Abstract
Mitochondria, the main site of cellular energy harvesting, are derived from proteobacteria that evolved within our cells in endosymbiosis. Mitochondria retained vestiges of their proteobacterial genome, the circular mitochondrial DNA (mtDNA), which encodes 13 subunits of the oxidative phosphorylation (OXPHOS) multiprotein complexes in the electron transport chain (ETC), while the remaining ~80 ETC components are encoded in the nuclear DNA (nDNA). A further ~1,400 proteins, which are essential for mitochondrial function are also encoded in nDNA. Thus the majority of mitochondrial proteins are translated in the cytoplasm, then imported, processed, and assembled in the mitochondria. An intricate protein quality control (PQC) network, constituted of chaperones and proteases that refold or degrade defective proteins, maintains mitochondrial proteostasis and ensures the cell and organism health. The mitochondrial unfolded protein response (UPRmt) is a relatively recently discovered PQC pathway, which senses the proteostatic disturbances specifically in the mitochondria and resolves the stress by retrograde signaling to the nucleus and consequent transcriptional activation of protective genes. This PQC system does not only transiently resolves the local stress, but can have long lasting effects on whole body metabolism, fitness and longevity. A delicate tuning of its activation levels might constitute a treatment of various diseases, such as metabolic diseases, cancer and neurodegenerative disorders.
Introduction
Mitochondria play a crucial role in the overall homeostasis of the cell. Mitochondria accommodate the enzymatic machinery capable of ATP production by oxidative phosphorylation (OXPHOS) and are the prime site of metabolic processing in unicellular organisms, plants and animals. As mitochondria evolved from endosymbiotic α-proteobacteria residing in the eukaryotic cell, they retained the vestiges of the circular bacterial DNA encoding for 13 proteins, and contain several proteins with strong similarities to bacterial proteins (Wallin, 1993). Most of the ~1500 mitochondrial proteins are, however, encoded by the nucleus and imported post-translationally by means of a specialized and highly conserved machinery (Chacinska et al., 2009; Neupert and Herrmann, 2007; Schmidt et al., 2010).
In the past years, this unique organelle received an increasing interest from the scientific community, as researchers have highlighted the implication of mitochondrial dysfunction in the ageing process and in common diseases such as cancer, diabetes and diverse neurological disorders (Nunnari and Suomalainen, 2012). Within this context, it is of particular interest to investigate the mechanisms that ensure optimal function of mitochondria. Here we give a brief overview of mitochondrial quality control systems, with a particular focus on the mitochondrial unfolded protein response (UPRmt) and its implications in animal physiology.
Mitochondrial quality control systems
As the mitochondrial proteome is continuously challenged by multiple factors, mitochondria have evolved an elaborate protein quality control (PQC) system that maintains proteostasis and mitochondrial function in response to various levels of proteotoxic damage (Fischer et al., 2012; Friedman and Nunnari, 2014; Rugarli and Langer, 2012).
Almost all mitochondrial proteins are transcribed and translated in the cytoplasm. They have to be imported through the double membrane of the mitochondria in their unfolded state, before they are folded and assembled within the mitochondria (Harbauer et al., 2014; Schmidt et al., 2010). As most ETC complexes are composed of subunits encoded by both the nuclear and mitochondrial genomes, they have to be present in well-defined stochiometrical ratios. A number of essential housekeeping proteins assist in processes, such as protein import, folding and supercomplex assembly. Among these proteins, chaperones of the heat shock protein (Hsp) family, such as mtHsp70, Hsp10 or Hsp60, fold the newly imported proteins or refold damaged proteins. Proteases such as HtrA2, Yme1l in the mitochondrial intermembrane space (IMS) and ClpP or Lon in the matrix furthermore guarantee the degradation of proteins that are irreversibly damaged. Several antioxidant enzymes indirectly contribute to the maintenance of proteostasis by clearing ROS.
Mitochondria do not behave as a multitude of isolated organelles, but are rather a connected and cooperative network that undergoes constant remodeling (Friedman and Nunnari, 2014). The dynamics of the mitochondrial network is regulated by proteins such as MFN1/2, OPA1 and DRP1, Mff, MiD49/51 that mediate fusion and fission, respectively (Andreux et al., 2013; Jin and Youle, 2013; Loson et al., 2013). Fusion of healthy mitochondria to mitochondria harboring damaged components constitutes a beneficial replacement and/or dilution process (Chan, 2012). Alternatively, fission promotes the segregation of dysfunctional mitochondria, favoring their subsequent elimination through mitophagy, governed amongst others by PINK1 and Parkin (Youle and van der Bliek, 2012). Depending on the level of damage, those mechanisms are gradually triggered to repair or eliminate mitochondrial proteins or mitochondrial units. In case of irreversible insults to the mitochondria that are beyond repair and hence jeopardize cellular survival, apoptosis will ensue (Friedman and Nunnari, 2014; Martinou and Youle, 2011).
As most of the PQC proteins are encoded in the nucleus, the state of mitochondrial health has to be communicated to the nucleus, in order to specifically adapt the PQC to proteostatic needs. The general term “retrograde signaling” defines all mitochondrial cues sent to the nucleus to respond to variations in the organelle homeostasis (Liu and Butow, 2006; Ryan and Hoogenraad, 2007).
