Abstract
Mutations in the WNT10A gene were first detected in the rare syndrome odonto-onycho-dermal dysplasia (OODD, OMIM257980) but have now also been found to cause about 35-50% of selective tooth agenesis (STHAG4, OMIM150400), a common disorder that mostly affects the permanent dentition. In our random sample of tooth agenesis patients, 40 percent had at least one mutation in the WNT10A gene. The WNT10A Phe228Ile variant alone reached an allele frequency of 0.21 in the tooth agenesis cohort, about 10 times higher than the allele frequency reported in large SNP data bases for Caucasian populations. Patients with bi-allelic WNT10A mutations have severe tooth agenesis while heterozygous individuals are either unaffected or have a mild phenotype. Mutations in the coding areas of the WNT10B gene which is co-expressed with WNT10A during odontogenesis, and the WNT6 gene which is located at the same chromosomal locus as WNT10A in humans, do not contribute to the tooth agenesis phenotype.
Keywords: WNT10A, gene mutations, selective tooth agenesis, ectodermal dysplasia, WNT10B, WNT6
INTRODUCTION
Adaimy et al. [2007] performed autozygosity mapping in three consanguineous Lebanese families with the rare odonto-onycho-dermal-dysplasia syndrome (OODD, OMIM 257980) which had been previously characterized phenotypically in the same population [Fadhil et al., 1983]. They found that all affected family members were homozygous for the same nonsense mutation in the WNT10A gene leading to the phenotypic features of severe hypodontia, onychodysplasia, smooth tongue as well as palmar and plantar hyperhidrosis and hyperkeratosis. The phenotype of heterozygous family members was not recorded. Two years later Bohring et al. [2009] reported that WNT10A mutations are not restricted to the rare OODD syndrome but also found in other ectodermal dysplasia entities like the Schöpf-Schulz-Passarge syndrome (OMIM 224750) which additionally features eyelid cysts and predisposition to adnexal skin tumors. Bohring et al. also described a high prevalence of apparently non-syndromic tooth agenesis among their homozygous patients as well as mild, predominantly dental symptoms in about half of the heterozygous family members. Three further reports about the high prevalence of WNT10A mutations in ectodermal dysplasia syndromes and in non-syndromic tooth agenesis followed in 2011 by Cluzeau et al., in 2012 by van den Boogaard et al., and in 2013 by Plaisancié et al.
The expression of Wnt10a along with Wnt10b, Shh, Bmps2 and 4 and other developmentally active gene products during mouse odontogenesis had been investigated as early as 1998 by Dassule and McMahon. They detected Wnt10a at around embryonic day 12 (E12) by in situ hybridization in the inner epithelial/ enamel knot area of the tooth bud, where it was co-expressed with Wnt10b. Since Wnt10b expression was recognizable a little earlier and more prominently than Wnt10a expression, further investigations in this study focused on the Wnt10b molecule. At later stages of tooth development (E14 to E18), Wnt10a can also be found in the mesenchymal preodontoblast layer where it contributes to or initiates odontoblast differentiation, possibly through the up-regulation of dentin sialophosphoprotein (Dspp) expression [Yamashiro et al., 2007].
WNT10A, which is located adjacent to WNT6 at 2q35 in humans, is also active during the development of hair follicles and limbs, and in hematopoiesis. In adult tissues it is expressed in lymph nodes, blood, adrenal gland, prostate, testis, ovary, retina, brain, lung and kidney; and may also play a role in several neoplastic disorders, notably ameloblastomas, keratocystic odontogenic tumors, lymphomas and leukemias but is also found up-regulated in several cancers. Functional studies showed that Wnt10a activates the canonical wnt pathway and regulates mesenchymal cell fate in that it inhibits adipogenesis and stimulates osteoblastogenesis [Cawthorn et al., 2012].
