Abstract
Large studies of extended families usually collect valuable phenotypic data that may have scientific value for purposes other than testing genetic hypotheses if the families were not selected in a biased manner. These purposes include assessing population-based associations of diseases with risk factors/covariates and estimating population characteristics such as disease prevalence and incidence. Relatedness among participants however, violates the traditional assumption of independent observations in these classic analyses. The commonly used adjustment method for relatedness in population-based analyses is to use marginal models, in which clusters (families) are assumed to be independent (unrelated) with a simple and identical covariance (family) structure such as those called independent, exchangeable and unstructured covariance structures. However, using these simple covariance structures may not be optimally appropriate for outcomes collected from large extended families, and may under- or over-estimate the variances of estimators and thus lead to uncertainty in inferences. Moreover, the assumption that families are unrelated with an identical family structure in a marginal model may not be satisfied for family studies with large extended families. The aim of this paper is to propose models incorporating marginal models approaches with a covariance structure for assessing population-based associations of diseases with their risk factors/covariates and estimating population characteristics for epidemiological studies while adjusting for the complicated relatedness among outcomes (continuous/categorical, normally/non-normally distributed) collected from large extended families. We also discuss theoretical issues of the proposed models and show that the proposed models and covariance structure are appropriate for and capable of achieving the aim.
Keywords: Correlated outcomes, Marginal models, Family study, Large and inter-related extended families
Introduction
In studies of large families, the family structures are usually complex and different, and the individuals may relate to each other in a range of ways. There may be considerable scientific value in assessing population-based associations of diseases with risk factors and estimating population characteristics such as disease prevalence and incidence in addition to testing genetic hypotheses. Since observations on family members are correlated, these results need to be adjusted. The commonly used adjustment method for relatedness in population-based studies is to use marginal models [1–3], in which clusters are assumed to be independent with a simple and identical covariance structure. In applying a marginal model to family data, clusters become families, and the assumption that clusters are independent with an identical covariance structure implies that families are unrelated with an identical family structure. For example, let yij denote an observed disease status (yij = 1, if diseased, and = 0, otherwise) of the jth member from the ith family, i = 1, 2, …, N; j = 1, 2, …, m. Then, the following marginal model is usually applied to assess population-based association of prevalence or cumulative incidence of the disease with its risk factors/covariates x1, …, xp,
where coefficients β0, …βp are unknown parameters, and the covariance/correlation structure among yijs from a family is assumed to be the same across all families and to follow some simple structures. The most often used covariance/correlation structures defined in the marginal model are the independent, exchangeable and unstructured structures [1, 2] (they have been accommodated in many statistical software packages such as SAS Process GENMOD, SAS software, SAS Institute, Cary, NC).
The independent and exchangeable correlation structures defined in the marginal model have the following forms:
On the other hand, if an extended family with, say, 8 members, is recruited based on “core sibs” in a family study and the 8 members are ordered as {father of core sibs, mother of core sibs, spouse of core sib 1, spouse of offspring 1 of core sib 1, core sib 1, core sib 2, offspring 1 of core sib 1, offspring 2 of core sib 1}, then the correlation matrix for outcomes collected from the 8 members should have the following form
This is because spouses are usually unrelated, and father and mother relate to their children (the *s in the matrix) and their grandchildren (the $s) differently. Therefore, if the independent correlation structure is applied to the data collected from this kind of extended family, then it implies that all members in the extended family are unrelated while in actuality all members are related except the spouse of offspring 1 of core sib 1. On the other hand, if the exchangeable correlation structure is used, it implies that all members in the extended family are related to the same degree, and thus ignores those unrelated (more than half of correlations are zero as shown in Rextended) and those related to different degrees (those *s and $s). It is well known and has been reported in the literature using either theoretical proofs or simulations that treating related as unrelated or vice versa may either under- or over-estimate the variances of estimated parameters, which in turn may lead to false inferences about associations of the relevant outcome with its risk factors/covariates [2, 4].
The unstructured covariance structure defined in the marginal model has the following form:
where m is family size, {σi,j}s are unknown variance/covariance components, σi,j = σj,i, but σ1,2 ≠ σ1,3 ≠ ⋯ ≠ σ1,m, σ2,3 ≠ σ2,4 ≠ ⋯≠ σ2,m, …, σm−2,m−1 ≠ σm−2,m, and . If the marginal model with the unstructured covariance structure is applied to the data collected from the abovementioned extended family (m = 8), then we need to estimate 36 (=8*(8+1)/2) unknown variance/covariance components in COV. This will be impractical if family size is large. Further, using the unstructured covariance structure in a marginal model also requires that we are able to uniquely order all members in each family in a way that is the same across all families (e.g., see the explanation about the “WITHIN” option in SAS Process GENMOD for details). However, it is difficult and sometimes impossible to order uniquely if families have different sizes or structures. Moreover, even when all recruited extended families have the same structure, if only a subset of the data is used in an analysis such as using the data collected from those aged 40–60 years only, then the numbers of the aged 40–60 members from different families may vary and so may the structures among the extracted members from different families.
Therefore, the current applicable covariance structures (independent, exchangeable and unstructured) used in a marginal model may not be optimal to describe the complicated relatedness, and the assumption of identical family structure may not be satisfied in larger extended families.