Mitochondria-to-nucleus signaling of the UPRmt
Accumulation of unfolded proteins leading to protein aggregation represents a dangerous threat not only for a specific subcellular compartment, but also for the rest of the cell. Chaperones assist protein folding and assembly and thus ensure proteostasis in the cell (Hartl et al., 2011). Proteotoxic stress, which exceeds protein the folding capacity by chaperones, is sensed and transduced to the nucleus to induce the transcription of genes implicated in proteostatic surveillance, a mechanism termed “unfolded protein response”. Heat was among the first identified stresses disrupting the protein folding homeostasis, which contributed to the name “Heat Shock Proteins” (Hsp) of many chaperones (Richter et al., 2010). Specific responses to a proteotoxic stress occurring in specific subcellular compartments, namely cytosol, endoplasmic reticulum (ER) and mitochondria, have been described. In the cytosol proteostasis is ensured by the heat shock factor (HSF) transcription factor family, which, among others, regulates Hsp70 and Hsp90 expression, whereas protein misfolding in the ER is assessed by the transmembrane proteins inositol-requiring 1 (IRE-1), activating transcription factor 6 (ATF6) and protein-like endoplasmic reticulum kinase (PERK), culminating with the induction of chaperones as BiP (GRP-78) (Buchberger et al., 2010; Mori, 2009; Walter and Ron, 2011).
The mitochondrial unfolded protein response (UPRmt) has been rather recently identified. In monkey COS-7 cells, overexpression of a mutant, aggregation-prone form of the mitochondrial protein ornithine transcarbamylase (OTC) triggered the accumulation of unfolded proteins in the mitochondria (Zhao et al., 2002). This led to an increase in mRNA and protein levels of Hsp60, Hsp10, the protease ClpP and the Hsp40 family chaperone mtDNAJ. Although initially discovered in mammalian cells, the molecular mechanism of this pathway has been more extensively characterized in the nematode C. elegans. Furthermore some studies in Drosophila and very recently in yeast have focused on the UPRmt. This paragraph summarizes the UPRmt signaling in those model systems, from the triggering stimuli initiating the mitochondria-to-nucleus signaling to the consequences on global protein synthesis.
Triggering the UPRmt
Any stress affecting proteostasis within the mitochondria, such as heat, could potentially activate the mitochondrial chaperones (Zhao et al., 2002). However, selective perturbations in the mitochondria enable a proper study of the UPRmt per se and specifically induce mitochondrial target proteins without affecting the expression of ER and cytoplasmic chaperones. The artificial accumulation of unfolded proteins was achieved by overexpression of mutant OTC in mammalian cells and in the Drosophila (Pimenta de Castro et al., 2012; Zhao et al., 2002). In C. elegans, the knock-down (KD) by RNAi feeding of mitochondrial proteases, such as spg-7, or mitochondrial chaperones, as hsp-6 and hsp-60 (orthologs of mtHsp70 and Hsp60 in mammals, respectively), strongly induces the UPRmt (Yoneda et al., 2004). As these proteins are essential components of PQC, impairment of either of them is sufficient to destabilize organelle proteostasis and trigger the UPRmt. The same effect can be observed by interfering with expression of prohibitin, a mitochondrial inner membrane complex that supervises ETC assembly, as reduced prohibitin levels result in active UPRmt in C. elegans as well as in yeast (Schleit et al., 2013; Yoneda et al., 2004). Exposing cells to toxic compounds, such as the ROS inducer paraquat, which subsequently increases protein damage, also activates the UPRmt (Yoneda et al., 2004).
Proteostasis is also challenged when missing or reduced expression of ETC subunits impedes the stoichiometry and/or assembly of the multi-protein OXPHOS complexes. Loss of function of mrps-5, a mitochondrial ribosomal protein (MRPs), or of other MRPs, potently activates the UPRmt, as impaired mitochondrial protein translation decreases the production of mitochondrial-encoded ETC subunits and results in an increased load of unassembled orphan ETC subunits encoded by the nucleus (Houtkooper et al., 2013). Our laboratory termed this concept “mito-nuclear protein imbalance”, which also occurs after knock-down of ETC subunits in the worm and in Drosophila (Durieux et al., 2011; Owusu-Ansah et al., 2013). Pharmacologically, doxycycline or chloramphenicol can reproduce this effect in the mouse and in the worm, as these antibiotics affect not only bacterial, but also mitochondrial translation, given that mitochondria are derived from bacterial ancestors (Houtkooper et al., 2013). Ethidium bromide, which causes a selective loss of the mitochondrial DNA (and hence mitochondrial protein production), also leads to mito-nuclear protein imbalance, activating the UPRmt in both the worm and mammalian cells (Martinus et al., 1996; Yoneda et al., 2004).
Interestingly, rapamycin, which inhibits cytosolic translation through inhibition of TOR signaling (Zid et al., 2009), also induces a mito-nuclear imbalance and the UPRmt, but in this case by generating an excess of orphan mitochondrial-encoded ETC subunits. In a similar fashion, several pharmacological treatments enhancing mitochondrial biogenesis, such as resveratrol or the activation of the worm sirtuin sir-2.1 by nicotinamide riboside (NR) or by PARP inhibitors, as well as sir-2.1 overexpression, also trigger the UPRmt (Mouchiroud et al., 2013b). It is therefore apparent that the ratio between nuclear and mitochondrial ETC subunits and not their absolute levels, is the predominant factor that causes mito-nuclear imbalance and triggers the UPRmt.
