Abstract
Purpose
SPINK1 over-expression has been described in prostate cancer and is linked with poor prognosis in many cancers. The objective of this study was to characterize the association between SPINK1 over-expression and prostate cancer specific survival.
Experimental Design
The study included 879 participants in the US Physicians’ Health Study and Health Professionals Follow–Up Study, diagnosed with prostate cancer (1983 – 2004) and treated by radical prostatectomy. Protein tumor expression of SPINK1 was evaluated by immunohistochemistry on tumor tissue microarrays.
Results
74/879 (8%) prostate cancer tumors were SPINK1 positive. Immunohistochemical data was available for PTEN, p-Akt, pS6, stathmin, androgen receptor (AR) and ERG (as a measure of the TMPRSS2:ERG translocation). Compared to SPINK1 negative tumors, SPINK1 positive tumors showed higher PTEN and stathmin expression, and lower expression of AR (p<0.01). SPINK1 over-expression was seen in 47 of 427 (11%) ERG negative samples and in 19 of 427 (4%) ERG positive cases (p=0.0003). We found no significant associations between SPINK1 status and Gleason grade or tumor stage. There was no association between SPINK1 expression and biochemical recurrence (p=0.56). Moreover, there was no association between SPINK1 expression and prostate cancer mortality (there were 75 lethal cases of prostate cancer during a mean of 13.5 years follow-up [HR 0.71 (95% confidence interval 0.29–1.76)]).
Conclusions
Our results suggest that SPINK1 protein expression may not be a predictor of recurrence or lethal prostate cancer amongst men treated by radical prostatectomy. SPINK1 and ERG protein expression do not appear to be entirely mutually exclusive, as some previous studies have suggested.
Keywords: SPINK1, ERG, prostate cancer, survival, radical prostatectomy
INTRODUCTION
SPINK1 encodes for a 56 amino acid peptide which is secreted in the prostate gland and whose function is to inhibit serine proteases such as trypsin (1). Recently, SPINK1 was identified in a meta-analysis as having outlier expression in ETS rearrangement negative prostate cancers and results indicated that SPINK1 was expressed exclusively in TMPRSS2:ERG negative prostate cancers (2). These data suggested that SPINK1 overexpression may represent a distinct prostate cancer subtype. Moreover, SPINK1 overexpression has been retrospectively associated with an increased risk of disease progression and biochemical recurrence in hormonally and surgically treated prostate cancer cohorts (2–3). Ateeq et al. demonstrated that SPINK1 positive cancers may potentially be targeted therapeutically through humanized SPINK-1 directed monoclonal antibodies and epidermal growth factor receptor (EGFR) inhibition (4). No study to date, however, has addressed the association between SPINK1 over-expression and prostate cancer specific survival in patients treated by radical prostatectomy.
MATERIALS AND METHODS
This study was based upon the analysis of men treated with radical prostatectomy for prostate cancer who were participants in the Physicians’ Health Study and Health Professionals Follow-up Study, and included 879 prostate cancer cases, diagnosed between 1983 and 2004, on whom archival formalin-fixed, paraffin-embedded tumor tissue specimens were available (5–6). Tumor tissue from radical prostatectomies was reviewed by our pathology team to provide uniform evaluation of Gleason score and to identify areas of high-density tumor for construction of tumor tissue microarrays. At least three tumor cores (0.6 mm) were sampled from each case (three cores were taken at a minimum from the same dominant tumor nodule with the highest Gleason score).
Immunohistochemistry was performed on 4–5μm sections of the tissue microarrays to assess protein expression of SPINK1 (mouse monoclonal, 1:100 dilution; H00006690-M01, Abnova, Taipei City, Taiwan), ERG (rabbit monoclonal, 1:200 dilution; EPR3864, Epitomics Inc., Burlingame, CA), PTEN (rabbit polyclonal, 1:200 dilution; PN37, Zymed Laboratories, San Francisco, CA), p-AKT (rabbit monoclonal, 1:50 dilution, D9E; Cell Signaling, Danvers, MA), pS6 (rabbit monoclonal, 1:50 dilution, Ser240/Ser244; Cell Signaling, Danvers, MA), stathmin (rabbit polyclonal, 1:50 dilution, Cell Signaling, Danvers, MA), Androgen Receptor (AR, rabbit polyclonal, 1:50 dilution; PG-21, Upstate Cell Signaling, Danvers, MA) and cell proliferation marker Ki67 (rabbit polyclonal, 1:2000 dilution; Vector Labs). To validate concomitant ERG/SPINK1 staining a dual stain for ERG (rabbit monoclonal, 1:1000 dilution, EPR3864, Abcam, Cambridge, MA) and SPINK1 (1:50 dilution) was performed on three whole tumor sections and the percentage of prostate tumor glands that co-express both proteins semiquantified. TUNEL assay was performed on 5μm sections of the tissue microarrays to identify the percentage of tumor cells undergoing apoptosis using the Apoptag Peroxidase In-Situ kit (Chemicon International) (7).