Let us take the Strong Heart Family Study (SHFS) [5] as an example. The SHFS was initiated in 1998 to identify genetic determinants of cardiovascular disease (CVD) in American Indians and to map and identify genes for CVD susceptibility. Families were recruited if they had a “core sibship” (a sibship with at least five living siblings, of whom three or more were original Strong Heart Study (SHS) [6] cohort members) and had at least 12 living offspring aged ≥15 years among the core sibship members. Each of the recruited families included the core sibship members, their parents (if alive), spouses, offspring, spouses of offspring, and grandchildren who were at least 15 years of age. A detailed description of the SHFS and SHS has been reported previously [5, 6] and published on the SHS web site (http://strongheart.ouhsc.edu). For example, 27 extended families were recruited in one of the centers of the SHFS. These extended families spanned four generations with the largest family size being 114 and the average size being 45. It is clear that family structures of these families are different and complicated. An application of the marginal model with either the independent or exchangeable covariance structure will lead to oversimplifying the complicated relationship among members in the extended families, while an application of the marginal model with the unstructured covariance structure will lead to estimating at least 6555 (=114*(114+1)/2) unknown variance/covariance components since the largest family size is 114. Moreover, because the families are recruited based on the “core sibship” and are not geographically distant from each other, it is not unusual for a recruited non-core member in a recruited family to be related to recruited members in other recruited families. This is often the case in family studies for populations such as American Indians, Alaska Natives or the Amish. Thus, the usual assumption that families (clusters) are unrelated (independent) in a marginal model may not be satisfied either.
Therefore, it is desirable and important to develop models to adjust for these kinds of complicated relatedness in order to assess the population-based associations and estimate population characteristics. This paper proposes such models for analyzing continuous or categorical, normally or non-normally distributed outcomes in “Models”, provides simulation and application examples in “Simulation and examples”, and discusses related theoretical issues in “Extension of the proposed models defined in (2.1) and (2.2)”, “Discussion”, and in the Appendix.
Models
Proposed model for a qualitative outcome
Let yi = 1, if person i has an outcome of interest such as a disease, and =0, otherwise. We propose the following marginal model for assessing association of the prevalence or cumulative incidence of the disease with its risk factors/covariates x1, …, xp.
| (2.1) |
where logit(․) is called the logit link function, ϕ(i, j) is the kinship coefficient [7] between two individuals i and j, ϕ(i, j) = P{At any given locus, two randomly picked alleles, one from person i and one from person j are identity by descent}, 1 > ra > 0, ra and coefficients β0, …βp are unknown parameters, and the observations are ordered sequentially by families, that is, i = 1, …, m1 denote the members in the 1st family, i = m1 + 1, …, m1 + m2 denote the members in the 2nd family, etc., and n denotes the total number of participants in the study.
The covariance structure of the model defined by (2.1) is based on the classic biometrical model [7], where ra denotes the component due to the additive effect, which is the major cause of resemblance between relatives. Following approach of the biometrical model, if the additive effects alone cannot adequately explain the data, a dominance component may be added [7]. That is, if necessary, we may use the following covariance structure in the model defined by (2.1)
| (2.1a) |
where ra > 0, rd > 0, ra + rd < 1, rd denotes the component due to the dominance effect, which represents variance due to non-linear interaction between transmissible alleles and is contributed by individuals who share both alleles identical- by-descent [7], and Δ7(i, j) is Jacquard’s coefficient [8], Δ7(i, j) = P{The two alleles of person i at any given locus are identical by descent with the two alleles of person j at the same locus}. Both ϕ(i, j)s and Δ7(i, j)s are directly calculated from the extended pedigrees of families ascertained in a family study since parents themselves may be related. For example, assuming parents are unrelated, it is well known that the kinship coefficient for two identical twins is 0.5; parent-offspring, 0.25; full sibs, 0.25; uncleniece, 0.125; etc. If there are no instances of inbreeding, no double cousins, and no twins in the pedigree, then Δ7(i, i) = 1, ∀i; Δ7(i, j) = 1/4, if i and j are full siblings; and zero otherwise. Many commercial software packages such as SAS Process INBREED (SAS software, SAS Institute, Cary, NC) and freely available software packages such as the PEDIG package [9] can be applied to calculate kinship coefficients between any two individuals based on their ascertained pedigree.
Compared to the independent, exchangeable and unstructured structures defined in the marginal model shown in the introduction, it is clear that the covariance structures in the proposed models defined by (2.1) or (2.1a) account appropriately for different degrees of relationship among family members. These covariance structures have at most two unknown parameters (ra and rd), and do not require that all members in each family be ordered in the same way across all families. The proposed covariance structure defines the relationship of outcomes collected from any two members in recruited families no matter how complicated and different the family structures are, or whether the families are related. The proposed model is also flexible for analyses that use only a subset of the data since the covariance structure for the subset can easily be composed from the corresponding sub-kinship matrix.
Proposed model for a continuous outcome
For a continuous outcome, y, such as cholesterol, the proposed model is
| (2.2) |
where ei’s denote random errors, 1 > ra > 0, ra denotes the component due to the additive effect, σ2, ra and coefficients β0, …, βp are unknown parameters, and the other notations are the same as those defined in (2.1). Similarly, if the dominance effect needs to be considered, we may use the following covariance structure in the model defined by (2.2)
| (2.2a) |
where ra > 0, rd > 0, ra + rd < 1, rd denotes the component due to the dominance effect.
Extension of the proposed models defined in (2.1) and (2.2)
To extend the models defined in (2.1) and (2.2) to cover different kinds of outcomes in a family study and to allow for additional random effects such as random environmental effects, we follow the procedure for generalized linear mixed models [4, 10, 11].