The general role of canonical wnt signaling during tooth development has been explored in more detail by stabilization of β-catenin or depletion of apc, a positive and a negative regulator of canonical wnt signaling respectively. Both procedures constitutively activate wnt signaling in tooth bud epithelium leading to the formation of many accessory tooth buds sprouting from the original tooth anlage. The resulting supernumerary teeth are often small but otherwise completely normal [Järvinen et al., 2006; Wang et al., 2009]. Interestingly, the expression of Pax9 and Msx1, two normally essential transcription factors in tooth bud mesenchyme, are not required for the formation of these supernumerary teeth. Furthermore, the inactivation of the Wnt secretion facilitator Wls was recently shown to prevent intraepithelial Wnt signaling leading to tooth developmental arrest [Zhu et al., 2013].
When we sequenced the WNT10A gene (in 2012) in the random collection of tooth agenesis patients who participate in our “missing tooth” study, we also found a large number of WNT10A mutations in our samples confirming the importance of WNT10A in tooth development.
MATERIALS AND METHODS
Patient recruitment
Tooth agenesis study participants were recruited via website and through collaboration with Drs. Alexandre Vieira (University of Pittsburgh) and Ophir Klein (University of California at San Francisco), following IRB approved protocols. People of all ages with any number of missing teeth except third molars were included, and only patients with overt ectodermal dysplasia symptoms were excluded. The final cohort consisted of 90 unrelated samples; half of them were from Caucasian Americans and the other half from patients from Turkey which are considered to be mostly of Mediterranean- European ancestry. Cheek swab or saliva samples were collected for the isolation of genomic DNA. The wild type sequence and allele frequencies of common variants in control populations were obtained from the NCBI SNP data bases as well as the NHLBI Exome sequencing project (ESP).
DNA Extraction from Buccal Swabs
DNA extraction was performed with the Puregene Buccal Cell Kit (Qiagen). Since buccal swabs do not yield sufficient material for the analysis of multiple genes, the samples were amplified by Whole Genome Amplification (WGA) with the REPLI-g WGA system (Qiagen) Successful genome amplification was verified by gel electrophoresis of amplified samples together with a quantitation marker. DNA samples received from our collaborators were also amplified by WGA.
Polymerase Chain Reaction and Sequencing of Products
Exons of the WNT10a, WNT10b and WNT6 genes were PCR amplified with GoTaq reagents (Promega) using a 96-well plate format for the 90 samples and the controls. Quality and quantity of PCR products was confirmed by gel-electrophoresis, followed by treatment with ExoSapIt (USB) and addition of the sequencing primers. Automated dideoxy chain terminator sequencing was done by GenScript, Piscataway, NJ.
Analysis of Sequencing Results
All sequences were visually inspected for heterozygous base changes and compared to the corresponding wild type sequences using the ‘BLAST’ program. Once a nucleotide change was found, the SNP (single nucleotide polymorphism) data base was consulted to determine if the SNP is a common polymorphism. For appropriate SNPs, the allele frequencies were compared between the experimental groups consisting of Caucasian and Turkish samples; and between the experimental and the Caucasian population control groups reported in NCBI and NHLBI data bases.