SPINK1 and ERG expression were classified as positive or negative by study pathologists as previously described (2, 8) (Fig. 1). Cases with SPINK1 staining in any cancerous epithelial cells were deemed SPINK1 positive (2). A case was called ERG positive if at least one core from an individual case had positive ERG staining observed within prostate cancer epithelial cells. For all cases, the presence of ERG staining in the vasculature endothelium served as a positive internal control, and assessment of ERG expression was restricted to cores in which the positive internal control was observed (8). Additionally each TMA contained internal controls including duplicate cores and normal prostate. All cases were double-read by a study pathologist to validate initial scores and non-informative cases were eliminated from the downstream analysis. Prior studies have shown that ERG overexpression is highly concordant with ERG rearrangement status as assessed by fluorescent in-situ hybridization (9, 10) and quantitative polymerase chain reaction (11). Expression of PTEN, p-AKT, pS6, stathmin, AR, and Ki-67 was quantified using the Ariol instrument SL-50 image analysis software (Applied Imaging, San Jose, CA) and results validated by manual quantification of scores in an estimated 5% of all tissue cores. Semi-automated assessment of staining intensity (scale: 0–255) and percent staining (scale: 0–100%) was performed using the MultiStain assay. The mean percent staining across cores was used as a measure of PTEN, pAkt, pS6 and stathmin expression. The mean nuclear staining intensity across cores was used as a measure of AR expression. Ki67 proliferation index was defined as the number of stained nuclei over the total number of tumor nuclei. For TUNEL, the Apoptag sum was calculated as the number of positive cells out of the total number of tumor cells. The whole area of each tumor tissue microarray core was evaluated for the sum. Areas of tumor were manually identified with masking of the stroma and normal/benign glands from image analysis as previously described (12).
Figure 1.
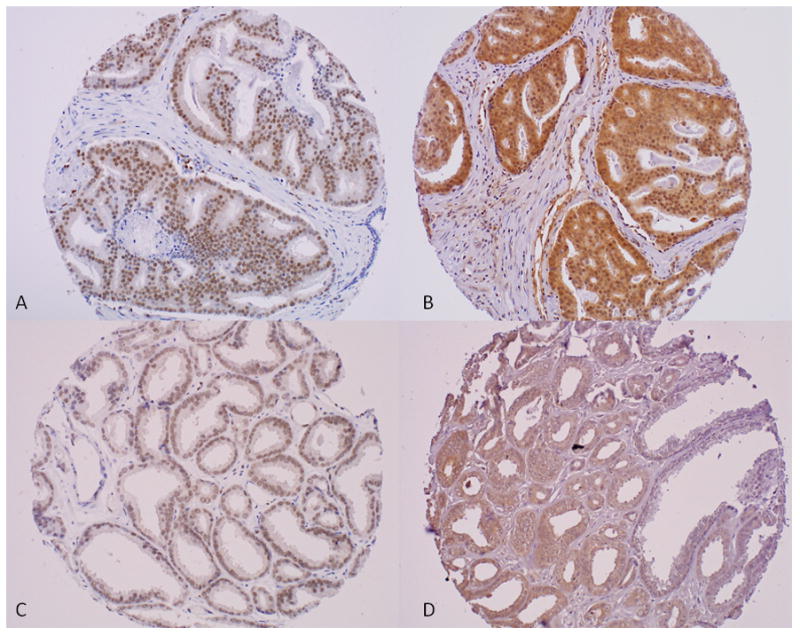
Immunohistochemistry for nuclear marker ERG (left hand panel) and cytoplasmic marker SPINK1 (right hand panel) showing diffuse positive staining in the exact same tumor cores from a case of prostate adenocarcinoma Gleason score 4+4 (A–B; x20) and from a case of prostate adenocarcinoma Gleason score 3+3 (C–D; x20).
