Abstract
Tau is a microtubule-associated protein that is genetically linked to dementia and linked to Alzheimer’s disease via its presence in intraneuronal neurofibrillary tangle deposits, where it takes the form of aggregated paired helical and straight filaments. Although the precise mechanisms by which tau contributes to neurodegeneration remain unclear, tau aggregation is commonly considered to be a critical component of tau-mediated pathogenicity. Nevertheless, the context in which tau aggregation begins in vivo is unknown. Tau is enriched in membrane-rich neuronal structures such as axons and growth cones, and can interact with membranes both via intermediary proteins and directly via its microtubule-binding domain (MBD). Membranes efficiently facilitate tau aggregation in vitro, and may therefore provide a physiologically relevant context for nucleating tau aggregation in vivo. Furthermore, tau-membrane interactions may potentially play a role in tau’s poorly understood normal physiological functions. Despite the potential importance of direct tau-membrane interactions for tau pathology and physiology, the structural mechanisms that underlie such interactions remain to be elucidated. Here, we employ electron spin resonance spectroscopy to investigate the secondary and long-range structural properties of the MBD of three-repeat tau isoforms when bound to lipid vesicles and membrane mimetics. We show that the membrane interactions of the tau MBD are mediated by short amphipathic helices formed within each of the MBD repeats in the membrane-bound state. To our knowledge, this is the first detailed elucidation of helical tau structure in the context of intact lipid bilayers. We further show, for the first time (to our knowledge), that these individual helical regions behave as independent membrane-binding sites linked by flexible connecting regions. These results represent the first (to our knowledge) detailed structural view of membrane-bound tau and provide insights into potential mechanisms for membrane-mediated tau aggregation. Furthermore, the results may have implications for the structural basis of tau-microtubule interactions and microtubule-mediated tau aggregation.
Introduction
Tau was originally identified as a microtubule-associated protein and shown to influence microtubule dynamics, reduce microtubule treadmilling, and stabilize microtubule structure and organization (1,2). In the human central nervous system, tau is found as six different isoforms that are generated by the alternative splicing of three exons (3,4). The longest human tau isoform, commonly referred to as htau40, contains all three alternative exons, whereas the shortest isoform, commonly referred to as tau352, contains none of the three (Fig. 1). Two of the exons code for protein sequences in the N-terminal projection domain of tau, whereas the third exon (exon 10) codes for an alternative additional repeat (R2) in the tandem repeat containing the microtubule-binding C-terminal domain of tau, which always contains the three other repeats (R1, R3, and R4). Tau is the principal protein component of the neurofibrillary tangle deposits that constitute one of two pathological hallmarks of Alzheimer’s disease (AD) (5–8). Within the intraneuronal tangle deposits, tau is found in an aggregated filamentous form that can assume various morphologies, including paired helical filaments (PHFs) and straight filaments (SFs), as well as ribbons and sheets. Whereas the familial forms of AD involve mutations in genes involved in the production of the amyloid-beta (Aβ) peptide, mutations in tau cause frontotemporal dementia (9–11), the second most common form of dementia after AD. Disease-linked alterations in the human tau gene MAPT lead either to mutant forms of tau protein or to changes in tau splicing that typically favor the formation of R2-containing four-repeat (4R) forms of the protein.
Figure 1.
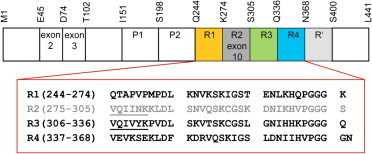
Cartoon representation of FL tau protein. Tau possesses a multidomain architecture that encompasses alternatively spliced exons (2,3,10), two proline-rich domains (P1 and P2), MBD repeats (R1–R4), and the pseudorepeat R′. The amino acid sequences of the tau MBD repeats are shown, each on a separate line. The alternatively spliced R2 repeat is shown in gray. The tau fragment K19 used in these studies includes repeats R1, R3, and R4 plus four C-terminal flanking residues (KKIE). The PHF6∗ and PHF6 hexapeptide aggregation-nucleating motifs are underlined in R2 and R3, respectively.