Transcriptional regulation of the UPRmt
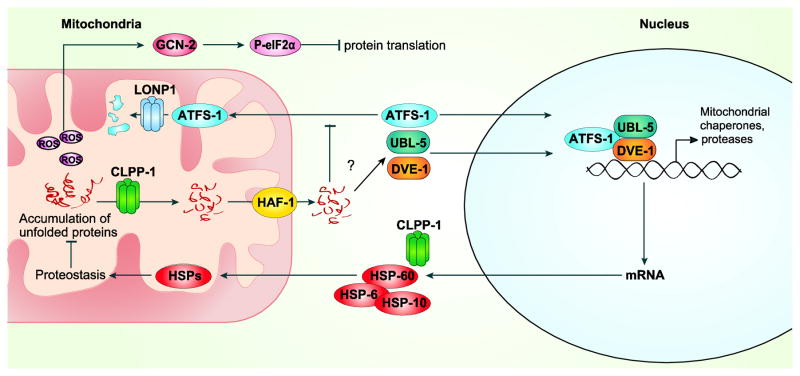
In C. elegans, the generation of reporter worm strains expressing GFP under the control of the promoter of either hsp-6 or hsp-60 (known as hsp-6::GFP and hsp-60::GFP strains) (Yoneda et al., 2004) greatly facilitated the study of the mitochondrial stress response at the transcriptional level. The use of these strains for RNAi based screens enabled the detailed characterization of the signaling components upstream of the transcriptional response (Benedetti et al., 2006; Haynes et al., 2007; Haynes et al., 2010). ubl-5, a gene encoding for a ubiquitin-like protein, was found to be required for the proper activation of these reporters during the UPRmt and for subsistence of the worms with stressed mitochondria (Benedetti et al., 2006). The UPRmt enhances UBL-5 levels and its nuclear localization, suggesting that it acts as a stress-responsive transcriptional regulator (Figure 1). Similarly, in later screenings, the bZIP family transcription factor atfs-1 and dve-1 were described as essential nuclear signaling components of the UPRmt (Haynes et al., 2007; Haynes et al., 2010). All three factors translocate to the nucleus upon mitochondrial stress. DVE-1 furthermore is reported to form a dimer with UBL-5, which together with ATFS-1 activates the transcription of the UPRmt genes (Haynes et al., 2010). However, how the folding stress is communicated to those nuclear players has not yet been fully characterized. clpp-1, a mitochondrial matrix protease, proved to be essential for ubl-5 induction, DVE-1 relocalization and activation of the UPRmt response (Haynes et al., 2007). The exact role of clpp-1 was revealed when the peptide efflux achieved by the inner membrane transporter HAF-1 was identified as a pivotal event in the signaling (Haynes et al., 2010). The model thus suggests that clpp-1 digests excess unfolded proteins in proteotoxic stress conditions. The resulting peptide fragments, which are transported to the cytosol by HAF-1, lead to the activation of the nuclear players DVE-1, UBL-5 and ATFS-1, ultimately inducing the reparative transcriptional response (Figure 1).
Figure 1. UPRmt signaling pathway in C.elegans.
Unfolded proteins, accumulating in the mitochondria, are digested by the protease CLPP-1 into short peptides. These peptides are exported into the cytoplasm through a transporter HAF-1 and by a yet unknown mechanism inhibit mitochondrial import. Impairment of the import allows the nuclear translocation of transcription factor ATFS-1, which in non-stress conditions moves into the mitochondria and is degraded by protease LONP-1. ATFS-1, together with other nuclear factors UBL-5 and DVE-1 activate the protective UPRmt target genes, which reconstitute the mitochondrial proteostasis. In parallel to ATFS-1 mediated transcriptional response, ROS, produced by stressed mitochondria, activate the kinase GCN-2, which phosphorylates eIF2α, which leads to down-regulation of global translation and thus reduces the load of new mitochondrial proteins to be folded.
Interestingly, in the absence of stress, ATFS-1 is imported into the mitochondria due to a mitochondrial localization signal (MLS) present at its N-terminus. Once within the mitochondria, ATFS-1 is constitutively degraded by the Lon protease (Nargund et al., 2012). However, ATFS-1 also contains a nuclear localization signal (NLS). During mitochondrial stress the mitochondrial import of ATFS-1 is reduced, ATFS-1 will accumulate in the nucleus, facilitating the activation of the downstream adaptive events that characterize the UPRmt response. These findings clarified part of the role of HAF-1 in signaling of the UPRmt, as this transporter was shown to reduce the import of ATFS-1 under mitochondrial stress conditions (Nargund et al., 2012).
In the mammals, fewer players in the UPRmt signaling have been identified. The main transcription factor implicated in the mammalian UPRmt is CHOP, which heterodimerizes with C/EBPβ upon overexpression of mutant OTC (Figure 2). As a result, the CHOP/C/EBPβ dimer binds to and activates the promoters of the UPRmt responsive genes (Horibe and Hoogenraad, 2007; Zhao et al., 2002). Although CHOP is also known to mediate the UPRER (Schroder, 2006), its specificity to the UPRmt might reside in the fact that both the CHOP and C/EBPβ promoters contain an AP1 site that is required for their induction upon mitochondrial stress but not upon UPRER (Horibe and Hoogenraad, 2007). The AP1 site is bound by the c-Jun transcription factor, which is regulated by JNK2. Promoter analysis of the UPRmt responsive genes revealed that they contain a CHOP binding site flanked by two mitochondrial unfolded protein response elements (MURE) (Aldridge et al., 2007). Among the 11 genes containing the MUREs and up-regulated upon mutant OTC expression are chaperones Hsp60, Hsp10, mtDnaJ (Hsp40 family), proteases ClpP and YME1L1, the import complex subunit Tim17A and mitochondrial enzymes such as thioredoxin 2 (Trx2), cytochrome C reductase, endonuclease G, NDUFB2.