Information on tumor stage, prostate specific antigen (PSA) level at diagnosis, and treatments were abstracted from medical records and pathology reports. Since 2000, newly diagnosed prostate cancer patients have been followed for biochemical recurrence and development of metastatic disease via mailed questionnaires. For men with prostate cancer in the Health Professionals Follow-up Study, their treating physicians were contacted to collect information about their clinical course and to confirm development of metastases. For men with prostate cancer in the Physicians’ Health Study, self-report of metastases by these physician participants was virtually always confirmed when records were available (among 80% of Physicians’ Health Study cases), so all metastatic cases were included as outcomes. Biochemical recurrence was participant reported, reported by the treating physician, or abstracted from medical records; defined as PSA above 0.2 ng/mL post-surgery sustained over two measures when abstracted from medical records. Cause of death is assigned following a centralized review of medical records and death certificates by study physicians. Follow-up for mortality is greater than 95% in both cohorts (in the Physicians’ Health Study mortality follow-up is greater than 99%).
We included men who had undergone radical prostatectomy and on whom we had SPINK1 status available (N=879; 364 men from the Physicians’ Health Study and 515 men from the Health Professionals Follow-up Study). We investigated whether age at diagnosis and follow-up time differed by SPINK1 status using t-tests. To test associations with Gleason score and pathological tumor stage, we used Chi-Square tests or Cochrane-Armitage trend tests. The association between SPINK1 status and PSA level at diagnosis, and between SPINK1 status and expression of Ki67, TUNEL, PTEN, pAKT, pS6, stathmin and AR, was tested using Wilcoxon rank-sum tests.
We used Cox proportional hazards models to calculate hazard ratios (HRs) and 95% confidence intervals (CIs) of the association between SPINK1 status and disease progression. Prostate cancer progression was defined as (1) time to lethal prostate cancer, defined as development of distant metastases or prostate cancer death, and (2) time to biochemical recurrence. Men who did not report a PSA rise but who reported lymph node metastases, distant metastases, or who died of prostate cancer were assigned a biochemical recurrence on the earliest date of any of these events. Men in the cohort were followed from the date of prostate cancer diagnosis until they experienced outcomes, until they were censored at death from other causes, or at end of follow-up, whichever occurred first. Follow-up for death extended through March 2011 for the Physicians’ Health Study and December 2011 the Health Professionals Follow-up Study. In both cohorts, follow-up for prostate cancer recurrence and metastases ended approximately two years before follow-up for death due to questionnaire timing. Men with missing information on pathological tumor stage (n=32) were assigned a missing indicator variable. We also conducted multivariable analyses limited to men with known tumor stage (n=847). The study was approved by the institutional review boards at the Harvard School of Public Health and Partners Healthcare.
RESULTS
Data on both SPINK1 and ERG expression were available for 854 men, SPINK1 and Ki-67 expression for 778 men, SPINK1 and TUNEL expression for 675 men, SPINK1 and PTEN expression for 761 men, SPINK1 and p-AKT expression for 741 men, SPINK1 and pS6 expression for 746 men, SPINK1 and stathmin expression for 743 men, and SPINK1 and AR expression for 802 men. The mean age at diagnosis was 65.4 years. The mean follow-up time was 13.5 years. In total, 75 men developed lethal prostate cancer, 213 men developed biochemical recurrence, and 260 men died of any cause during follow-up.
Table 1 presents clinical characteristics amongst the men with prostate cancer overall, as well as stratified by SPINK1 status. Eight percent of the men in the cohort had SPINK1 positive tumors. We found no significant associations between SPINK1 status and clinico-pathologic features including cell proliferation marker Ki-67 and TUNEL (apoptotic marker). There was no significant association between SPINK1 status and biochemical recurrence or lethal prostate cancer (Table 2). These results did not vary significantly by cohort, and did not materially change when we restricted the multivariate analyses to men with known pathological stage.
Table 1.