The precise mechanisms by which tau leads to neurodegeneration in frontotemporal dementia, and presumably in AD as well, remain unclear, but tau assembly and aggregation into PHFs and other filamentous species are commonly considered to play a key role. Although tau can be induced to aggregate in vitro in a number of ways, the context for the nucleation of tau aggregation in vivo is not known. Because lipid membranes are known to enhance tau aggregation in vitro (12–15) and tau has been reported to interact both indirectly and directly with cellular membranes (16–20), membrane surfaces may provide an effective locus for tau aggregation in vivo. This conjecture is supported by observations of membrane-associated tau filaments in tissues from AD brains (21,22). Despite the potential importance of membrane-mediated tau aggregation, little is known about the structural consequences of tau membrane binding or how membrane binding may enhance tau aggregation. Here, we use electron spin resonance (ESR) spectroscopy to explore the conformations of regions within the tau microtubule-binding domain (MBD) in the context of a membrane-bound tau MBD fragment as well as a full-length (FL) human tau isoform. We find that within each repeat, a short segment that was previously identified as exhibiting a weak preference for helical structures adopts a fully helical amphipathic structure. Thus, tau-membrane interactions are mediated by short amphipathic helices. We show that the helices are located at the surface of the membrane and do not penetrate deeply into the hydrophobic membrane interior. Distance measurements between helical segments located in different repeats indicate that the individual helices behave independently of one another and that the intervening regions likely behave as highly dynamic and flexible linkers. The implications for tau structure and membrane-induced tau aggregation are discussed.
Materials and Methods
Protein mutagenesis, expression, and purification
Double and single tau mutants were produced using a site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) as directed by the manufacturer. Recombinant proteins were expressed in E. coli BL21/DE3 cells (Novagen, San Diego, CA) transfected with plasmids encoding the tau fragment K19 or the FL tau isoform tau352 under the control of a T7 promoter. The cells were lysed by sonication and lysates were cleared from cell debris by ultracentrifugation at 150,000 × g. Tau variants were further purified by cation-exchange chromatography followed by reverse-phase high-performance liquid chromatography on a C4 column (GRACE, Columbia, MD). Protein purity was confirmed by SDS-PAGE. Purified proteins were lyophilized and stored at −20°C.
Preparation of lipid vesicles and membrane mimetics
Lipids were purchased from Avanti Polar Lipids (Alabaster, AL). Deuterated compounds were obtained from Cambridge Isotope Laboratories (Tewksbury, MA). For continuous-wave (CW)-ESR measurements, aliquots of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-(phospho-L-serine) (POPS) in chloroform were mixed in proportions to yield a final 1:1 molar ratio of noncharged/charged lipids. A separate sample of the same molar proportion of POPC and POPS plus spin-labeled lipid 1-palmitoyl-2-stearoyl-(5-doxyl)-sn-glycero-3-phosphocholine (16:0-5 Doxyl PC or 5PC) added to a 1:690 nonlabeled/labeled lipids molar ratio was prepared. The chloroform mixtures were dried under nitrogen gas and then kept under vacuum for at least 4 h. Dried lipids were dissolved in 10 mM NaH2PO4, 10 mM NaCl pH 7.4. Small unilamellar vesicles (liposomes) of 1:1 POPC/POPS were prepared by repeated sonication. To remove large multilamellar vesicles and titanium particles, the ready liposome solutions were centrifuged at 3000 × g for 30 min at room temperature. Throughout all experiments, only freshly made liposomes were used. The typical vesicle diameters were ∼75 nm as determined by dynamic light scattering (not shown). Membrane-mimetic micelles composed of lyso-1-palmitoyl-phosphatidylglycerol (LPPG) were prepared at a concentration of 40 mM. Spheroidal or rod-like SDS micelles were prepared using SDS-d25 at concentrations of 40 or 450 mM, respectively.
ESR sample preparation
Nitroxide spin-labeled proteins were produced by dissolving K19 and tau352 cysteine mutants in buffer, adding S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSSL; Toronto Research Chemicals, Toronto, Canada) to a 30:1 MTSSL/protein molar ratio, and agitating for 2 hr at room temperature. Excess spin label was removed using Micro Bio-Spin columns (Bio-Rad Laboratories, Hercules, CA). Samples for CW-ESR experiments were prepared in NaH2PO4, 10 mM NaCl pH 7.4, and samples for distance measurements were prepared in NaH2PO4, 100 mM NaCl using D2O instead of H2O, pD 7.4 (regular pH reading without pD correction). Select duplicate distance measurements carried out in the buffer used for CW-ESR experiments produced virtually identical time-domain pulse-ESR dipolar spectroscopy (PDS) data and distance distributions. For membrane or membrane-mimetic-containing samples, singly and doubly spin-labeled cysteine mutants of K19 or tau352 were incubated with liposomes at molar ratios ranging from 1:1200 to 1:1600, to ensure close to 100% binding as confirmed by CW-ESR. The final protein concentrations ranged from 40 μM to 52 μM. Doubly spin-labeled cysteine mutants were mixed with membrane mimetics in the following proportions: 60 μM (or 30 μM) protein/40 mM SDS, 100 μM protein/450 mM SDS, and 60 μM protein/40 mM LPPG.