Figure 2. UPRmt signaling in mammals in the matrix and intermembrane space (IMS).
Accumulation of unfolded proteins in the mitochondrial matrix leads to activation of JNK2, which triggers c-Jun binding to AP-1 elements to up-regulate CHOP and C/EBPβ transcription. Dimer of CHOP and C/EBPβ transcription factors binds to specific UPRmt promoter element and activates the target genes. Additionally, PKR decreases global translation rate by phosphorylating eIF2α and mitochondrial import is attenuated by down-regulation of TIM17A. Under proteotoxic stress in mitochondrial IMS, increased levels of unfolded proteins and ROS trigger activation of AKT, which phosphorylates ERα. Activated ERα upregulates the transcription of PQC protease OMI, which restores IMS proteostasis.
Interestingly, the folding capacity of the mitochondrial intermembrane space (IMS) can be specifically affected by the overexpression of a mutant form of the IMS located endonuclease G (Radke et al., 2008). This triggers a different stress response, activating other genes than those of the “canonical” mitochondrial matrix UPRmt, such as the IMS protease OMI and the proteasome. Unliganded estrogen receptor α mediates this IMS-UPR in a manner dependent on ROS generation and activation of AKT signaling (Papa and Germain, 2011).
Effects of the UPRmt on translation and mitochondrial protein import
Besides the induction of transcriptional targets of UPRmt targets, other mechanisms aimed at restoring proteostasis and mitochondrial integrity occur in the course of the UPRmt. Notably, the further generation of new mitochondrial proteins is reduced by impeding global protein synthesis in the cytosol (Baker et al., 2012). During mitochondrial stress conditions in the worm, general control non-derepressible-2 kinase (GCN-2) phosphorylates translation initiation factor eIF2α in a ROS-dependent manner and thus slows down cytosolic translation (Baker et al., 2012). GCN-2 and ATFS-1 effects are dissociable and they signal in different arms of the UPRmt. Similarly, dsRNA-activated protein kinase (PKR), mediates phosphorylation of eIF2α, thus attenuating protein translation in the cytosol during the UPRmt in mammals (Rath et al., 2012). These findings link the UPRmt to the integrated stress response (ISR), a pathway comprising kinases that act negatively on translation through eIF2α phosphorylation following oxidative stress, ER stress, viral infections and other cellular attacks (Wek and Cavener, 2007; Wek et al., 2006).
In addition to stalling of the protein synthesis upon mitochondrial stress, the import of new mitochondrial proteins is also affected, in order to limit the load of proteins to be folded within mitochondria. In HEK293 cells treated with arsenite (As(III)), induction of the ISR decreases total protein levels of the TIM17A subunit of the mitochondrial protein import complex TIM23, by increasing its degradation and repressing its translation (Aldridge et al., 2007; Rainbolt et al., 2013). Conversely, when import is artificially repressed by the knock-down of TIM23 subunits in HEK293 cells and in C. elegans, the UPRmt is activated in an haf-1/atfs-1-dependent manner in the worm, enhancing also its resistance to paraquat (Rainbolt et al., 2013). The importance of mitochondrial import regulation is also evident in C. elegans, as a general decrease of mitochondrial import is required for ATFS-1 nuclear translocation and consequent activation of transcriptional program of the UPRmt (Nargund et al., 2012). Of note is the fact that in mammalian cells, transcription of TIM17A gene was shown to be induced as a target of the UPRmt due to the presence of MURE sites in its promoter, indicating a recovery of mitochondrial import upon resolution of the stress (Aldridge et al., 2007).
Physiological implications of the UPRmt
Extension of lifespan and cell-non-autonomous signaling of the UPRmt
Studies of the effects of the UPRmt on whole body metabolism and overall fitness have started in simple model organisms such as C. elegans and D. melanogaster; however, recent studies suggest that the UPRmt may have a similar important role in mammals.
KD of ETC components in C. elegans reduce developmental rates and body size. Interestingly, this also leads to a robust extension of lifespan (Dillin et al., 2002). A simple interpretation, coherent with the “ROS theory of aging” (Harman, 1956), would attribute the increased lifespan to the reduced respiration rates, which leads to generation of less ROS byproducts. However, later studies identified UPRmt activation as causative for the lifespan extension after ETC disruption, as exemplified by the longevity of the cytochrome c oxidase cco-1 mutant (Durieux et al., 2011). Similarly, a study in Drosophila showed that mild perturbation of the ETC in muscle has positive effects on muscle function, locomotor activity and lifespan due to UPRmt activation (Owusu-Ansah et al., 2013). More recently, our laboratory established that the UPRmt subsequent to the presence of a mito-nuclear imbalance also robustly extends worm lifespan (Houtkooper et al., 2013). In line with these findings, low expression of mouse Mrps5 (or other MRPs) triggers the UPRmt and correlates with a long lifespan in the BXD mice genetic reference population, demonstrating the evolutionary conservation of this mechanism in mammals (Argmann et al., 2005; Peirce et al., 2004). Mito-nuclear imbalance also contributes to the lifespan extension driven by the activation of C. elegans sirtuin, sir-2.1 (Mouchiroud et al., 2013b). Pharmacological or genetic manipulations leading to NAD+ accumulation or enhanced sir-2.1 expression levels in C. elegans boost mitochondrial metabolism, induce mito-nuclear imbalance and activate the UPRmt, which in parallel to an antioxidant program leads to a significant lifespan extension (Mouchiroud et al., 2013a).