Clinical Characteristics for all men and by SPINK1 expression status among 879a men treated with radical prostatectomy for prostate cancer, Physicians’ Health Study and Health Professionals Follow-up Study cohorts
| Characteristic | All Men | SPINK1 Negative | SPINK1 Positive | Pb |
|---|---|---|---|---|
| Number | 879 | 805 | 74 | |
| Mean Follow-Up Time, years (SDc) | 13.5 (4.6) | 13.4 (4.6) | 13.6 (4.5) | 0.78 |
| Mean Age at Diagnosis, years (SD) | 65.4 (6.0) | 65.4 (6.0) | 65.0 (6.2) | 0.54 |
| Median PSA at Diagnosis, ng/ml (IQRd) | 7.0 (5.6) | 7.0 (5.6) | 7.0 (7.5) | 0.88 |
| Tumor Stage, n (%) | ||||
| pT2 N0/Nx | 599 (71) | 547 (71) | 52 (71) | |
| pT3 N0/Nx | 222 (26) | 204 (26) | 18 (25) | |
| pT4/N1/M1 | 26 (3) | 23 (3) | 3 (4) | 1.00 |
| Gleason Score, n (%) | ||||
| 2–6 | 185 (21) | 173 (21) | 12 (16) | |
| 3+4 | 325 (37) | 298 (37) | 27 (36) | |
| 4+3 | 214 (24) | 194 (24) | 20 (27) | |
| 8–10 | 155 (18) | 140 (17) | 15 (20) | 0.25 |
| Lethal Prostate Cancere, n (%) | ||||
| No | 804 (91) | 735 (91) | 69 (93) | |
| Yes | 75 (9) | 70 (9) | 5 (7) | 0.57 |
| Biochemical Recurrence, n (%) | ||||
| No | 666 (76) | 612 (76) | 54 (73) | |
| Yes | 213 (24) | 193 (24) | 20 (27) | 0.56 |
| All Cause Mortalityf, n (%) | ||||
| No | 619 (70) | 568 (71) | 51 (69) | |
| Yes | 260 (30) | 237 (29) | 23 (31) | 0.77 |
| Ki67 expression, median (IQR)g | 0. 12 (0.45) | 0. 12 (0.45) | 0.11 (0.39) | 0.70 |
| TUNEL, median (IQR)h | 0.5 (2.0) | 0.5 (2.0) | 0.5 (2.0) | 0.11 |
| ERG negative, n | 427 | 380 | 47 | |
| ERG positive, n | 427 | 408 | 19 | 0.0003 |
Numbers may not add up to 879 because men with missing information for a characteristic are not included in that characteristic.
P-values are based on the following tests: t-test for follow-up time and age at diagnosis; Wilcoxon rank sums test for PSA at diagnosis, Ki67 expression, and TUNEL; exact Cochran-Armitage trend test for tumor stage, Cochran-Armitage trend test for Gleason sum; Chi-square test for lethal prostate cancer, biochemical recurrence, all-cause mortality, and ERG status.
SD: standard deviation.
IQR: inter-quartile range.
Lethal prostate cancer includes metastases to distant organs, and prostate cancer death.
All-cause mortality includes prostate cancer death and death due to any other cause
Ki67 expression was available for 778 men with known SPINK1 status.
TUNEL was available for 675 men with known SPINK1 status.
Table 2.
Hazard ratios (HRs) and 95% confidence intervals (CIs) for prostate cancer recurrence and death by SPINK1 expression status among 879 men treated with radical prostatectomy for prostate cancer, Physicians’ Health Study and Health Professionals Follow-up Study cohorts
| Characteristic | Reduced modela | Full modelb | ||
|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | |
| Lethal Prostate Cancer (n=75)c | ||||
| SPINK1 − | 1.00 | 1.00 | ||
| SPINK1 + | 0.71 | (0.29–1.76) | 0.60 | (0.24–1.50) |
| Biochemical Recurrence (n=213) | ||||
| SPINK1 − | 1.00 | 1.00 | ||
| SPINK1 + | 1.01 | (0.63–1.60) | 1.03 | (0.65–1.65) |
Adjusted for age at diagnosis (<60, 60–64, 65–69, 70+), and cohort (PHS, HPFS)
Adjusted for age at diagnosis (<60, 60–64, 65–69, 70+), cohort (PHS, HPFS), tumor stage (pT2 N0/NX, pT3 N0/NX, pT4/N1/M1, unknown), and Gleason score (≤6, 3+4, 4+3, ≥8)
Lethal prostate cancer includes metastases to distant organs, and prostate cancer death
Expression of PTEN, stathmin and AR differed significantly according to SPINK1 status. Compared to SPINK1 negative tumors, SPINK1 positive tumors showed higher PTEN and stathmin expression (p<0.01), and lower expression of AR (p<0.01) (Table 3). There was no significant association between SPINK1 status and p-AKT expression (p=0.22) or pS6 expression (p=0.23). SPINK1 expression was seen in 47 of 427 (11%) ERG negative samples and in 19 of 427 (4%) ERG positive cases (p=0.0003; via chi-square test). In 12 of the 19 (63%) cases staining positive for both SPINK1 and ERG, dual SPINK1/ERG staining was seen in at least two of three same tumor cores (Table 4). Concomitant ERG/SPINK1 staining was validated on three whole tumor sections using a dual stain for ERG and SPINK1 (Fig. 2). In 2 of these 3 cases the percentage of prostate tumor glands that co-expressed both proteins was less than 5% of the overall tumor volume in that nodule; in the third case it was less than 10%.
Table 3.
