automated system took OD measurements at 600 nm for each flask at four timepoints, targeted to evenly span an OD range of 0.05–0.3 based on the most recently calculated growth rate and the starting OD of the flask. Growth rates were determined by taking the slope of a least-squares linear regression line fit to the logarithm of the OD measurements. Once reaching the target OD of 0.3, 10 µl of culture was passed into a new flask, and in the even numbered experiments this passage volume was changed to 100 µl after 20 days of evolution. At the OD of 0.3, glucose concentration only dropped from 4 to approximately 3.5 g/l (determined by high-performance liquid chromatography [HPLC] measurement of the cultures), so exponential growth in excess glucose conditions was constantly maintained.
Growth rates for each flask were discarded as untrustworthy if fewer than four OD points were sampled, if the points spanned a range of fewer than 0.2 or more than 0.4 OD units, or if the R2 correlation was below 0.99. To reduce noise, the data were smoothed by averaging each point with its five adjacent neighbors on either side after applying weights following a normal distribution (σ = 2) centered on the point in question. The evolution trajectory curves were obtained by fitting a monotonically increasing piecewise cubic spline to the smoothed data. Fitting to the unsmoothed data resulted in negligible changes to the spline. The CCD was calculated as outlined previously (Lee et al. 2011).
Glucose M9 minimal media consisted of 4 g/l glucose, 0.1 mM CaCl2, 2.0 mM MgSO4, 1× trace element solution, and 1× M9 salts. The 4,000× trace element stock solution consisted of 27 g/l FeCl3*6H2O, 2 g/l ZnCl2*4H2O, 2 g/l CoCl2*6H2O, 2 g/l NaMoO4*2H2O, 1 g/l CaCl2*H2O, 1.3 g/l CuCl2*6H2O, 0.5 g/l H3BO3, and Concentrated HCl dissolved in ddH2O and sterile filtered. The 10× M9 salts stock solution consisted of 68 g/l Na2HPO4 anhydrous, 30 g/l KH2PO4, 5 g/l NaCl, and 10 g/l NH4Cl dissolved in ddH2O and autoclaved.
Cross-Mixing Analysis
The sequencing results of the evolved endpoint strains highlighted the fact that there was unintentional cross-mixing between the populations over the course of the ALE. Strains 6 (41 total mutations) and 8 (55 total mutations) shared 24 identical mutations which did not occur in any of the other isolated strains, including the SNP which caused a truncated form of MutD (DnaQ) likely responsible for the observed hypermutator phenotype (Echols et al. 1983). Although less immediately apparent, it seems likely that strains 2 (34 total mutations) and 3 (30 total mutations) suffered from the same cross-mixing—they share three identical mutations (a 1,222-bp deletion, a 1-bp deletion, and an SNP) which do not occur in any of the other strains, including the mutL mutation which likely causes their hypermutator phenotype (Ban and Yang 1998). For these reasons, we did not consider the mutations shared only between strains 6 and 8 or 2 and 3 to have arisen independently when performing mutational recurrence analysis.
Given these occurrences of cross-mixing it was important to establish that the nonmutator strains did not likewise share a partially evolved ancestor, which would complicate the determination of recurring mutations. If it were the case that all identical mutations resulted from cross-mixing and did not arise independently, then we would expect to see a number of identical mutations in two or more strains, but these strains would not share identical mutations with any other strains. This was decidedly not the case (supplementary fig. S5, Supplementary Material online). For example, consider the identical rpoC mutation shared between strains 5, 7, and 10. If we posit that this is the result of the same rpoC mutant invading and fixing within all three populations, then it must be that the remaining shared mutations all occurred independently (hfq in strains 1 and 7, ygaH/mprA in strains 4 and 5, and pykF and mlaE in strains 4, 5, and 7) given that the other strains do not share the rpoC mutation. Thus in the “worst case” scenario, only one of these four sets of shared mutations can be explained away by cross-mixing.