Let Y = (y1, …, yn)τ, xi = (1, xi1, …, xip)τ, X = (x1, …, xn)τ, B = (β0, β1, …, βp)τ, zi = (zi1, …, ziq)τ, Z = (z1, …, zn)τ, U = (u1, u2, …, uq)τ, and e = (e1, e2, …, en)τ, where yi denotes the outcome variable observed in individual i with fixed covariates xi and explanatory variables zi that are associated with the random effects U, and U and the errors, e, are assumed to be independent. Assume that U has a mean zero O and covariance matrix Cov(U) = G (θ) that depends on an unknown vector θ; and given U, yi will have means , variances Var(yi/U), and covariance components
| (2.3) |
or
| (2.3a) |
if the dominance effect is added, where 0 < ra < 1 (or 0 < ra, rd < 1 and ra + rd < 1 if the dominance effect is added). Assume that is related to by a link function , with inverse h = g−1. Then and
| (2.4) |
It is clear that if there are no random effects involved, the model defined by (2.1) is a special case of (2.4) when y is binomially distributed and the logit link function is used. The model defined by (2.2) is the one in which the identity link function is used and Var(yi/U) = σ2. Models in which outcomes have other distributions such as those belonging to the linear exponential family with respective link functions are also accommodated in (2.4) [4, 10, 11].
If all families are unrelated and have an identical structure, the estimation procedures for σ2, ra, rd, θ, B and U in the models defined by (2.1), (2.2) or (2.4), the proofs for asymptotic properties of the estimators, and their statistical inferences follow those used in the generalized linear mixed models [4, 10, 11]. In the case where families in a study can be classified into unrelated subgroups, and families in each subgroup are unrelated and have identical structure, we show in the Appendix that similar approaches and asymptotic properties also hold. When families in a study are related and large and have different sizes and structures, and the number of families may be only moderate, it is difficult to theoretically prove the asymptotic properties of the estimated parameters from our models because families may be related. However, we showed by a simulation that the usual asymptotic properties also hold in this case no matter whether the families are related or with different family sizes or structures if the total number of members from all families is large enough even though the number of families may be only moderate (Appendix).
Simulation and examples
As mentioned in the introduction, it has been reported in the literature by either theoretical proofs or simulation methods that treating related observations as unrelated or vice versa may either under- or over-estimate the variances of estimated parameters, and thus may lead to false inferences about the significances of associations of a relevant outcome with its risk factors/covariates [2, 4]. To further confirm these uncertainties, we use a simulation to compare power of the proposed models with that of the marginal model with either the exchangeable or independent correlation structure in detecting significant associations of an outcome measure with its risk factors/covariates when correlation structure in a data is neither the exchangeable nor independent.
Let FPGij denote measured fasting plasma glucose (FPG) of the jth member in the ith family. The following proposed model (2.2) is used to simulate association of FPG with its seven risk factors/covariates: age, waist circumference (WAIST), body mass index (BMI), urinary albumin to creatinine ratio (UACR), insulin, low density lipoprotein (LDL), and triglycerides (TG),
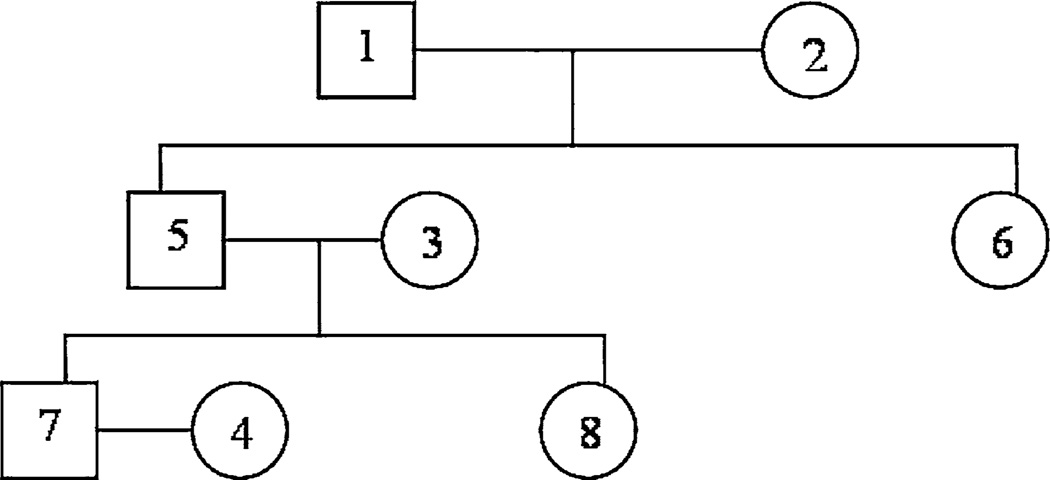
The families are those 50 unrelated extended families in which each family had 8 members ordered as {father of core sibs, mother of core sibs, spouse of core sib 1, spouse of offspring 1 of core sib 1, core sib 1, core sib 2, offspring 1 of core sib 1, offspring 2 of core sib 1}. Figure 1 shows the pedigree for the ordered 8 members. Assume FPGs observed from a family are normally distributed with Var(FPGij) = 1000 (or standard deviation of 31.6) and ra = 0.5, then based on the definition of the covariance in (2.2) and the calculated kinship matrix Φ from the pedigree of the ordered 8 members, FPGs have the following correlation matrix
where FPGi. =(FPGi1, …, FPGi8)τ denotes FPG collected from 8 members of the ith family, i=1, …, 50, and
Fig. 1.