Tumor protein expression of Ki67, TUNEL, PTEN, pAKT, pS6, Stathmin, and AR by SPINK1 protein expression status among men treated with radical prostatectomy for prostate cancer, Physicians’ Health Study and Health Professionals Follow-up Study cohorts
| Marker | No. | Range | SPINK1 − Median (IQRa) |
SPINK1 + Median (IQR) |
Correlation coefficient (r)b | Pc |
|---|---|---|---|---|---|---|
| PTEN | 761 | 0–0.91 | 0.14 (0.22) | 0.27 (0.35) | +0.17 | <0.01 |
| pAKT | 741 | 0–0.73 | 0.02 (0.12) | 0.03 (0.14) | +0.04 | 0.22 |
| pS6 | 746 | 0–0.97 | 0.08 (0.24) | 0.04 (0.16) | −0.06 | 0.09 |
| Stathmin | 743 | 0–0.82 | 0.01 (0.02) | 0.02 (0.04) | +0.21 | <0.01 |
| AR | 802 | 91–152 | 118 (11) | 114 (11) | −0.10 | <0.01 |
IQR: Inter-quartile range
Spearman Correlation Coefficient
P-values from Wilcoxon rank sums test
Table 4.
Dual ERG/SPINK1 positive tumor cases among men treated with radical prostatectomy for prostate cancer, Physicians’ Health Study and Health Professionals Follow-up Study cohorts
| * One Core + | * Two Cores + | * Three Cores + | Different Replicate Cores + | Total Number of Cases |
|---|---|---|---|---|
| 7(37) | 6(31.5) | 6(31.5) | 0(0) | 19(100) |
Exact same tumor cores
Figure 2.
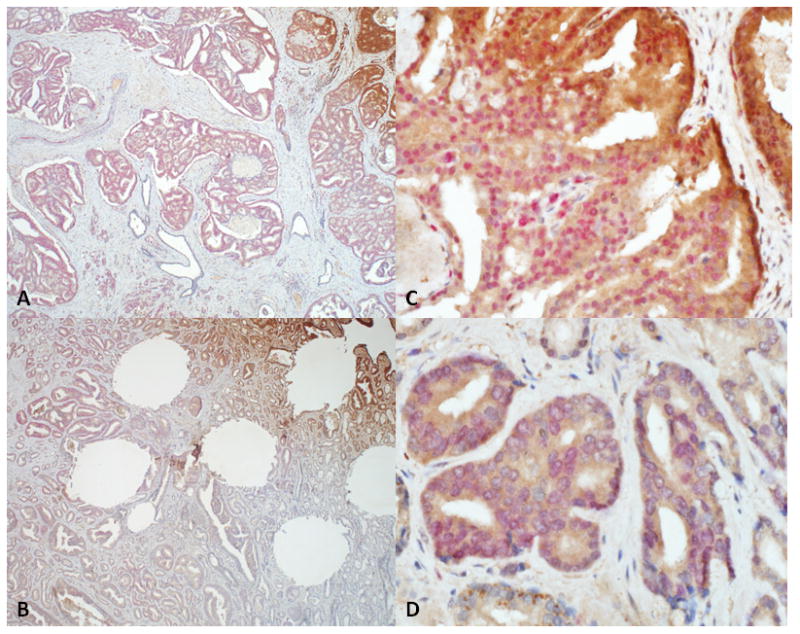
Dual immunohistochemical staining for nuclear marker ERG (red) and cytoplasmic marker SPINK1 (brown) showing positive staining in discrete foci of whole tumor sections from two separate cases (A–C; B–D) of prostate adenocarcinoma (x4 (left hand column); x40(right hand column)). Note in panel B there areas of dual positivity for SPINK1 and ERG (top) and areas of tumor negative for both markers (bottom).
DISCUSSION
Overexpression of SPINK1 has been associated with prognosis in many cancers. Initial work in ETS rearrangement negative prostate cancers indicated that SPINK1 positive prostate cancers were a distinct cancer subtype with an aggressive phenotype (2). In this study, we used the same criteria and antibody procedures as Tomlins et al. (2) to define SPINK1 positivity but we found no positive association between SPINK1 status and clinic-pathologic factors or prostate cancer-specific survival, despite the frequency of SPINK1 positive tumors in our cohort being similar to what has been previously published (8%) (2–3). Leinonen et al. investigated the association between SPINK1 status and clinic-pathologic factors and progression-free survival among 186 men primarily treated hormonally for prostate cancer (3). They observed no statistically significant associations between SPINK1 status and Gleason score, clinical stage, or Ki67 expression, but observed a significantly shorter progression-free survival amongst men with SPINK1 positive compared to SPINK1 negative tumors (RR 2.3, 95 % CI: 1.1–4.6). In the Lippolis et al. study, including 3,385 men treated with radical prostatectomy for prostate cancer, SPINK1 was weakly associated with pathological tumor stage, but otherwise the findings were consistent with the current report with no association between SPINK1 positivity and biochemical recurrence or development of metastatic disease (13). Grupp et al found no association between SPINK1 status and clinico-pathologic factors or biochemical recurrence among more than 8,000 (presumably including the 3,385 men in the Lippolis study) surgically treated prostate cancer patients. Taken together, these data suggest that SPINK1 protein expression is not a strong predictor of biochemical recurrence or lethal prostate cancer amongst men treated by radical prostatectomy.