RESULTS
Wnt10A but neither WNT6 nor WNT10B contribute to tooth agenesis
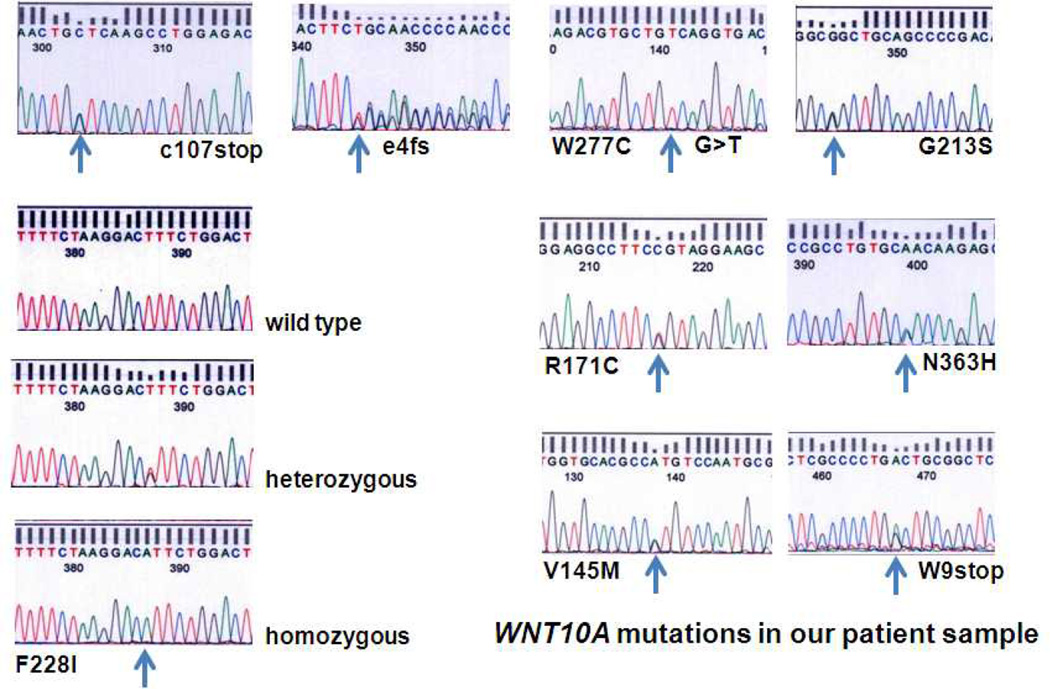
About 40 percent of our random group of tooth agenesis patients had at least one missense, nonsense or frameshift mutation in the WNT10A gene. The different mutations encountered in our patient samples are shown in Figure 1. Most common was the mono- or bi-allelic Phe228Ile mutation with a prevalence of about 31 percent and an allele frequency of 0.21 compared to an allele frequency of about 0.02 in large Caucasian control populations and only 0.007 in African American controls (Table I). The allele frequency of Phe228Ile was also calculated separately for our Caucasian (0.216) and Turkish participants (0.20) to exclude any influence that ethnic background differences could have had on the allele frequency.
Figure 1.
WNT10A mutations found in our tooth agenesis patient cohort; e4=exon4 of WNT10A, fs=frame shift, F=Phenylalanine, I=Isoleucine, W=Tryptophan, N=Asparagine, R=Arginine, V=Valine, M=Methionine, H=Histidine, C=Cysteine, G=Glycine, S=Serine.
Table I.
The WNT10A mutation Phe228Ile is unlikely to be a common polymorphism since its allele frequency is about 10 times higher in the tooth agenesis group than it is in several large control groups.
| Source | Ethnicity/ study group |
Number of detected alleles |
F228I allele frequency |
|---|---|---|---|
| Our Tooth Agenesis (TA) study | Caucasians with TA | 127 Phe and 33 IIle | 0.206 |
| NCBI SNP data base | Caucasians | 3348 Phe and 67 Ile | 0.02 |
| NHLBI Exome Sequencing Project (ESP) | Caucasians | 8389 Phe and 209 Ile | 0.024 |
| NHLBI Exome Sequencing Project (ESP) | African Americans | 4370 Phe and 32 Ile | 0.007 |
| Bohring et al. 2009 Am J Hum Genet 85: 97 | Caucasians without TA | 396 Phe and 2 Ile | 0.005 |
Since Phe228Ile is so much more common in Caucasian tooth agenesis patients, it either is the causative factor or is closely linked to the causative mutation. Since WNT10A is located only a few kb telomeric of WNT6 on chromosome 2q35 we included the latter in our analysis but did not find any mutations or polymorphisms that were syntenic with the nucleotide change leading to WNT10A Phe228Ile. The common WNT6 variant Pro155Arg occurred in our tooth agenesis population at a frequency similar to normal control populations and only one of the 10 patients who had the WNT6 Pro155Arg variant also had WNT10A Phe228Ile.