There are temporal and spatial requirements for UPRmt activation, in order for it to have beneficial effects on lifespan. In C. elegans, UPRmt induction by RNAi against cco-1 or mrps-5 only during the larval stage is sufficient to ensure a lasting effect on lifespan (Dillin et al., 2002; Durieux et al., 2011). Conversely, cco-1 or mrps-5 RNAi starting in adulthood is neither able to activate the UPRmt nor impact on longevity (Dillin et al., 2002). In addition to the prerequisite of a specific time frame, only mitochondrial stress in selected worm tissues, i.e. intestine and neurons, but not muscle, can extend longevity (Durieux et al., 2011). Interestingly, perturbations of the mitochondrial homeostasis in one tissue can be sensed and communicated to other tissues by cell-non-autonomous cues that were termed “mitokines”. Knocking down the signaling component ubl-5 selectively in neurons can block this inter-tissue UPRmt signal, suggesting that the retrograde signaling arm is required only in the tissue emitting the mitokine (Durieux et al., 2011). In D. melanogaster, ImpL2, the ortholog of the Insulin binding protein 7 (IGFBP7) that is secreted in response to KD of ETC components in the muscle, participates in the organismal adaptation to mitochondrial perturbation and mediates lifespan extension (Owusu-Ansah et al., 2013). Interestingly, in human patients with mitochondrial myopathy due to ETC deficiencies, the cytokine FGF-21 was shown to be secreted from muscle tissue suggesting that the ETC dysfunction may mimic fasting and induce the release of the fasting hormone FGF-21 (Suomalainen et al., 2011). This muscle release of FGF-21 drives inter-organ communication that results in enhanced ketogenesis in the liver and lipid mobilization from the fat, suggesting FGF-21 to be a human mitokine (Suomalainen et al., 2011). In another study, interference with autophagy and the resulting mitochondrial dysfunction in mice also led to the secretion of the Fgf21 mitokine (Kim et al., 2013). Interestingly, the ISR was implicated in this response, as loss of autophagy led to phosphorylation of eIF2α and increased the expression of activating transcription factor 4 (Atf4). In this context, Fgf21 secretion improved insulin sensitivity and protected mice from obesity, although the direct link with the UPRmt has not yet been examined (Kim et al., 2013).
UPRmt-induced mitohormesis
UPRmt activation upon mitochondrial stress is intrinsically linked to a certain level of mitochondrial dysfunction, questioning what is the balance between harmful and beneficial effects of the UPRmt. If the mitochondrial insult is mild, the adaptive stress response that ensues can overcome the initial insult and have a beneficial, long-lasting impact. This phenomenon resembles the concept of “mitohormesis” caused by ROS (Ristow and Zarse, 2010). Treatment with low doses of inducers of oxidative stress, such as paraquat, generates low levels of ROS and the resulting adaptive response extends lifespan, whereas treatment with high doses resulting in excessive levels of ROS is toxic (Ristow and Zarse, 2010). However, in the case of cco-1 (Durieux et al., 2011) or mrps-5 (Houtkooper et al., 2013) RNAi in the developing worm, ROS does not play a role in the lifespan extension, hence representing a unique case of mitohormesis only driven by the UPRmt.
Whereas the UPRmt improves the fitness of an organism and extends its lifespan, if the level of stress inflicted to the mitochondria is too high, the ensuing UPRmt might be insufficient to counteract the damage inflicted, and hence an adaptive response will turn into a detrimental response. This explains why worms exhibit a shortened lifespan after hsp-6 RNAi (Haynes et al., 2007), although the UPRmt is strongly activated by this genetic manipulation (Yoneda et al., 2004). Similarly, in the fly, overexpression of a mutant OTC protein negatively impacts on lifespan and phenocopies mutations in PINK1 and Parkin (Pimenta de Castro et al., 2012), as it causes a too severe level of mitochondrial dysfunction. Interestingly, the UPRmt and the mitophagy quality control systems were recently found to be triggered concomitantly, as PINK1 recruitment on the mitochondrial membrane was enhanced by accumulation of unfolded proteins in the mitochondria, as well as by the knock-down of the LONP1 protease, showing that these responses are connected to some degree (Jin and Youle, 2013).
This mitohormetic action of the UPRmt is well illustrated in a recent report in yeast (Schleit et al., 2013). Although dietary restriction (DR) has been shown to extend lifespan in diverse species (Kennedy et al., 2007), the effect of DR is highly dependent on the genotype of the organism. Among the yeast strains presenting the highest increase of replicative lifespan upon DR is the Δphb2 strain, a mutant of a subunit of the prohibitin complex (Schleit et al., 2013). KD of prohibitin in yeast activates the UPRmt, as also observed in C.elegans (Yoneda et al., 2004). Interestingly, Δphb2 mutation improves lifespan only in the context of DR, while in nutrient-rich medium it shortens the lifespan. The difference between these two conditions is that a general reduction of translation rates occurs upon DR, which attenuates the UPRmt (Schleit et al., 2013). Thus, DR lowers the mitochondrial stress to a level, which enables the positive effects of mitohormesis mediated by UPRmt activation. Similarly, phb-2 RNAi strongly activates the UPRmt and shortens lifespan in wild type worms, while in mutant worms with reduced translation rates, the UPRmt is induced to a lower extent, leading to longevity (Schleit et al., 2013). These observations suggest that slowing-down the translation rate might be beneficial in some cases of mitochondrial dysfunction associated with high UPRmt activation. This mechanism could have interesting therapeutic implications that warrant further study.