Recent work by Ateeq et al. suggests that the PI3K pathway is one of the few key signaling pathways downstream of the SPINK1-EGFR axis. As such we looked at the associations between SPINK1 status and pAKT, pS6, stathmin and PTEN expression (4). We found higher expression of wild-type PTEN (p<0.01), stathmin (p<0.01) and pAKT (p=0.22) in SPINK1 positive compared to negative tumors, though the latter association was not statistically significant. Higher levels of pAKT and stathmin may indicate that there is activation of the PI3K pathway in SPINK1 overexpressing tumors. PTEN deletion is associated with subsequent activation of the PI3K pathway, which promotes cell proliferation, survival and other cellular pathways (14–15). These data suggest that any interaction of SPINK1 with the PI3K pathway appears to be downstream of PTEN. Indeed, a recent small study of 59 patients with castration-resistant prostate cancer found an association between PTEN deletion and SPINK1 overexpression (16). Previous studies have shown reduced PTEN expression in tumors overexpressing ERG (17–20). If SPINK1 protein is mutually exclusive from ERG, then SPINK1 in theory could be associated with higher wild-type PTEN; further work is needed to explore this potential relationship.
SPINK1 protein is expressed in the androgen independent but androgen responsive 22RV1 xenograft prostate cancer cell line (1), and appears to be regulated by androgens. As such we looked at the association between SPINK1 status and AR protein expression in prostate tumors. Herein we found lower expression of AR in SPINK1 positive tumors compared to SPINK1 negative cases (p<0.01). In contrast, Bismar et al. found no AR amplification in SPINK1 overexpressing tumors (16). A possible explanation for this discrepancy in results is that decreased levels of AR in tumor overexpressing SPINK1 reflects a compensatory AR downregulation as a result of decreased levels of one or more AR regulated genes. ERG overexpressing tumors seem to have higher levels of AR versus tumors not overexpressing ERG (21).
Some studies suggest that SPINK1 is expressed exclusively in TMPRSS2:ERG negative prostate cancers (2, 13, 16, 22, 23). However, additional studies have demonstrated the presence of TMPRSS2:ERG in a small but significant percentage of SPINK1 positive prostate cancers (3, 23, 24). Our results corroborate these latter studies, with SPINK1 expression being more frequent in ERG negative (11%) than in ERG positive cancers (4%; P = 0.0003). The discrepancy in study results may reflect differences in methodology employed to elucidate the presence of both SPINK1 (at the protein or mRNA level) and the TMPRSS2:ERG fusion, intra-tumoral heterogeneity or may be related to the power of the studies in question. However, it appears that co-expression of both SPINK and ERG is a focal event in prostate tumors and this most likely explains why previously co-expression of both markers was felt to be mutually exclusive. Interestingly, Jhavar et al found that higher expression of SPINK1 mRNA was restricted to cancers that lack ERG rearrangement, but a poor correlation was found between SPINK1 mRNA and SPINK1 protein expression and ERG and SPINK1 were not mutually exclusive when measured at the protein level (23). In this study, we used ERG immunohistochemistry as a surrogate marker for the presence of the fusion in contrast to 2 and 3-color fluorescent in situ hybridization (FISH) using in several prior studies (2–3, 23). Leinonen et al found SPINK1 expression (same antibody as current study) in 12/110 (11%) of TMPRSS2:ERG negative (assessed by three-color FISH) cases and in 7/60 (12%) TMPRSS2:ERG positive cases from prostate needle biopsies. In 123 prostatectomy treated patients, they found SPINK1 expression in 11/78 (14%) of TMPRSS2:ERG negative cases and 2/46 (4%) of TMPRSS2:ERG positive cases. In contrast, in the study by Lippolis et al (n=3,385) (13), ERG and SPINK1 were completely mutually exclusive however different scoring methodologies were employed in antibody assessment and importantly only one tumor core from each patient was represented on their TMA which may fail to highlight intra-tumoral heterogeneity which has been observed in SPINK1 staining (22). In the study by Grupp et al (n=8,642), presumably including all cases in Lippolis et al study and an additional 5,257 cases, SPINK1 (same antibody as current study) was almost exclusively expressed in ERG negative cases; SPINK1 was seen in 506/4,861 (10.4%) ERG negative cases and 13/3,781 (0.3%) ERG positive cases. As in the study by Lippolis et al, different scoring methodologies were employed in antibody assessment and only one tumor core from each patient was represented on their TMA. In the Bhalla et al study, ERG and SPINK1 expression (same antibody as current study) were essentially mutually exclusive, however 2 TMA cores showed dual ERG and SPINK1 staining (2% of ERG positive cases) albeit only one core showed concomitant expression in the same tumor focus. Interestingly, in our study a similar but slightly higher percentage of ERG positive cases showed concomitant SPINK1 staining, i.e. 4% (22). The study by Bhalla et al is a smaller cohort (n = 284) compared to this present study and has significant differences in terms of sample selection where metastatic tumors and rare morphological variants were included in their cohort. Further studies are needed to assess the clinical significance of this potentially rare dual SPINK and ERG positive molecular subtype.