Since the WNT10A and WNT10B proteins are co-expressed in the inner dental epithelium of developing teeth and share 62 percent identity, it was conceivable that WNT10B mutations may also cause missing teeth. But sequencing of the whole coding area of the WNT10B gene did not reveal any nucleotide changes that could possibly be implicated in the tooth agenesis phenotype.
Phenotypes associated with WNT10A mutations
We did not receive any reports about missing primary teeth although some patients remembered having relatively small deciduous teeth. The number of missing teeth in the permanent dentition depended strongly on whether the affected individual was heterozygous or homozygous/ compound heterozygous for WNT10A mutations. Heterozygous patients were missing up to 6 permanent teeth while homozygous patients were generally missing from 6 to 26, most often 16 teeth.
While all patients with bi-allelic mutations had oligodontia, many heterozygous relatives of study participants were not affected suggesting incomplete penetrance (Fig 2). Syndromic ectodermal dysplasia manifestations were not encountered in our study population because they constituted exclusion criteria for participation in the study. One study participant however had a history of benign skin tumors, possibly bearing some resemblance to the Schöpf-Schulz-Passarge syndrome and another one reported mild heat intolerance.
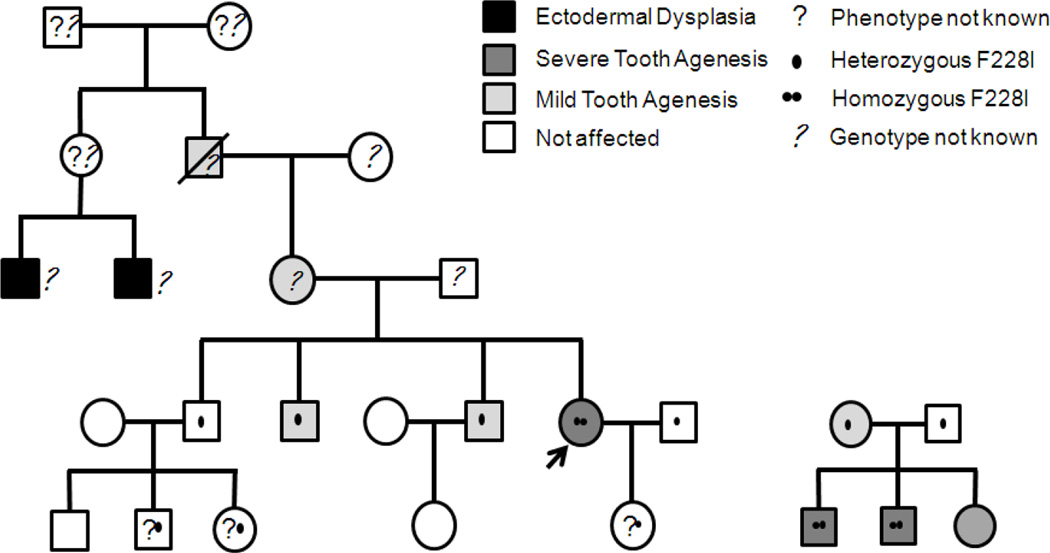
Figure 2.
Two pedigrees showing incomplete penetrance and variability of the phenotype depending on the number of WNT10A alleles affected. The fifth generation of the pedigree on the left is still too young for phenotype evaluation. An additional feature in this family is that the husband of the homozygous index patient (arrow) also carries the F228I variant, a 1 in 50 chance. The pedigree on the right also shows that two different tooth agenesis genes may contribute to the phenotypes, because the daughter has several missing teeth but no F228I mutation while her two homozygous brothers have severe tooth agenesis. F=Phenylalanine, I=Isoleucine
The tooth agenesis pattern in heterozygous patients parallels that of common mild tooth agenesis with a predominant absence of lower second premolars and upper lateral incisors; but mandibular incisors are also frequently absent, occasionally even a canine or a first premolar. The pattern is similar to EDA pathway associated selective tooth agenesis in that the anterior teeth are more often affected [Tarpey et al. 2007; Mues et al. 2009, 2010]. Patients with homozygous WNT10a mutations have also posterior tooth agenesis similar to syndromic EDA pathway mutation phenotypes [Lexner et al., 2006].