Pedigree for the ordered eight members
For example, ϕ(father of core sibs, offspring 1 of core sib 1) = ϕ17 = 0.125. When ra = 0.8, FPGs have the following correlation matrix that has larger correlation coefficients
The parameters used in the simulation model are obtained from the SHFS data in order to make the model realistic and meaningful. It is clear that R1 or R2 contains three levels of correlations (e.g., 0, 0.125 and 0.25 in R1), which is neither the exchangeable correlation (that assumes all correlation coefficients are the same and non-zero) nor independent correlation (that assumes all correlation coefficients are zero) structure defined in the marginal model. One thousand replicated simulated data sets, each including 400 simulated observations from the 400 members in the 50 unrelated extended families, were generated. For each of the 1,000 simulated data sets, the proposed model (2.2) and the respective marginal model with the exchangeable or independent correlation structure were applied separately to estimate the regression coefficients for the seven risk factors/covariates of FPG in these models. To estimate power of a model in detecting significant association of FPG with a given risk factor/covariate, say, WAIST, after adjusting for the other risk factors/covariates in the model, we tested the null hypothesis: H0: “the coefficient for WAIST in the model” = 0, at a given significant level (type 1 error rate) α based on each of 1,000 simulated data. The empirical power of the model at the given significance level α in detecting the significant association of FPG with WAIST after adjusting for the other risk factors/covariates in the model was estimated as “the total number of the tests based on the 1,000 replicated simulated data sets where H0 was rejected at the given significance level α “divided by 1,000. The same procedure was applied simultaneously for each of the seven risk factors/covariates in the model. Table 1 displays estimates of the empirical powers of the proposed model (2.2) and the marginal model with the exchangeable or independent correlation structure in detecting the significant association of FPG with each of its risk factors/covariates after adjusting for the other risk factors/covariates in the model at two different significance levels (α = 0.05 or 0.01) based on the 1,000 simulated data sets generated by using two different correlation matrices R1 and R2. For example, at the assumed correlation matrix R1 and the given significance level α = 0.05, the estimated empirical powers of the proposed model (2.2) and the marginal model with the exchangeable or independent correlation structure in detecting the significant association of FPG with WAIST after adjusting for the other risk factors/covariates in the model are 0.611, 0.598 and 0.556, respectively. The power of the proposed model (2.2) is almost always greater than the power of the marginal model with either the exchangeable or independent correlation structure, and the differences between the powers increased with the correlation coefficients (those based on R2 are larger than those based on R1). Our simulation results indicated that using either simplified exchangeable or independent correlation structure in analyzing correlated data with more complicated correlation structures (e.g., those defined by R1 or R2) may either under- or overestimate the variances of parameters, and thus may lead to incorrect inferences about the significance of associations of an outcome with its risk factors/covariates. It may be expected that such uncertainty will be greater if simplified exchangeable or independent correlation structure is used in analyzing correlated data collected from the SHFS extended families that span four generations with the largest family size being 114 and the average size being 45.
Table 1.
Empirical powers of the proposed model (2.2) and the marginal model with the exchangeable (M-E) or independent (M-I) correlation structure in detecting the significant association of FPG with each of its risk factors/covariates after adjusting for the other risk factors/covariates in the model at two different significance levels (type 1 error rates) based on the 1,000 simulated data sets generated by using two different correlation matrixes R1 and R2
| Correlation matrix | Risk factor/covariate | Significance level α = 0.05 | Significance level α = 0.01 | ||||
|---|---|---|---|---|---|---|---|
| Model (2.2) | M-E | M-I | Model (2.2) | M-E | M-I | ||
| R1 | Age (year) | 0.806 | 0.799 | 0.752 | 0.622 | 0.595 | 0.533 |
| Waist (cm) | 0.611 | 0.598 | 0.556 | 0.392 | 0.373 | 0.318 | |
| BMI (kg/m2) | 0.461 | 0.446 | 0.409 | 0.254 | 0.250 | 0.212 | |
| Log(TG) (mg/dL) | 0.994 | 0.992 | 0.989 | 0.982 | 0.969 | 0.967 | |
| Log(UACR) | 1.000 | 0.999 | 0.999 | 0.999 | 0.999 | 0.999 | |
| LDL (mg/dL) | 0.282 | 0.272 | 0.237 | 0.113 | 0.115 | 0.098 | |
| Log(Insulin) (µU/mL) | 0.379 | 0.389 | 0.342 | 0.173 | 0.184 | 0.144 | |
| R2 | Age (year) | 0.946 | 0.849 | 0.776 | 0.815 | 0.662 | 0.541 |
| Waist (cm) | 0.770 | 0.612 | 0.529 | 0.535 | 0.365 | 0.297 | |
| BMI (kg/m2) | 0.581 | 0.443 | 0.388 | 0.344 | 0.253 | 0.182 | |
| Log(TG) (mg/dL) | 1.000 | 0.996 | 0.986 | 0.997 | 0.969 | 0.946 | |
| Log(UACR) | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 0.999 | |
| LDL (mg/dL) | 0.392 | 0.311 | 0.242 | 0.165 | 0.139 | 0.101 | |
| Log(Insulin) (µU/mL) | 0.565 | 0.443 | 0.369 | 0.346 | 0.224 | 0.166 | |
BMI body mass index, LDL low density lipoprotein, TG triglycerides, UACR urinary albumin and creatinine ratio, Waist waist circumference
Table 2 shows results when applying the proposed model (2.2) and the respective marginal model with either independent or exchangeable correlation structure to one of the 1000 simulated data sets that used the correlation structure defined in R1. We can interpret the results from the proposed model (2.2) in the same way as those from a conventional linear regression model since the proposed model (2.2) is a marginal model and the coefficients from a marginal model represent population averaged results [2, 4]. For example, based on the results from the proposed model (2.2) in Table 2, age, WAIST, log(UACR), log(insulin) and log(TG) were significantly and positively associated with FPG; while BMI and LDL were significantly and negatively associated with FPG. However, the results from the respective marginal models show that, besides the differences in the estimated coefficients, which maybe minor if the sample size is large [2, 4], some of the significant risk factors/covariates of FPG in the proposed model were no longer significant in the respective marginal models with either independent (age, log(insulin), and LDL) or exchangeable (age and LDL) covariance structures, reflecting uncertainty in inferences due to the simplification in correlation structures.