In conclusion, our results suggest that SPINK1 protein expression may not be a predictor of recurrence or lethal prostate cancer amongst men treated by radical prostatectomy. In addition, SPINK1 and ERG protein overexpression do not appear to be entirely mutually exclusive as some previous studies have suggested.
TRANSLATIONAL RELEVANCE.
SPINK1 over-expression has been described in prostate cancer and is linked with poor prognosis in many cancers. The objective of this study was to characterize the association between SPINK1 over-expression and prostate cancer specific survival. The study included 879 participants in the US Physicians’ Health Study and Health Professionals Follow–Up Study, diagnosed with prostate cancer (1983 – 2004) and treated by radical prostatectomy. Immunohistochemical data was available for SPINK1, PTEN, p-Akt, pS6, stathmin, androgen receptor (AR) and ERG. Our results suggest that SPINK1 protein expression may not be a predictor of recurrence or lethal prostate cancer amongst men treated by radical prostatectomy. SPINK1 and ERG protein expression do not appear to be entirely mutually exclusive, as some previous studies have suggested.
Acknowledgments
Grant Support
This work was supported by the Dana-Farber/Harvard Cancer Center Specialized Programs of Research Excellence program in prostate cancer (5P50CA090381-08); and the National Cancer Institute (T32 CA009001 to K.L.P., CA55075, CA141298, CA13389, CA34944, CA40360, CA097193, EDRN U01 CA113913 and PO1 CA055075); and the National Heart, Lung, and Blood Institute (HL26490 and HL3595); and the Prostate Cancer Foundation (to LAM, NEM and KLP); and the Swedish Research Council (to AP); and the Royal Physiographic Society in Lund (to AP).
We are grateful to the participants and staff of the Physicians’ Health Study and Health Professionals Follow-up Study for their valuable contributions. In addition, we would like to thanks the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The Dana-Farber/Harvard Cancer Center Tissue Microarray Core Facility constructed the tissue microarrays in this project.
Footnotes
Conflict of Interest: The authors have no conflict of interest to declare.