DISCUSSION
Traditionally we distinguished between syndromic and non-syndromic tooth agenesis. Syndromic tooth agenesis was most often encountered as part of an ectodermal dysplasia phenotype while nonsyndromic tooth agenesis, also called isolated or selective tooth agenesis/hypodontia (STHAG1-6 and X1 in OMIM), should only affect the dentition without any systemic manifestations. Inherent in the syndromic versus nonsyndromic classification was the assumption that the two disorders had fundamentally different genetic causations.
However, the more we learn about the pathogenesis of tooth agenesis the more we realize that there is an extensive overlap between the genetic basis of syndromic and nonsyndromic forms of tooth agenesis [Nieminen, 2009] and the phenotypic distinction may not have been helpful for the search of additional TA genes. From a genetic point of view, nonsyndromic and syndromic tooth agenesis are often caused by the same genes, but the development of some teeth seems to be more sensitive to gene dosage and thus are the first organs to be affected, while other ectodermal appendices may still form normally [Mues et al., 2010].
The distinction between syndromic and nonsyndromic TA may also be problematic with respect to the new, biologically based diagnostic and therapeutic approaches for which dental professionals who are traditionally the only health care providers for nonsyndromic TA patients, may still be little prepared. Classification of the STHAGs as ectodermal dysplasia entities may therefore be desirable from a clinical point of view.
The high prevalence of WNT10A mutations is truly astounding, especially since this gene is hardly ever mentioned in the extensive literature about the molecular genetics of tooth development. Even more surprising is the large number of Caucasian tooth agenesis patients with one particular mutation, WNT10A Phe228Ile. Pathogenic mutations are usually lost from the gene pool of a population unless they have some kind of survival advantage like for example heterozygous mutations in the hemoglobin genes which are of benefit in areas with high malaria incidence. The survival advantage is often lost in individuals with homozygous mutations like in sickle cell disease, but since homozygously affected individuals are quite rare, the mutation has an overall positive effect on population growth. It will certainly be interesting to find a cause for the high prevalence of WNT10A Phe228Ile mutations.
Interesting is also that the tooth agenesis pattern of patients with WNT10A mutations resembles that of EDA pathway associated hypodontia. Both WNT and EDA pathways are known to operate predominantly in the epithelial layer of the developing tooth and repeated interactions between the two pathways have been observed during ectodermal appendage formation and in vitro [Laurikkala et al., 2001; Durmowicz et al., 2002; Zhang et al., 2009]. It is therefore possible that the new, biological replacement therapies for EDA pathway mutation associated disorders may also be of value in the much more common WNT10A disorder. It would certainly be worth testing this possibility because EDA replacement therapies have shown great efficacy in ameliorating disease symptoms in animals and are currently tested in humans, while the generation of WNT10A specific therapeutics would be quite complicated not only from a chemical engineering point of view but also because of the lack of a one to one correspondence of ligands and receptors in the WNT pathway, potentially leading to severe adverse effects, and even more importantly, a lack of a wnt10a deficient, diphyodont animal model for the testing of these therapeutics.
ACKNOWLEDGMENTS
We thank all patients for their participation in this study which was supported by NIH/NIDCR awards R03DE019554-01 and R01DE019471-01 and-02. At the Pittsburgh site, we would like to thank Kathleen Deeley, Megan Weber, and Jacqueline Noel for providing support in sample handling and internal quality control. Some of the samples were collected with the support of Asli Patir and Mine Yildirim. The University of Pittsburgh Dental Registry and DNA Repository is supported by the University of Pittsburgh School of Dental Medicine and NIH Grant UL1 RR024153.