DNA Sequencing
After colonies were isolated and selected on Lysogeny Broth (LB) agar plates, genomic DNA was extracted using Promega’s Wizard DNA Purification Kit. The quality of DNA was assessed with ultraviolet absorbance ratios using a Nano drop. DNA was quantified using Qubit dsDNA High Sensitivity assay. Paired-end resequencing libraries were generated using a Nextera XT kit from Illumina (San Diego, CA) with 1 ng of input DNA total. Sequences were obtained using an Illumina Miseq with a PE500v2 kit. The breseq pipeline (Barrick et al. 2009) version 0.22 with bowtie2 (Langmead and Salzberg 2012) was used to map sequencing reads and identify mutations relative to the E. Coli K12 MG1655 genome (NCBI accession number NC_000913.2). All samples had an average mapped coverage of at least 25×.
RNA Sequencing
RNA-sequencing data were generated under conditions of exponential-phase, aerobic growth in glucose M9 minimal media. Cells were washed with Qiagen RNA-protect Bacteria Reagent and pelleted for storage at −80 °C prior to RNA extraction. Cell pellets were thawed and incubated with Readylyse Lysozyme, SuperaseIn, Protease K, and 20% sodium dodecyl sulfate for 20 min at 37 °C. Total RNA was isolated and purified using the Qiagen RNeasy Mini Kit columns, following vendor procedures. An on-column DNase-treatment was performed for 30 min at room temperature. RNA was quantified using a Nano drop and quality assessed by running an RNA-nano chip on a bioanalyzer. Paired-end, strand-specific RNA-seq was performed following a modified dUTP method (Latif et al. 2013). The rRNA was isolated using Epicentre’s Ribo-Zero rRNA removal kit for Gram Negative Bacteria. Sequences were run on an Illumina Miseq using a PE50v2 kit. Reads were mapped to the E. coli K12 Genome (NC_000913.2) using bowtie2, allowing for up to two mismatches and enforcing paired-end constraints. Differentially expressed genes were determined by cuffdiff (Trapnell et al. 2010) with upper-quartile normalization and setting a maximum false discovery rate of 0.05. Principal component analysis was performed on the RNA-seq FPKM (fragments per kilobase of exon per million fragments mapped) values using the pca function in MATLAB.
Physiological Measurements
Growth curves for selected clones were started from stationary phase overnight cultures and grown under conditions identical to the ALE experiment, but with sampling occurring every 20 min. At each sampling time, the OD600 was taken along with a small volume of the growing culture that was then filter-sterilized. The filtrate was injected into an HPLC column (Aminex HPX-87H Column #125-0140). Compound concentrations at each time point were determined by comparison to a standard curve of know concentrations, and were used to determine rates of glucose uptake and acetate secretion by the cells. No other compounds were detected in the filtrate.
Multiplex Automated Genome Engineering
For recombineering, cells containing the pMA7 plasmid (manuscript under preparation) were grown in 15-ml LB media, with shaking at 37 °C to an OD600 of 0.4. Recombineering was mediated by the lambda Red beta protein and induced through the ParaBAD promoter for 10 min by adding arabinose to a final concentration of 0.2%. After induction, cells were placed on ice for at least 15 min before being harvested, washed, and finally resuspended in a total volume of 200 µl ice-cold sterile water. A 50-µl volume of the cells was mixed with 5 pmol oligo of each oligo and electroporated in 0.1-mm gap cuvettes; 1.8 kV, 200 Ω, 25 µF. Immediately after electroporation, 1 ml LB was added to the cells. Cells were transferred to a 50-ml Falcon tube to a total volume of 5 ml LB and grown for at least 3 h at 37 °C to allow full segregation of chromosomal DNA. A total of nine consecutive rounds of recombineering were performed. A total number of 29 oligos were designed (supplementary table S2, Supplementary Material online) to recreate the majority of the mutations identified in ALE lines 1, 4, 5, 7, 9, and 10. Eight recombineering experiments were performed in parallel: Six using the oligos corresponding to each of the nonmutator ALE lines, one using all 29 oligos, and one control using oligo P9 malK (supplementary table S2, Supplementary Material online). Introduction of the malK oligo does not alter fitness in the glucose environments used, but makes the cells turn purple when grown on MacConkey agar maltose plates. In this control MAGE experiment, the recombinant formation frequency was found to be on average 10% by plating the recombineered population and determining the proportion of purple colonies. Recombinant formation frequency is influenced largely by oligo length (Wang et al. 2009), which, along with all other experimental variables, was kept constant for all recombineering, thus allelic biases should not be present.