References
- 1.Paju A, Hotakainen K, Cao Y, Laurila T, Gadaleanu V, Hemminki A, et al. Increased expression of tumor-associated trypsin inhibitor, TATI, in prostate cancer and in androgen-independent 22Rv1 cells. Eur Urol. 2007;52:1670–9. doi: 10.1016/j.eururo.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 2.Tomlins SA, Rhodes DR, Yu J, Varambally S, Mehra R, Perner S, et al. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell. 2008;13:519–28. doi: 10.1016/j.ccr.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leinonen KA, Tolonen TT, Bracken H, Stenman UH, Tammela TL, Saramäki OR, et al. Association of SPINK1 expression and TMPRSS2:ERG fusion with prognosis in endocrine-treated prostate cancer. Clin Cancer Res. 2010;16:2845–51. doi: 10.1158/1078-0432.CCR-09-2505. [DOI] [PubMed] [Google Scholar]
- 4.Ateeq B, Tomlins SA, Laxman B, Asangani IA, Cao Q, Cao X, et al. Therapeutic targeting of SPINK1-positive prostate cancer. Sci Transl Med. 2011;3:72ra17. doi: 10.1126/scitranslmed.3001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaziano JM, Glynn RJ, Christen WG, Kurth T, Belanger C, MacFadyen J, et al. Vitamins E and C in the prevention of prostate and total cancer in men: the Physicians’ Health Study II randomized controlled trial. JAMA. 2009;301:52–62. doi: 10.1001/jama.2008.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hennekens CH, Eberlein K. A randomized trial of aspirin and beta-carotene among U.S. physicians. Prev Med. 1985;14:165–8. doi: 10.1016/0091-7435(85)90031-3. [DOI] [PubMed] [Google Scholar]
- 7.Dhillon PK, Barry M, Stampfer MJ, Perner S, Fiorentino M, Fornari A, et al. Aberrant cytoplasmic expression of p63 and prostate cancer mortality. Cancer Epidemiol Biomarkers Prev. 2009;18:595–600. doi: 10.1158/1055-9965.EPI-08-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pettersson A, Graff RE, Bauer SR, Pitt MJ, Lis RT, Stack EC, et al. The TMPRSS2:ERG Rearrangement, ERG Expression, and Prostate Cancer Outcomes: a Cohort Study and Meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21:1497–509. doi: 10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaux A, Albadine R, Toubaji A, Hicks J, Meeker A, Platz EA, et al. Immunohistochemistry for ERG expression as a surrogate for TMPRSS2-ERG fusion detection in prostatic adenocarcinomas. Am J Surg Pathol. 2011;35:1014–20. doi: 10.1097/PAS.0b013e31821e8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park K, Tomlins SA, Mudaliar KM, Chiu YL, Esgueva R, Mehra R, et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590–8. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Leenders GJ, Boormans JL, Vissers CJ, Hoogland AM, Bressers AA, Furusato B, et al. Antibody EPR3864 is specific for ERG genomic fusions in prostate cancer: implications for pathological practice. Mod Pathol. 2011;24:1128–38. doi: 10.1038/modpathol.2011.65. [DOI] [PubMed] [Google Scholar]
- 12.Fiore C, Bailey D, Conlon N, Wu X, Martin N, Fiorentino M, et al. Utility of multispectral imaging in automated quantitative scoring of immunohistochemistry. J Clin Pathol. 2012;65:496–502. doi: 10.1136/jclinpath-2012-200734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lippolis G, Edsjo A, Stenman U-H, Bjartell A. A high-denisty tissue microarray from patients with clinically localized prostate cancer reveals ERG and TATI exclusivity in tumour cells. Prostate Cancer and Prostatic Disease. 2013;16:145–150. doi: 10.1038/pcan.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–14. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 16.Bismar TA, Yoshimoto M, Duan Q, Liu S, Sircar K, Squire JA. Interactions and relationships of PTEN, ERG, SPINK1 and AR in castration-resistant prostate cancer. Histopathology. 2012;60:645–52. doi: 10.1111/j.1365-2559.2011.04116.x. [DOI] [PubMed] [Google Scholar]
- 17.Bismar TA, Yoshimoto M, Vollmer RT, Duan Q, Firszt M, Corcos J, et al. PTEN genomic deletion is an early event associated with ERG gene rearrangements in prostate cancer. BJU Int. 2011;107:477–85. doi: 10.1111/j.1464-410X.2010.09470.x. [DOI] [PubMed] [Google Scholar]
- 18.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol. 2009;22:1083–93. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–6. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minner S, Enodien M, Sirma H, Luebke AM, Krohn A, Mayer PS, et al. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clin Cancer Res. 2011;17:5878–88. doi: 10.1158/1078-0432.CCR-11-1251. [DOI] [PubMed] [Google Scholar]
- 22.Bhalla R, Kunju LP, Tomlins SA, Christopherson K, Cortez C, Carskadon S, et al. Novel dual-color immunohistochemical methods for detecting ERG-PTEN and ERG-SPINK1 status in prostate carcinoma. Mod Pathology. 2013;26:835–848. doi: 10.1038/modpathol.2012.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jhavar S, Brewer D, Edwards S, Kote-Jarai Z, Attard G, Clark J, et al. Integration of ERG gene mapping and gene-expression profiling identifies distinct categories of human prostate cancer. BJU Int. 2009;103:1256–69. doi: 10.1111/j.1464-410X.2008.08200.x. [DOI] [PubMed] [Google Scholar]
- 24.Grupp K, Diebel F, Sirma H, Simon R, Breitmeyer K, Steurer S, et al. SPINK1 expression is tightly linked to 6q15- and 5q21-deleted ERG-fusion negative prostate cancers but unrelated to PSA recurrence. Prostate. 2013;73:1690–8. doi: 10.1002/pros.22707. [DOI] [PubMed] [Google Scholar]