Implications of the UPRmt in disease
As increased UPRmt can be both beneficial and harmful, depending on the level of the UPRmt, it is conceivable that it can be either a cause or a potential treatment strategy for disease. Although there are no studies as of yet that show the direct implication of the UPRmt in disease, several reports suggest that the UPRmt may be linked to a specific set of disorders.
Metabolism and diabetes
As discussed above, impaired prohibitin function activates the UPRmt in several model systems (Schleit et al., 2013; Yoneda et al., 2004). In the mouse, a pancreatic β-cell-specific knockout of Phb2 contributes to progressive development of diabetes due to β-cell dysfunction (Supale et al., 2013). Although Opa1 proteolysis and impaired mitochondrial dynamics were identified as potential mechanisms behind the β-cell dysfunction, it will be interesting to test, to which extent the activation of the UPRmt could contribute or inversely limit the pathogenesis of diabetes in this context.
Also linked to diabetes and metabolic disease, the hypothalamic knockout of Hsp60 revealed an implication of this chaperone in the development of insulin resistance (Kleinridders et al., 2013). Expression of Hsp60 in hypothalamus was shown to be dependent on leptin. As insulin and leptin resistance are known to be linked, this may explain why diabetic patients have decreased HSP60 levels in the brain. Loss of Hps60 by itself causes mitochondrial dysfunction and ROS overproduction and consequently leads to hypothalamic insulin resistance and diabetes. Hsp60 was thus proposed to be the effector of leptin protective actions on mitochondria and act as the integrator of insulin signaling (Kleinridders et al., 2013).
In Drosophila, knockdown of an ETC complex I component in the muscle induces the UPRmt and leads to secretion of ImpL2, which non-autonomously represses insulin signaling by binding IGF and other insulin-like molecules (Owusu-Ansah et al., 2013). Although mitochondrial perturbation occurs only in the muscle, the entire fly is smaller, demonstrating that a growth-inhibiting signal is communicated from the muscle to all tissues. As mentioned above, Imp2L could hence be considered as a fly mitokine, achieving part of the organismal adaption to the stress by repressing systemic insulin signaling. Consistent with this hypothesis, overexpression of Imp2L in flies increases lifespan and enhances lysosome biogenesis, which could contribute to mitophagy as a mechanism to enhance mitochondrial function upon aggregate accumulation (Owusu-Ansah et al., 2013).
Another line of support for the existence of a link between the UPRmt and metabolism came from studies in the worm where UPRmt activation was shown to lead to the up-regulation of the expression of some glycolytic enzymes (Nargund et al., 2012). This suggests that a metabolic remodeling happens concurrently with the occurrence of UPRmt and energy production may shift from oxidative phosphorylation towards glycolysis when mitochondria are stressed.
Neurological disorders
Drosophila that are overexpressing a mutant OTC protein develop mitochondrial dysfunction phenotypes similar to mutants of PINK1 and Parkin (Pimenta de Castro et al., 2012), two mitophagy regulators that are found mutated in familial forms of Parkinson’s diseases (Andreux et al., 2013). This also suggests a link between the UPRmt and neurodegenerative disorders, associated with mitochondrial dysfunction such as Parkinson’s, Alzheimer’s and Huntington’s disease (de Castro et al., 2011).
Notably, the DR-driven attenuation of translation in Δphb2 yeast (Schleit et al., 2013) (discussed above) suggests that interfering with translation and mitochondrial protein import may restore mitochondrial function in the context of neurodegenerative diseases. In line with this premise, repression of cytosolic translation showed beneficial effects on mitochondrial function in yeast (Wang et al., 2008) and protected Drosophila against PINK-induced pathogenesis (Liu and Lu, 2010) although the underlying mechanisms have yet to be characterized.
Cancer
The mitochondrial stress response could also be connected with the control of cell proliferation and cancer. Cancer is associated with extensive remodeling of cellular metabolism, required to sustain proliferation. This was first highlighted by the fact that cancer cells display enhanced rates of glycolysis and lactate production, a phenomenon called now the “Warburg effect” (Warburg, 1956). Although this could suggest that mitochondria are impaired or not used in cancer cells, it is now commonly accepted that mitochondrial function is necessary for cancer cell viability and tumorigenicity (Wallace, 2012). Antibiotics targeting mitochondrial translation, such as the actinonins, have been successfully used as anti-proliferative agents (Lee et al., 2004; Skrtic et al., 2011). Part of actinonin’s mechanism of action involves stalling of mitochondrial ribosomes, followed by a decay of the MRPs and of mitochondrial RNA, culminating with fractionation of the mitochondrial network (Richter et al., 2013). This initiates a retrograde signaling to the nucleus that results in a block of cell proliferation. Consistent with this, actinonin was recently shown to induce the expression of some UPRmt genes in Burkitt’s lymphoma cells (Sheth et al., 2014). It is also tempting to speculate that anti-cancer activity of the inhibition of cytochrome c oxidase in complex IV, induced by treatment with the copper chelator, tetrathiomolybdate, may involve the induction of the UPRmt (Rath et al., 2012), in a manner analogous to that achieved in the worm by cco-1 RNAi targeting the complex IV component, COX4 (Durieux et al., 2011). The potential involvement of the UPRmt in these processes would be interesting to investigate, knowing that they likely involve a mito-nuclear protein imbalance. Although the previous examples suggests that UPRmt activation could be potentially be used as a cancer treatment strategy, mitochondrial chaperones Hsp60 and Hsp10 are comprised in a signature of the statistically 67 most frequent genes induced in tumors versus normal tissue (Rhodes et al., 2004), suggesting that the UPRmt is activated in cancer. Thus, future studies linking cancer and the UPRmt will be required for a better understanding.