All CW-ESR measurements on spin-labeled lipids and on spin-labeled tau mutants bound to POPC/POPS liposomes or free in solution were performed on a Bruker ELEXIS E500 (Bruker, Billerica) spectrometer equipped with a Bruker ER 4122SHQE resonator. The spectra were recorded at 25°C using a Bruker ER4131VT temperature controller. The full extent of the nitroxide CW-ESR spectrum was recorded using an incident microwave power of 1.26 mW, with a field modulation of 2.3 G for 5PC and lipid-bound protein, and 1.2 G for free protein in solution. All samples for CW-ESR measurements were placed into 50 μL precision microcapillaries with sealed bottoms (Kimble Glass, Vineland, NJ).
Nitroxide spectrum microwave power saturation (23) was employed to measure accessibility to the commonly used fast-relaxing agents oxygen (O2) and Ni(II)-diammine-2,2'-(ethane-1,2-diyldiimino)diacetic acid complex (NiEDDA). Measurements were performed on regular samples in the presence of O2, deoxygenated and argon-filled samples, and deoxygenated and argon-filled samples containing NiEDDA. Sample deoxygenation was performed on a vacuum line by repeatedly bringing the capillary tube with the sample to a soft vacuum level and filling it with argon gas. Finally, the capillary tube was filled with argon to ∼0.9 bar and flame sealed. In all cases, the sample volume was ∼6.7 μL (i.e., 10 mm capillary length). The final concentration of NiEDDA was 5 mM, and that for O2 was obtained by equilibration in air at 25°C. To obtain the half-saturation parameter, P1/2 (23,24), the central line in the nitroxide CW-ESR spectrum (∼20 G width for lipid samples and ∼8 G for protein without lipid) was recorded as a function of the microwave power (varied from 0.5 mW to 200 mW in 20 steps; see Fig. S1 in the Supporting Material). The measured intensity of the central line was plotted as a function of the square root of the microwave power applied (Figs. S1 and S2) and the data were fitted to the equation
| (1) |
where P is the microwave power applied and ε is a line-homogeneity parameter. We obtained the best fits using ε = 1.5 (which is typical of a highly homogeneous spectrum).
In the calculations of the accessibility parameter, Π, for both O2 and NiEDDA, we used the data for K347C in deoxygenated solution as a reference (Fig. S2 b). A soluble protein was used as such a reference in a previous study on a membrane protein (25). Indeed, the reference used affects the absolute accessibility values (vide infra), but the insertion depth (or contrast) parameter, Φ, defined as a logarithm of the ratio of accessibilities to O2 and NiEDDA (defined above) is, of course, reference invariant. The accessibilities to O2 and NiEDDA were calculated using the expression
| (2) |
where ΔH (or ΔHref) is the line width and superscript ref indicates the reference. All analyses and fittings of CW-ESR power saturation data were performed in OriginLab software (OriginLab, Northampton, MA). To ensure data reproducibility, the measurements were performed twice on all singly labeled residues in helixes 3 and 4 in K19 and on 5PC-labeled samples.
DEER distance measurements and distance modeling
Distance measurements were performed on doubly spin-labeled tau mutants free in solution (100 μM protein) and in the presence of spheroidal and rod-like SDS micelles, LPPG micelles, and POPC/POPS liposomes. Glycerol-d8 was added to 40% (w/v) to samples of protein in buffer and to 30% (w/v) to samples of protein bound to SDS or LPPG. The liposome samples did not contain glycerol. All samples for pulse-ESR measurements were placed into custom-sized 1.8 mm i.d. borosilicate glass sample tubes (Wilmad-LabGlass, Vineland, NJ) and plunge-frozen in LN2. All distance measurements were performed at 60 K as previously described (26,27) using an in-house-built Ku-band pulse ESR spectrometer (28) operating at 17.3 GHz. A standard four-pulse double electron-electron resonance (DEER) sequence (29) with π/2-π-π pulse widths of 16 ns, 32 ns, and 32 ns, respectively, and a 32 ns pump π pulse was used. The frequency separation between the detection and pump pulses was 70 MHz. The detection pulses were positioned at the low-field edge of the nitroxide spectrum. The dipolar evolution times ranged from 1.8 to 8.2 μs depending on the distance measured and the spin-label phase relaxation time. The data recording times were in the range of 2–16 h depending on the distance measured, sample composition, and protein concentration. The liposome samples usually required 8–16 h and shorter times were used for other samples.