Table 2.
Marginal models for fasting plasma glucose on its risk factors/covariates based on the simulated data
| Variable | Proposed model (2.2) | Marginal model with the exchangeable covariance |
Marginal model with the independent covariance |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Coeff. | SE | P | Coeff. | SE | P | Coeff. | SE | P | |
| Intercept | −15.448 | −10.101 | −12.443 | ||||||
| Age (year) | 0.217 | 0.101 | 0.0318 | 0.130 | 0.105 | 0.2200 | 0.170 | 0.112 | 0.1291 |
| Waist (cm) | 0.612 | 0.173 | 0.0005 | 0.555 | 0.187 | 0.0032 | 0.630 | 0.192 | 0.0011 |
| BMI (kg/m2) | −1.380 | 0.449 | 0.0023 | −1.304 | 0.386 | 0.0008 | −1.357 | 0.495 | 0.0064 |
| Log(UACR) | 5.442 | 1.124 | <0.0001 | 5.984 | 1.130 | <0.0001 | 6.057 | 1.259 | <0.0001 |
| Log(Insulin) (µU/mL) | 7.025 | 2.212 | 0.0016 | 5.514 | 2.167 | 0.0114 | 4.587 | 2.499 | 0.0672 |
| LDL (mg/dL) | −0.098 | 0.042 | 0.0200 | −0.058 | 0.048 | 0.2298 | −0.062 | 0.047 | 0.1895 |
| Log(TG) (mg/dL) | 15.149 | 3.022 | <0.0001 | 14.974 | 3.282 | <0.0001 | 14.438 | 3.369 | <0.0001 |
Coeff., regression coefficient; P, P value; SE, the standard error
Discussion
As indicated earlier, the proposed models are appropriate, capable, flexible and efficient for population-based association studies while adjusting for complicated relatedness in data collected from family studies with large extended families.
If outcomes are continuous and normally distributed, and families in a study are unrelated to each other and have an identical family structure, the proposed model defined in (2.4) with the identity link function is the variance components model used in pedigree analyses [7, 12, 13]. However, if outcomes are discrete or not normally distributed, the proposed model defined in (2.4) is different from those proposed models for correlated data in the literature. For example, for binary outcomes, the proposed Model (2.1) is different from those threshold models [14–16] or the generalized linear mixed model (GLMM) with a logit link function [17] or the mixed models on which family-based association tests (FBAT) are based [18, 19].
The threshold model for the disease outcome yi (yi = 1, if person i has the disease; and =0, otherwise) and its risk factors/covariates x1, …, xp is defined as follows. Let the random effects U = (u1, u2, …, un)τ be normally distributed with
Assume given U, yi, i = 1, …, n, are mutually independent, and
where ϑ is the underlying threshold, zi, i = 1, …, n, are the underlying random variables, and Φ−1(․) stands for the inverse of the standardized cumulative normal distribution function, which is also called the probit link function in the GLMM.
The respective GLMM with a logit link function in this case is defined as follows. Assume given the same random effects U = (u1, u2, …, un)τ defined above, yi, i = 1, …, n, are mutually independent, and
The mixed model on which FBAT is based for the dichotomous phenotype yij of the jth offspring in the ith family is defined as follows. Assume given the random variable uij that codes for genotype of the jth offspring in the ith family at a particular allele, i = 1, …,m; j = 1, …, ni, yij’s are independent and
where x1, …, xp usually denote demographic or environmental effects, and uij are dependent on parental genotypes. FBAT based on this model tests the null genetic hypothesis H0 : “no association in the presence of linkage”; or “no linkage and no association between a marker and the phenotype”, which is equivalent to testing whether the coefficient γ of the random variables equals zero, that is, H0 : γ = 0.
It is clear that the proposed Model (2.1) is a marginal model while the threshold model and the GLMM model as well as the FBAT based model are mixed random effects models. It is known that a marginal model and a mixed random effects model can lead to different estimations and interpretations for the regression coefficients [2, 4]. In the proposed Model (2.1), the logit of the expectation E(I(yi = 1)) = P(yi = 1) was directly modeled with its risk factors/covariates x1, …, xp, and the covariance is directly defined through yi’s themselves. Therefore, like the example of the proposed Model (2.2) shown in “Simulation and examples”, the estimated coefficients β̂0, β̂1, …, β̂p for the risk factors/covariates from the proposed Model (2.1) can be interpreted in the same way as those estimated coefficients from a conventional logistic regression model since the coefficients from a marginal model represent population averaged results [2, 4]. While in the previous mentioned mixed random effects models, say, the GLMM model, the logit of the conditional (conditioning on the random effects U) expectation E(I(yi = 1)/U) = p(yi = 1/U) was modeled with both the fixed effects x1, …, xp and the random effect ui, and the covariance is partially defined by the random effects U. Therefore, the estimated coefficients β̃0, β̃1, …, β̃p for the fixed effects, x1, …, xp from these models are dependent on individual random effects uis and the distribution of uis by the definition of the conditional expectation and thus are individual- specific not population averaged [2, 4, 10, 17]. The Gibbs sampling method [20, 21] was usually used in fitting these mixed random effects models. Similar differences also exist between the proposed Model (2.4) and the GLMM models. Marginal models are appropriate when inferences about the population-average are the focus, which are what we focused in this paper and are usually the focus in population-based epidemiological studies; while a random effects model is most useful when the objective is to make inference about individuals rather than the population average [2, 4]. We adopted a marginal rather than random effects model approach for our covariance structure since we are interested only in assessing population-based associations of diseases with their risk factors/covariates and estimating population characteristics in epidemiological studies.