Perspectives
Throughout this review, we have tried to give a glimpse of the relevance of the UPRmt in mitochondrial homeostasis in mammals. We emphasized how this pathway crucially impacts on lifespan and fitness of lower species, such as C.elegans and D.melanogaster. Moreover, the cell-non-autonomous nature of the UPRmt suggests that this stress response can be communicated among distant tissues and determine the aging rate of the whole organism. The fact that Mrps5 has been identified as a longevity gene in mice (Houtkooper et al., 2013) and the lethality of Hsp60 knockout (Kleinridders et al., 2013) indicates that the UPRmt is also essential for mitochondrial function and whole body homeostasis in mammals. However, a more fundamental understanding of the UPRmt pathway in mammals is urgently required. Future research efforts will not only need to map the tissues and the physiological conditions in which the UPRmt is triggered, but also need to provide a deep mechanistic insight into the mammalian UPRmt signaling and to elucidate the physiological and pathophysiological consequences of the UPRmt activation. The fact that mitochondrial fusion often accompanies the UPRmt should prompt researchers to investigate how the UPRmt communicates with and/or orchestrate the triggering of other mitochondrial, stress responses, such as fission, fusion, mitophagy and apoptosis (Jin and Youle, 2013). On top of that, if the homeostatic nature of the UPRmt, which has been well established in lower species, is conserved in mammals, modulating this stress response might constitute a therapeutic strategy to treat diseases characterized by mitochondrial dysfunction.
Acknowledgments
JA is the Nestlé Chair in Energy Metabolism. Work in the laboratory is supported by the Ecole Polytechnique Fédérale de Lausanne, the National Institutes of Health (R01AG043930), the Swiss National Science Foundation (31003A-124713) and Systems X (51RTP0-151019).
References
- Aldridge JE, Horibe T, Hoogenraad NJ. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PloS one. 2007;2:e874. doi: 10.1371/journal.pone.0000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreux PA, Houtkooper RH, Auwerx J. Pharmacological approaches to restore mitochondrial function. Nature reviews Drug discovery. 2013;12:465–483. doi: 10.1038/nrd4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argmann CA, Chambon P, Auwerx J. Mouse phenogenomics: the fast track to “systems metabolism”. Cell metabolism. 2005;2:349–360. doi: 10.1016/j.cmet.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Baker BM, Nargund AM, Sun T, Haynes CM. Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS genetics. 2012;8:e1002760. doi: 10.1371/journal.pgen.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174:229–239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Molecular cell. 2010;40:238–252. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annual review of genetics. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- de Castro IP, Martins LM, Loh SH. Mitochondrial quality control and Parkinson’s disease: a pathway unfolds. Molecular neurobiology. 2011;43:80–86. doi: 10.1007/s12035-010-8150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer F, Hamann A, Osiewacz HD. Mitochondrial quality control: an integrated network of pathways. Trends in biochemical sciences. 2012;37:284–292. doi: 10.1016/j.tibs.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbauer AB, Zahedi RP, Sickmann A, Pfanner N, Meisinger C. The Protein Import Machinery of Mitochondria-A Regulatory Hub in Metabolism, Stress, and Disease. Cell metabolism. 2014;19:357–372. doi: 10.1016/j.cmet.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. Journal of gerontology. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Developmental cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Molecular cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horibe T, Hoogenraad NJ. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PloS one. 2007;2:e835. doi: 10.1371/journal.pone.0000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013;9:1750–1757. doi: 10.4161/auto.26122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Steffen KK, Kaeberlein M. Ruminations on dietary restriction and aging. Cellular and molecular life sciences : CMLS. 2007;64:1323–1328. doi: 10.1007/s00018-007-6470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim YN, Kim SS, Kim do H, Hur KY, Kim HK, et al. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nature medicine. 2013;19:83–92. doi: 10.1038/nm.3014. [DOI] [PubMed] [Google Scholar]
- Kleinridders A, Lauritzen HP, Ussar S, Christensen JH, Mori MA, Bross P, Kahn CR. Leptin regulation of Hsp60 impacts hypothalamic insulin signaling. The Journal of clinical investigation. 2013 doi: 10.1172/JCI67615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MD, She Y, Soskis MJ, Borella CP, Gardner JR, Hayes PA, Dy BM, Heaney ML, Philips MR, Bornmann WG, et al. Human mitochondrial peptide deformylase, a new anticancer target of actinonin-based antibiotics. The Journal of clinical investigation. 2004;114:1107–1116. doi: 10.1172/JCI22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Lu B. Reduction of protein translation and activation of autophagy protect against PINK1 pathogenesis in Drosophila melanogaster. PLoS genetics. 2010;6:e1001237. doi: 10.1371/journal.pgen.1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Butow RA. Mitochondrial retrograde signaling. Annual review of genetics. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- Loson OC, Song Z, Chen H, Chan DC. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Molecular biology of the cell. 2013;24:659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinou JC, Youle RJ. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Developmental cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinus RD, Garth GP, Webster TL, Cartwright P, Naylor DJ, Hoj PB, Hoogenraad NJ. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. European journal of biochemistry/FEBS. 1996;240:98–103. doi: 10.1111/j.1432-1033.1996.0098h.x. [DOI] [PubMed] [Google Scholar]