REFERENCES
- Adaimy L, Chouery E, Megarbane H, Mroueh S, Delague V, Nicolas E, Belguith H, de Mazancourt P, Megarbane A. Mutation in WNT10A is associated with an autosomal recessive ectodermal dysplasia: the odonto-onycho-dermal dysplasia. Am J Hum Genet. 2007;81:821–828. doi: 10.1086/520064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohring A, Stamm T, Spaich C, Haase C, Spree K, Hehr U, Hoffmann M, Ledig S, Sel S, Wieacker P, Ropke A. WNT10A mutations are a frequent cause of a broad spectrum of ectodermal dysplasias with sex-biased manifestation pattern in heterozygotes. Am J Hum Genet. 2009;85:97–105. doi: 10.1016/j.ajhg.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthorn WP, Bree AJ, Yao Y, Du B, Hemati N, Martinez-Santibañez G, MacDougald OA. Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a β-catenin-dependent mechanism. Bone. 2012;50(2):477–489. doi: 10.1016/j.bone.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluzeau C, Hadj-Rabia S, Jambou M, Mansour S, Guigue P, Masmoudi S, Bal E, Chassaing N, Vincent MC, Viot G, Clauss F, Manière MC, Toupenay S, Le Merrer M, Lyonnet S, Cormier-Daire V, Amiel J, Faivre L, de Prost Y, Munnich A, Bonnefont JP, Bodemer C, Smahi A. Only four genes (EDA1, EDAR, EDARADD, and WNT10A) account for 90% of hypohidrotic/anhidrotic ectodermal dysplasia cases. Hum Mutat. 2010;32:70–77. doi: 10.1002/humu.21384. [DOI] [PubMed] [Google Scholar]
- Dassule HR, McMahon AP. Analysis of epithelial-mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Dev Biol. 1998;202(2):215–227. doi: 10.1006/dbio.1998.8992. [DOI] [PubMed] [Google Scholar]
- Durmowicz MC, Cui C-Y, Schlessinger D. The EDA gene is a target of but does not regulate Wnt signaling. Gene. 2002;285:203–211. doi: 10.1016/s0378-1119(02)00407-9. [DOI] [PubMed] [Google Scholar]
- Fadhil M, Ghabra TA, Deeb M, Der Kaloustian VM. Odontoonychodermal dysplasia: a previously apparently undescribed ectodermal dysplasia. Am J Med Genet. 1983;14:335–346. doi: 10.1002/ajmg.1320140213. [DOI] [PubMed] [Google Scholar]
- Järvinen E, Salazar-Ciudad I, Birchmeier W, Taketo MM, Jernvall J, Thesleff I. Continuous tooth generation in mouse is induced by activated epithelial Wnt/beta-catenin signaling. PNAS. 2006;103:18627–18632. doi: 10.1073/pnas.0607289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurikkala J, Mikkola M, Mustonen T, Aberg T, Koppinen P, Pispa J, Nieminen P, Galceran J, Grosschedl R, Thesleff I. TNF signaling via the ligand-receptor pair ectodysplasin and edar controls the function of epithelial signaling centers and is regulated by Wnt and activin during tooth organogenesis. Dev Biol. 2001;229(2):443–455. doi: 10.1006/dbio.2000.9955. [DOI] [PubMed] [Google Scholar]
- Lexner MO, Bardow A, Hertz JM, Nielsen LA, Kreiborg S. Anomalies of tooth formation in hypohidrotic ectodermal dysplasia. Internat J Paed Dent. 2006;17:10–18. doi: 10.1111/j.1365-263X.2006.00801.x. [DOI] [PubMed] [Google Scholar]