Following recombineering, the mixed cell populations were cultured and serially passed for 3 days, under the same conditions as the ALE experiment, to select for the strains most fit for growth at 42 °C. These enriched MAGE populations were then streaked on minimal M9 glucose agar plates and single colonies were isolated, screened for improved growth rates at 42 °C in a microtiter plate reader, and subjected to whole-genome DNA resequencing.
Supplementary Material
Supplementary data sets S1–S3, figures S1–S5, and tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Richard Szubin for his assistance with conducting experiments. This work was supported by the Novo Nordisk Foundation. T.E.S. is supported through the National Science Foundation Graduate Research Fellowship (grant DGE1144086).
References
- Ban C, Yang W. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell. 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- Blaby IK, Lyons BJ, Wroclawska-Hughes E, Phillips GC, Pyle TP, Chamberlin SG, Benner SA, Lyons TJ, Crecy-Lagard V, Crecy E. Experimental evolution of a facultative thermophile from a mesophilic ancestor. Appl Environ Microbiol. 2012;78:144–155. doi: 10.1128/AEM.05773-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank D, Wolf L, Ackermann M, Silander OK. The predictability of molecular evolution during functional innovation. Proc Natl Acad Sci U S A. 2014;111:3044–3049. doi: 10.1073/pnas.1318797111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordbar A, Monk JM, King ZA, Palsson BO. Constraint-based models predict metabolic and associated cellular functions. Nat Rev Genet. 2014;15:107–120. doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- Camps M, Naukkarinen J, Johnson BP, Loeb LA. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc Natl Acad Sci U S A. 2003;100:9727–9732. doi: 10.1073/pnas.1333928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SM, Marx CJ. Evolution after introduction of a novel metabolic pathway consistently leads to restoration of wild-type physiology. PLoS Genet. 2013;9:e1003427. doi: 10.1371/journal.pgen.1003427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charusanti P, Conrad TM, Knight EM, Venkataraman K, Fong NL, Xie B, Gao Y, Palsson BO. Genetic basis of growth adaptation of Escherichia coli after deletion of pgi, a major metabolic gene. PLoS Genet. 2010;6:e1001186. doi: 10.1371/journal.pgen.1001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaulk SG, Smith Frieday MN, Arthur DC, Culham DE, Edwards RA, Soo P, Frost LS, Keates RA, Glover JN, Wood JM. ProQ is an RNA chaperone that controls ProP levels in Escherichia coli. Biochemistry. 2011;50:3095–3106. doi: 10.1021/bi101683a. [DOI] [PubMed] [Google Scholar]
- Cheng KK, Lee BS, Masuda T, Ito T, Ikeda K, Hirayama A, Deng L, Dong J, Shimizu K, Soga T, et al. Global metabolic network reorganization by adaptive mutations allows fast growth of Escherichia coli on glycerol. Nat Commun. 2014;5:3233. doi: 10.1038/ncomms4233. [DOI] [PubMed] [Google Scholar]
- Conrad TM, Frazier M, Joyce AR, Cho BK, Knight EM, Lewis NE, Landick R, Palsson BO. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci U S A. 2010;107:20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad TM, Joyce AR, Applebee MK, Barrett CL, Xie B, Gao Y, Palsson BO. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 2009;10:R118. doi: 10.1186/gb-2009-10-10-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TF, Rozen DE, Lenski RE. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc Natl Acad Sci U S A. 2003;100:1072–1077. doi: 10.1073/pnas.0334340100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragosits M, Mattanovich D. Adaptive laboratory evolution—principles and applications for biotechnology. Microb Cell Fact. 2013;12:64. doi: 10.1186/1475-2859-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Figueroa NV, Pina-Escobedo A, Schroeder I, Simons RW, Garcia-Mena J. Polynucleotide phosphorylase interacts with ribonuclease E through a betabetaalphabetabetaalpha domain. Biochimie. 2006;88:725–735. doi: 10.1016/j.biochi.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Echols H, Lu C, Burgers PM. Mutator strains of Escherichia coli, mutD and dnaQ, with defective exonucleolytic editing by DNA polymerase III holoenzyme. Proc Natl Acad Sci U S A. 1983;80:2189–2192. doi: 10.1073/pnas.80.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folichon M, Arluison V, Pellegrini O, Huntzinger E, Regnier P, Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–7310. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong SS, Joyce AR, Palsson BO. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states. Genome Res. 2005;15:1365–1372. doi: 10.1101/gr.3832305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H, Bennett BD, Amini S, Reaves ML, Hottes AK, Rabinowitz JD, Tavazoie S. Regulatory and metabolic rewiring during laboratory evolution of ethanol tolerance in E. coli. Mol Syst Biol. 2010;6:378. doi: 10.1038/msb.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zhou T, Wilke CO. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput Biol. 2010;6:e1000664. doi: 10.1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera TS, Csonka LN, Paliy O. Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J Bacteriol. 2008;190:3712–3720. doi: 10.1128/JB.01990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyaneshwar P, Paliy O, McAuliffe J, Jones A, Jordan MI, Kustu S. Lessons from Escherichia coli genes similarly regulated in response to nitrogen and sulfur limitation. Proc Natl Acad Sci U S A. 2005;102:3453–3458. doi: 10.1073/pnas.0500141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harano Y, Kinoshita M. Translational-entropy gain of solvent upon protein folding. Biophys J. 2005;89:2701–2710. doi: 10.1529/biophysj.104.057604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindre T, Knibbe C, Beslon G, Schneider D. New insights into bacterial adaptation through in vivo and in silico experimental evolution. Nat Rev Microbiol. 2012;10:352–365. doi: 10.1038/nrmicro2750. [DOI] [PubMed] [Google Scholar]
- Ibarra RU, Edwards JS, Palsson BO. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420:186–189. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- Jain C, Deana A, Belasco JG. Consequences of RNase E scarcity in Escherichia coli. Mol Microbiol. 2002;43:1053–1064. doi: 10.1046/j.1365-2958.2002.02808.x. [DOI] [PubMed] [Google Scholar]
- Jensen KF. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto T, Iijima L, Tatsumi M, Ono N, Oyake A, Hashimoto T, Matsuo M, Okubo M, Suzuki S, Mori K, et al. Transition from positive to neutral in mutation fixation along with continuing rising fitness in thermal adaptive evolution. PLoS Genet. 2010;6:e1001164. doi: 10.1371/journal.pgen.1001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Marcuschamer D, Santos CN, Yu H, Stephanopoulos G. Mutagenesis of the bacterial RNA polymerase alpha subunit for improvement of complex phenotypes. Appl Environ Microbiol. 2009;75:2705–2711. doi: 10.1128/AEM.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landini P, Zehnder AJ. The global regulatory hns gene negatively affects adhesion to solid surfaces by anaerobically grown Escherichia coli by modulating expression of flagellar genes and lipopolysaccharide production. J Bacteriol. 2002;184:1522–1529. doi: 10.1128/JB.184.6.1522-1529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latif H, Lerman JA, Portnoy VA, Tarasova Y, Nagarajan H, Schrimpe-Rutledge AC, Smith RD, Adkins JN, Lee DH, Qiu Y, et al. The genome organization of Thermotoga maritima reflects its lifestyle. PLoS Genet. 2013;9:e1003485. doi: 10.1371/journal.pgen.1003485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Feist AM, Barrett CL, Palsson BO. Cumulative number of cell divisions as a meaningful timescale for adaptive laboratory evolution of Escherichia coli. PLoS One. 2011;6:e26172. doi: 10.1371/journal.pone.0026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Molla MN, Cantor CR, Collins JJ. Bacterial charity work leads to population-wide resistance. Nature. 2010;467:82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. 1. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138:1315–1341. [Google Scholar]
- Michel B. Replication fork arrest and DNA recombination. Trends Biochem Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- Murphy H, Cashel M. Isolation of RNA polymerase suppressors of a (p)ppGpp deficiency. Methods Enzymol. 2003;371:596–601. doi: 10.1016/S0076-6879(03)71044-1. [DOI] [PubMed] [Google Scholar]