It is easy to verify that when families in a study are those unrelated pedigrees that contain only identical twins or only siblings without twins, the proposed models are equivalent to the marginal model with the exchangeable covariance structure that is often adopted in populationbased analyses [22].
Therefore, if families in a family study are unrelated and have identical structure, the covariance structure defined in the proposed models can be treated as an additional appropriate covariance structure to the existing covariance structures used in marginal models. If families in a family study are large and related, and have different sizes and structures like the extended families in the SHFS, the proposed models are most appropriate for and capable of handling this type of data for population-based analyses. The SAS macros for applying the proposed models will be available upon request.
Other marginal models for family data in the literature include the odds-ratio regression models [23] that model on each specific pair such as siblings or a parent-sibling pair, and the frailty models [24, 25] that take care of censored observations.
Acknowledgments
This study was supported by cooperative agreement grants (No. U01-HL41642, U01-HL41652, U01-HL41654, U01-HL65520, and U01-HL65521) from the National Heart, Lung, and Blood Institute. The authors wish to thank the Indian Health Service hospitals and clinics at each center, the directors of the Strong Heart Study clinics, Betty Jarvis, Dr. Tauqeer Ali, Marcia O’Leary, Dr. Marie Russell, Bert Lewis, and their staffs. The opinions expressed in this paper are those of the authors and do not necessarily reflect those of the Indian Health Service.
Abbreviations
- BMI
Body mass index
- CVD
Cardiovascular disease
- FBAT
Family-based association tests
- FPG
Fasting plasma glucose
- GLMM
Generalized linear mixed model
- LDL
Low density lipoprotein
- SHFS
Strong heart family study
- SHS
Strong heart study
- TG
Triglycerides
- UACR
Urinary albumin to creatinine ratio
- WAIST
Waist circumference
Appendix
Case 1: Families in a study are classified into unrelated subgroups, and families in each subgroup are unrelated and have identical structure
This is the case for a family study in which we combined those related study families together to form an expanded family and then classified all such expanded families into different subgroups based on their structures.
Let Φl and nl be the kinship coefficient matrix and the number of families associated to the subgroup l, respectively, l = 1, 2, …, S, S is the total number of subgroups of families in the study, and . Let denote the observed outcomes from the νth family in the subgroup l, ν = 1, 2, …, nl, and fl,ξ denote the density function of ylν, where ξ denotes all unknown parameters. Suppose that S remains fixed, each of nl tends to infinity, and nl/M → λl > 0, ∀l. Let Il (ξ) denote the information matrix corresponding to fl,ξ and let
The likelihood L(ξ) is given by
and the likelihood equations by
where w denotes the number of all unknown parameters. Then we have the following theorem:
Theorem 1 Suppose fl,ξ for each l satisfies some general regular conditions [26], then with a probability approaching 1, there exist solutions ξ̂M of the likelihood equations such that
ξ̂M is consistent for estimating ξ.
is asymptotically normal with (vector) mean zero and covariance matrix [I(ξ)]−1.
The proof of this theorem for the model defined by (2.4) follows those of Theorem 6.1 in Lehmann [26] and those of the generalized linear mixed models [4, 10, 11].
The conditions used in the proof are reasonable because in a family study, the size of the largest expanded family must be finite since only several generations and certain individuals will be included. Therefore the number of different structures in recruited families in the study should be finite also. Assuming the finite family structures are distributed homogenously, it is expected that the number of families with each of these finite family structures will all be large if the study population size is large enough.
Case 2: Families in a study may be related and large, and have different sizes and structures, and the number of families may be only moderate.
It is difficult to theoretically prove the asymptotic properties of the estimated parameters from our models in this case because families may be related. However, we believe that the asymptotic properties also hold in this case if the total number of members from all families is large enough. To show that the asymptotic properties may still hold in this case, the following simulation study was conducted. We simulated data using the following model, which was derived from the SHFS data in order to make the coefficients realistic and meaningful, for the association of age and waist circumference (WAIST) with fasting plasma glucose (FPG):
where ei‘s are normally distributed with mean zero and the covariance structure defined by the model in (2.2) with σ2 = 2,043 and ra = 0.3553 (or equivalently parameterized as ), and the kinship matrix that is derived from 653 individuals aged 30–65 in the 27 recruited families in the SHFS (since it is difficult to choose an appropriate 653-dimensional covariance matrix for related families with different family structures and sizes). We will show, by successively adding three more families in the simulation, that the asymptotic properties of the estimated parameters from our proposed model will hold no matter whether the included families are related or with different family sizes or structures if the total number of members from included families is large enough even though the number of families may be only moderate. Step 1: First, we simulated data for all members in the first three families based on the model and the sub-kinship matrix corresponding to these members, and then obtained a set of estimated parameters based on the simulated data. Next, we repeated this procedure 100 times and thus obtained 100 sets of the estimated parameters, then tested whether the 100 estimations of each parameter came from a normal distribution by using the Kolmogorov–Smirnov test. Similarly, in each successive step (Steps 2–9), we added three more families and performed the same simulation and analyses. The results from this simulation study are shown in Table 3. For example, at the last step, Step 9, we used the kinship matrix for all 653 members aged 30–65 in all 27 families. Means from the 100 estimates of intercept, coefficients of age and WAIST, and were 14.24, 0.986, 0.521, 717.68 and 1330.75, respectively. All of these means were quite close to the respective true parameters, 14.13, 0.98, 0.53, 726 and 1317 in the above model used in the simulation (actually, the true parameters all fell within the one-standard-error confidence intervals of the respective means). These estimated parameters were considered to have normal distributions based on the results from the Kolmogorov–Smirnov tests (all P > 0.1). Similar results also held even at the earlier steps such as Step 8 that included only 24 families but with 598 members. These results show that the asymptotic properties also hold in this case.