- Mues G, Tardivel A, Willen L, Kapadia H, Seaman R, Frazier-Bowers S, Schneider P, D'Souza RN. Functional analysis of Ectodysplasin-A mutations causing selective tooth agenesis. Eur J Hum Genet. 2010;18(1):19–25. doi: 10.1038/ejhg.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mues G, Griggs R, Hartung AJ, Whelan G, Best LG, Srivastava AK, D’Souza R. From Ectodermal Dysplasia to Selective Tooth Agenesis. Am J Med Genet. 2009;149A(9):2037–2041. doi: 10.1002/ajmg.a.32801. [DOI] [PubMed] [Google Scholar]
- Nieminen P. Genetic basis of tooth agenesis. J Exp Zool (Mol Dev Evol) 2009;312B:320–342. doi: 10.1002/jez.b.21277. [DOI] [PubMed] [Google Scholar]
- Plaisancié J, Bailleul-Forestier I, Gaston V, Vaysse F, Lacombe D, Holder-Espinasse M, Abramowicz M, Coubes C, Plessis G, Faivre L, Demeer B, Vincent-Delorme C, Dollfus H, Sigaudy S, Guillén-Navarro E, Verloes A, Jonveaux P, Martin-Coignard D, Colin E, Bieth E, Calvas P, Chassaing N. Mutations in WNT10A are frequently involved in oligodontia associated with minor signs of ectodermal dysplasia. Am J Med Genet. 2013;161A:671–678. doi: 10.1002/ajmg.a.35747. [DOI] [PubMed] [Google Scholar]
- Tarpey P, Pemberton TJ, Stockton DW, Das P, Ninis V, Edkins S, Futreal PA, Wooster R, Kamath S, Nayak R, Stratton MR, Patel PI. A novel Gln358Glu mutation in ectodysplasin A associated with X-linked dominant incisor hypodontia. Am J Med Genet. 2007;143A:390–394. doi: 10.1002/ajmg.a.31567. [DOI] [PubMed] [Google Scholar]
- Van den Boogaard MJ, Creton M, Bronkhorst Y, van der Hout A, Hennekam E, Lindhout D, Cune M, Ploos van Amstel HK. Mutations in WNT10A are present in more than half of isolated hypodontia cases. J Med Genet. 2012;49:327–331. doi: 10.1136/jmedgenet-2012-100750. [DOI] [PubMed] [Google Scholar]
- Wang XP, O'Connell DJ, Lund JJ, Saadi I, Kuraguchi M, Turbe-Doan A, Cavallesco R, Kim H, Park PJ, Harada H, Kucherlapati R, Maas RL. Apc inhibition of Wnt signaling regulates supernumerary tooth formation during embryogenesis and throughout adulthood. Development. 2009;136(11):1939–1949. doi: 10.1242/dev.033803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro T, Zheng L, Shitaku Y, Saito M, Tsubakimoto T, Takada K, Takano-Yamamoto T, Thesleff I. Wnt10a regulates dentin sialophosphoprotein mRNA expression and possibly links odontoblast differentiation and tooth morphogenesis. Differentiation. 2007;75:452–462. doi: 10.1111/j.1432-0436.2006.00150.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Tomann P, Andl T, Gallant NM, Huelsken J, Jerchow B, Birchmeier W, Paus R, Piccolo S, Mikkola ML, Morrisey EE, Overbeek PA, Scheidereit C, Millar SE, Schmidt-Ullrich R. Reciprocal requirements for EDA/EDAR/NF-kappaB and Wnt/beta-catenin signaling pathways in hair follicle induction. Dev Cell. 2009;17:49–61. doi: 10.1016/j.devcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Zhao P, Liu Y, Zhang X, Fu J, Yu HM, Qiu M, Chen Y, Hsu W, Zhang Z. Intra-epithelial requirement of canonical Wnt signaling for tooth morphogenesis. J Biol Chem. 2013;288:12080–12089. doi: 10.1074/jbc.M113.462473. [DOI] [PMC free article] [PubMed] [Google Scholar]