- Navarro Llorens JM, Tormo A, Martinez-Garcia E. Stationary phase in gram-negative bacteria. FEMS Microbiol Rev. 2010;34:476–495. doi: 10.1111/j.1574-6976.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- Perwez T, Hami D, Maples VF, Min Z, Wang BC, Kushner SR. Intragenic suppressors of temperature-sensitive rne mutations lead to the dissociation of RNase E activity on mRNA and tRNA substrates in Escherichia coli. Nucleic Acids Res. 2008;36:5306–5318. doi: 10.1093/nar/gkn476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock LJ. Multidrug-resistance efflux pumps—not just for resistance. Nat Rev Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. An alternative route for recycling of N-acetylglucosamine from peptidoglycan involves the N-acetylglucosamine phosphotransferase system in Escherichia coli. J Bacteriol. 2009;191:5641–5647. doi: 10.1128/JB.00448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy VA, Bezdan D, Zengler K. Adaptive laboratory evolution—harnessing the power of biology for metabolic engineering. Curr Opin Biotechnol. 2011;22:590–594. doi: 10.1016/j.copbio.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Rao VSH, Rao PRS. Global stability in chemostat models involving time delays and wall growth. Nonlinear Anal Real World Appl. 2004;5:141–158. [Google Scholar]
- Riehle MM, Bennett AF, Long AD. Genetic architecture of thermal adaptation in Escherichia coli. Proc Natl Acad Sci U S A. 2001;98:525–530. doi: 10.1073/pnas.021448998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringquist S, Shinedling S, Barrick D, Green L, Binkley J, Stormo GD, Gold L. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol Microbiol. 1992;6:1219–1229. doi: 10.1111/j.1365-2958.1992.tb01561.x. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Gelfand MS, Mironov AA, Rakhmaninova AB. Comparative approach to analysis of regulation in complete genomes: multidrug resistance systems in gamma-proteobacteria. J Mol Microbiol Biotechnol. 2001;3:319–324. [PubMed] [Google Scholar]
- Rodriguez-Verdugo A, Gaut BS, Tenaillon O. Evolution of Escherichia coli rifampicin resistance in an antibiotic-free environment during thermal stress. BMC Evol Biol. 2013;13:50. doi: 10.1186/1471-2148-13-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen DE, Philippe N, Arjan de Visser J, Lenski RE, Schneider D. Death and cannibalism in a seasonal environment facilitate bacterial coexistence. Ecol Lett. 2009;12:34–44. doi: 10.1111/j.1461-0248.2008.01257.x. [DOI] [PubMed] [Google Scholar]
- Shachrai I, Zaslaver A, Alon U, Dekel E. Cost of unneeded proteins in E. coli is reduced after several generations in exponential growth. Mol Cell. 2010;38:758–767. doi: 10.1016/j.molcel.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Sinensky M. Homeoviscous adaptation—a homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc Natl Acad Sci U S A. 1974;71:522–525. doi: 10.1073/pnas.71.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- Soutourina OA, Bertin PN. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev. 2003;27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- Stoebel DM, Hokamp K, Last MS, Dorman CJ. Compensatory evolution of gene regulation in response to stress by Escherichia coli lacking RpoS. PLoS Genet. 2009;5:e1000671. doi: 10.1371/journal.pgen.1000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Rodriguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS. The molecular diversity of adaptive convergence. Science. 2012;335:457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt. Activities at the Universal Protein Resource (UniProt) Nucleic acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri GN, Altman E, Sangurdekar DP, Khodursky AB, Eiteman MA. Overflow metabolism in Escherichia coli during steady-state growth: transcriptional regulation and effect of the redox ratio. Appl Environ Microbiol. 2006;72:3653–3661. doi: 10.1128/AEM.72.5.3653-3661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayendran C, Barsch A, Friehs K, Niehaus K, Becker A, Flaschel E. Perceiving molecular evolution processes in Escherichia coli by comprehensive metabolite and gene expression profiling. Genome Biol. 2008;9:R72. doi: 10.1186/gb-2008-9-4-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang FF, Qian W. Evolutionary rewiring and reprogramming of bacterial transcription regulation. J Genet Genomics. 2011;38:279–288. doi: 10.1016/j.jgg.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Wiser MJ, Ribeck N, Lenski RE. Long-term dynamics of adaptation in asexual populations. Science. 2013;342:1364–1367. doi: 10.1126/science.1243357. [DOI] [PubMed] [Google Scholar]
- Wood TE, Burke JM, Rieseberg LH. Parallel genotypic adaptation: when evolution repeats itself. Genetica. 2005;123:157–170. doi: 10.1007/s10709-003-2738-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc Natl Acad Sci U S A. 2006;103:9107–9112. doi: 10.1073/pnas.0602917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.