Table 3.
Results from the simulations for the asymptotic properties of the estimated parameters in the simulated model
| Step | Families | No. of members in each family |
Families used in each step |
Total No. of members |
Estimated σa (True σa = 726) | Estimated σε (True σε = 1,317) | ||||
| Mean | SE | P | Mean | SE | P | |||||
| 1 | 1,2,3 | 21,56,50 | 1–3 | 127 | 726.83 | 50.82 | 0.010 | 1303.48 | 42.00 | 0.060 |
| 2 | 4,5,6 | 22,9,26 | 1–6 | 184 | 710.14 | 39.58 | 0.141 | 1294.31 | 37.16 | 0.150 |
| 3 | 7,8,9 | 32,7,13 | 1–9 | 236 | 720.68 | 37.01 | 0.150 | 1332.18 | 33.05 | 0.150 |
| 4 | 10,11,12 | 14,14,42 | 1–12 | 306 | 709.98 | 41.20 | 0.150 | 1342.56 | 32.26 | 0.056 |
| 5 | 13,14,15 | 39,19,41 | 1–15 | 405 | 701.25 | 30.77 | 0.010 | 1332.58 | 28.06 | 0.150 |
| 6 | 16,17,18 | 17,2,10 | 1–18 | 434 | 723.13 | 28.79 | 0.150 | 1300.78 | 24.68 | 0.150 |
| 7 | 19,20,21 | 16,23,26 | 1–21 | 499 | 730.49 | 21.76 | 0.150 | 1319.40 | 20.04 | 0.150 |
| 8 | 22,23,24 | 50,14,35 | 1–24 | 598 | 712.68 | 22.51 | 0.150 | 1323.36 | 19.01 | 0.100 |
| 9 | 25,26,27 | 13,21,21 | 1–27 | 653 | 717.68 | 19.31 | 0.150 | 1330.75 | 15.57 | 0.150 |
| Step | Families used in each step |
Estimated intercept (True intercept = 14.13) |
Estimated Coeff. of age (True coeff. = 0.98) |
Estimated Coeff. of Waist (True coeff. = 0.53) |
||||||
| Mean | SE | P | Mean | SE | P | Mean | SE | P | ||
| 1 | 1–3 | 19.863 | 2.905 | 0.087 | 0.893 | 0.039 | 0.084 | 0.509 | 0.024 | 0.150 |
| 2 | 1–6 | 13.364 | 2.494 | 0.016 | 1.000 | 0.034 | 0.150 | 0.527 | 0.019 | 0.088 |
| 3 | 1–9 | 16.923 | 2.135 | 0.150 | 0.961 | 0.034 | 0.134 | 0.506 | 0.016 | 0.094 |
| 4 | 1–12 | 16.758 | 1.792 | 0.150 | 0.958 | 0.022 | 0.150 | 0.515 | 0.015 | 0.150 |
| 5 | 1–15 | 13.585 | 1.539 | 0.150 | 0.960 | 0.017 | 0.150 | 0.542 | 0.013 | 0.065 |
| 6 | 1–18 | 13.209 | 1.572 | 0.150 | 0.980 | 0.023 | 0.150 | 0.542 | 0.012 | 0.134 |
| 7 | 1–21 | 15.184 | 1.329 | 0.150 | 0.966 | 0.019 | 0.150 | 0.524 | 0.011 | 0.147 |
| 8 | 1–24 | 15.138 | 1.457 | 0.150 | 0.974 | 0.020 | 0.150 | 0.525 | 0.012 | 0.150 |
| 9 | 1–27 | 14.240 | 1.156 | 0.125 | 0.986 | 0.016 | 0.150 | 0.521 | 0.010 | 0.150 |
P, P value from Kolmogorov–Smirnov test for testing whether the 100 estimations of the parameter based on 100 times simulated data are from a normal distribution; Mean, mean of an estimated parameter based on the 100 estimations of the parameter; SE, standard error of an estimated parameter based on the 100 estimations of the parameter; Step, simulation step
Footnotes
Conflict of interest Nothing to declare.
Contributor Information
Wenyu Wang, Email: wenyu-wang@ouhsc.edu, Center for American Indian Health Research, College of Public Health, University of Oklahoma, Health Sciences Center, P. O. Box 26901, Oklahoma City, OK 73190, USA.
Elisa T. Lee, Center for American Indian Health Research, College of Public Health, University of Oklahoma, Health Sciences Center, P. O. Box 26901, Oklahoma City, OK 73190, USA
Barbara V. Howard, MedStar Health Research Institute, Washington, DC, USA
Richard R. Fabsitz, Epidemiology and Biometry Program, National Heart, Lung, and Blood Institute, Bethesda, MD, USA
Richard B. Devereux, Cornell University Medical Center, New York, NY, USA
Jean W. MacCluer, Department of Genetics, Southwest Foundation for Biomedical Research, San Antonio, TX, USA
Sandra Laston, Department of Genetics, Southwest Foundation for Biomedical Research, San Antonio, TX, USA.