- Mori K. Signalling pathways in the unfolded protein response: development from yeast to mammals. Journal of biochemistry. 2009;146:743–750. doi: 10.1093/jb/mvp166. [DOI] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Auwerx J. NAD(+) metabolism: a therapeutic target for age-related metabolic disease. Critical reviews in biochemistry and molecular biology. 2013a;48:397–408. doi: 10.3109/10409238.2013.789479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013b;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annual review of biochemistry. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa L, Germain D. Estrogen receptor mediates a distinct mitochondrial unfolded protein response. Journal of cell science. 2011;124:1396–1402. doi: 10.1242/jcs.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC genetics. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta de Castro I, Costa AC, Lam D, Tufi R, Fedele V, Moisoi N, Dinsdale D, Deas E, Loh SH, Martins LM. Genetic analysis of mitochondrial protein misfolding in Drosophila melanogaster. Cell death and differentiation. 2012;19:1308–1316. doi: 10.1038/cdd.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radke S, Chander H, Schafer P, Meiss G, Kruger R, Schulz JB, Germain D. Mitochondrial protein quality control by the proteasome involves ubiquitination and the protease Omi. The Journal of biological chemistry. 2008;283:12681–12685. doi: 10.1074/jbc.C800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainbolt TK, Atanassova N, Genereux JC, Wiseman RL. Stress-Regulated Translational Attenuation Adapts Mitochondrial Protein Import through Tim17A Degradation. Cell metabolism. 2013;18:908–919. doi: 10.1016/j.cmet.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath E, Berger E, Messlik A, Nunes T, Liu B, Kim SC, Hoogenraad N, Sans M, Sartor RB, Haller D. Induction of dsRNA-activated protein kinase links mitochondrial unfolded protein response to the pathogenesis of intestinal inflammation. Gut. 2012;61:1269–1278. doi: 10.1136/gutjnl-2011-300767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Molecular cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Richter U, Lahtinen T, Marttinen P, Myohanen M, Greco D, Cannino G, Jacobs HT, Lietzen N, Nyman TA, Battersby BJ. A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation. Current biology : CB. 2013;23:535–541. doi: 10.1016/j.cub.2013.02.019. [DOI] [PubMed] [Google Scholar]
- Ristow M, Zarse K. How increased oxidative stress promotes longevity and metabolic health: The concept of mitochondrial hormesis (mitohormesis) Experimental gerontology. 2010;45:410–418. doi: 10.1016/j.exger.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Rugarli EI, Langer T. Mitochondrial quality control: a matter of life and death for neurons. The EMBO journal. 2012;31:1336–1349. doi: 10.1038/emboj.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MT, Hoogenraad NJ. Mitochondrial-nuclear communications. Annual review of biochemistry. 2007;76:701–722. doi: 10.1146/annurev.biochem.76.052305.091720. [DOI] [PubMed] [Google Scholar]
- Schleit J, Johnson SC, Bennett CF, Simko M, Trongtham N, Castanza A, Hsieh EJ, Moller RM, Wasko BM, Delaney JR, et al. Molecular mechanisms underlying genotype-dependent responses to dietary restriction. Aging cell. 2013;12:1050–1061. doi: 10.1111/acel.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nature reviews Molecular cell biology. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- Schroder M. The unfolded protein response. Molecular biotechnology. 2006;34:279–290. doi: 10.1385/MB:34:2:279. [DOI] [PubMed] [Google Scholar]
- Sheth A, Escobar-Alvarez S, Gardner J, Ran L, Heaney ML, Scheinberg DA. Inhibition of human mitochondrial peptide deformylase causes apoptosis in c-myc-overexpressing hematopoietic cancers. Cell death & disease. 2014;5:e1152. doi: 10.1038/cddis.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrtic M, Sriskanthadevan S, Jhas B, Gebbia M, Wang X, Wang Z, Hurren R, Jitkova Y, Gronda M, Maclean N, et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer cell. 2011;20:674–688. doi: 10.1016/j.ccr.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen A, Elo JM, Pietilainen KH, Hakonen AH, Sevastianova K, Korpela M, Isohanni P, Marjavaara SK, Tyni T, Kiuru-Enari S, et al. FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet neurology. 2011;10:806–818. doi: 10.1016/S1474-4422(11)70155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supale S, Thorel F, Merkwirth C, Gjinovci A, Herrera PL, Scorrano L, Meda P, Langer T, Maechler P. Loss of prohibitin induces mitochondrial damages altering beta-cell function and survival and is responsible for gradual diabetes development. Diabetes. 2013;62:3488–3499. doi: 10.2337/db13-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondria and cancer. Nature reviews Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin IE. Symbionticism in the light of recent cytological investigations--Ivan E. Wallin, 1969. Bio Systems. 1993;31:181–183. doi: 10.1016/0303-2647(93)90047-g. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wang X, Zuo X, Kucejova B, Chen XJ. Reduced cytosolic protein synthesis suppresses mitochondrial degeneration. Nature cell biology. 2008;10:1090–1097. doi: 10.1038/ncb1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Wek RC, Cavener DR. Translational control and the unfolded protein response. Antioxidants & redox signaling. 2007;9:2357–2371. doi: 10.1089/ars.2007.1764. [DOI] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochemical Society transactions. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. Journal of cell science. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. The EMBO journal. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]