Anthony G. Comuzzie, Department of Genetics, Southwest Foundation for Biomedical Research, San Antonio, TX, USA
Nawar M. Shara, MedStar Health Research Institute, Washington, DC, USA
Thomas K. Welty, Aberdeen Area Tribal Chairmen’s Health Board, Rapid City, SD, USA
References
- 1.Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 2.Diggle PJ, Heagerty P, Liang KY, Zeger SL. Analysis of longitudinal data. New York: Oxford University Press Inc.; 2002. [Google Scholar]
- 3.Heagerty PJ, Zeger SL. Marginalized multilevel models and likelihood inference. Stat Sci. 2000;15:1–26. [Google Scholar]
- 4.McCulloch CE, Searle SR. Generalized, linear, and mixed models. New York: Wiley; 2001. [Google Scholar]
- 5.North KE, Howard BV, Welty TK, Best LG, Lee ET, Yeh JL, Fabsitz RR, Roman MJ, MacCluer JW. Genetic and environmental contributions to cardiovascular disease risk in american indians: the strong heart family study. Am J Epidemiol. 2003;157:303–314. doi: 10.1093/aje/kwf208. [DOI] [PubMed] [Google Scholar]
- 6.Lee ET, Welty TK, Fabsitz R, Cowan LD, Le N-A, Oopik AJ, Cucchiara AJ, Savage PJ, Howard BV. The strong heart study—a study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol. 1990;132:1141–1155. doi: 10.1093/oxfordjournals.aje.a115757. [DOI] [PubMed] [Google Scholar]
- 7.Khoury MJ, Beaty TH, Cohen BH. Fundamentals of genetic epidemiology. New York: Oxford University Press; 1993. [Google Scholar]
- 8.Jacquard A. The genetic structure of population. New York: Springer; 1974. [Google Scholar]
- 9.Boichard D. PEDIG: a fortran package for pedigree analysis suited for large populations. Proceedings of the 7th world congress on genetics applied to livestock production; 2002-08-19/23; Montpellier. 2002. pp. 525–528. [Google Scholar]
- 10.Breslow NE, Clayton DG. Approximate inference in generalized linear mixed models. J Am Stat Assoc. 1993;88:9–25. [Google Scholar]
- 11.Wolfinger R, O’Connell M. Generalized linear mixed models: a pseudo-likelihood approach. J Stat Comput Simul. 1993;48:233–243. [Google Scholar]
- 12.Lange K, Westlake J, Spence MA. Extensions to pedigree analysis. III. Variance components by the scoring method. Ann Hum Genet. 1976;39:485–491. doi: 10.1111/j.1469-1809.1976.tb00156.x. [DOI] [PubMed] [Google Scholar]
- 13.Hopper JL, Mathews JG. Extensions to multivariate normal models for pedigree analysis. Ann Hum Genet. 1982;46:373–383. doi: 10.1111/j.1469-1809.1982.tb01588.x. [DOI] [PubMed] [Google Scholar]
- 14.Hopper JL. Variance components for statistical genetics: applications in medical research to characteristics related to human diseases and health. Stat Methods Med Res. 1993;2:199–223. doi: 10.1177/096228029300200302. [DOI] [PubMed] [Google Scholar]
- 15.Duggirala R, Williams JT, Williams-Blangero S, Blangero J. A variance component approach to dichotomous trait linkage analysis using a threshold model. Gen Epidemiol. 1997;14:987–992. doi: 10.1002/(SICI)1098-2272(1997)14:6<987::AID-GEPI71>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 16.Neale MC, Cardon LR. Methodology for genetic studies of twins and families. London: Kluwer; 1992. [Google Scholar]
- 17.Burton PR, Tiller KJ, Gurrin LC, Cookson WO, Musk AW, Palmer LJ. Genetic variance components analysis for binary phenotypes using generalized linear mixed models (GLMMs) and Gibbs sampling. Genet Epidemiol. 1999;17:118–140. doi: 10.1002/(SICI)1098-2272(1999)17:2<118::AID-GEPI3>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 18.Laird NM, Horvath S, Xu X. Implementing a unified approach to family based tests of association. Genet Epidemiol Suppl. 2000;19:S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 19.Lunetta KL, Faraone SV, Biederman J, Lair-d NM. Family-based tests of association and linkage that use unaffected sibs, covariates, and interactions. Am J Hum Genet. 2000;66:605–614. doi: 10.1086/302782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeger SL, Karim MR. Generalized linear models with random effects; a Gibbs sampling approach. J Am Stat Assoc. 1991;86:79–86. [Google Scholar]
- 21.Smith AFM, Roberts GO. Bayesian computation via the Gibbs sampler and related Markov Chain Monte Carlo Methods. J R Stat Soc B. 1993;55:3–23. [Google Scholar]
- 22.Cupples LA, Arruda HT, Benjamin EJ, D’Agostino RB, Sr, Demissie S, Destefano AL, et al. The Framingham Heart Study 100 K SNP genome-wide association study resource: Overview of 17 phenotype working group reports. BMC Med Genet. 2007;8(Suppl 1):S1. doi: 10.1186/1471-2350-8-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang KY, Beaty TH. Measuring familial aggregation using odds ratio regression models. Genet Epi. 1991;8:361–370. doi: 10.1002/gepi.1370080602. [DOI] [PubMed] [Google Scholar]
- 24.Liang KY. Estimating effects of probands’ characteristics on familial risk: I. Adjustment for censoring and correlated ages at onset. Genet Epidemiol. 1991;8:329–338. doi: 10.1002/gepi.1370080505. [DOI] [PubMed] [Google Scholar]
- 25.Pulver AE, Liang KY. Estimating effects of probands’ characteristics on familial risk: II. The association between ages at onset and familial risk in the Maryland schizophrenia sample. Genet Epidemiol. 1991;8:339–350. doi: 10.1002/gepi.1370080506. [DOI] [PubMed] [Google Scholar]
- 26.Lehmann EL. Theory of point estimation. New York: Wiley; 1983. [Google Scholar]