The background (baseline) was removed from raw time-domain signals, and distances were reconstructed from the baseline-corrected and normalized signals using the Tikhonov regularization method (30) and refined by the maximum-entropy method (31). In most cases, the background signal in the raw time-domain DEER data was removed by fitting to a straight line in a semilogarithmic plot, i.e., to a homogeneous background. However, in the case of liposome samples, backgrounds deviated slightly from a straight line and were fit using second-degree polynomials (Fig. S3).
The experimental distance distributions were modeled by fitting them to Gaussian functions in the OriginLab software. The quality of the fit parameter, R2, was in the range of 0.7–0.99, with R2 > 0.9 being the case for more than 90% of the fits.
Results
To characterize the structural properties of tau MBD of human tau protein bound to lipid membranes, we employed site-directed spin labeling (SDSL) and ESR spectroscopy (32), a method that can provide detailed information about protein secondary and higher-order structure, as well as report on dynamics (23,24,26,27,32–43). Furthermore, ESR has proven to be a powerful tool for structural studies of membrane-bound intrinsically disordered proteins (26,27,44) and their amyloid forms (45,46). We conducted experiments using a tau fragment (K19) encompassing residues 244–274 and 306–372 of the longest human tau isoform, htau40. K19 corresponds to the MBD repeats of the alternatively spliced three-repeat (3R) forms of tau that contain repeats R1, R3, and R4, whereas repeat R2 is spliced out (Fig. 1). In addition, we also performed a subset of experiments using the shortest FL tau isoform, tau352, to assess whether the behavior of the K19 fragment mirrors that of the FL protein. We employed CW-ESR microwave power-saturation experiments (23–25) to obtain local structural information at the single-residue level. PDS measurements based on DEER spectroscopy (29,35,42,47) provided information regarding the long-range tertiary organization of the tau MBD repeats when bound to membrane vesicles and membrane mimetics.
Tau K19 binds to unilamellar 1:1 POPC/POPS lipid vesicles, and the segments comprising residues 253–261, 315–323, and 346–355 adopt a highly helical structure
The tau MBD interacts directly with biological membranes (16,19,20,48,49), and recent studies employing solution-state NMR demonstrated that regions within tau K19 adopt a highly helical conformation upon interaction with membrane-mimetic detergent and acidic lysophospholipid micelles (50). Circular dichroism (CD) measurements suggested that helical structure is also formed within K19 in the presence of 1:1 POPA/POPC liposomes. Helical structure in the micelle-bound protein was localized to residues 253–261 (helix 1), 315–323 (helix 3), and 346–355 (helix 4), located within repeats 1, 3, and 4 of K19, respectively (50), but whether these regions are helical in the vesicle-bound state is not known.
To directly probe the conformation of these segments in liposome-bound tau K19, we generated 27 single cysteine mutants positioned at residues 252–261, 315–322, and 345–354 within the K19 fragment, as well as nine single cysteine mutants positioned at residues 253–261 in the shortest FL tau isoform, tau352. We labeled these tau variants with MTSSL and monitored binding to liposomes by recording the MTSSL CW-ESR spectrum as a function of the spin-label position in the protein polypeptide chain in buffer solution and in the presence of POPC/POPS vesicles at lipid/protein molar ratios of at least 1200:1, well above values associated with lipid-induced tau aggregation (13,51). To ensure complete binding, we used a higher proportion of acidic phospholipid than is typically found in biological membranes. At lower anionic lipid content, tau membrane affinity is decreased, leading to a greater percentage of unbound protein at a given protein/lipid molar ratio, but the helical character of the membrane-bound fraction is preserved (50). Furthermore, in certain disease states, including AD, anionic phospholipid content is increased in cell membranes (52).
The nitroxide CW-ESR spectra for both K19 and FL tau protein in the absence of lipids are characterized by narrow peaks, indicating a highly mobile spin label (Fig. 2, green), which is typical for nitroxide spin label attached to unstructured proteins, such as the previously characterized α-synuclein (53,54), or other unstructured protein segments (55). These data are in agreement with NMR data indicating that tau K19 and FL tau are highly dynamic and unstructured in solution (50,56–60).
Figure 2.
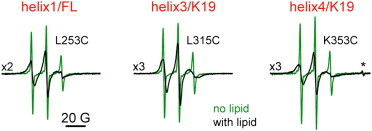
Representative CW-ESR spectra of spin-labeled residues in the MBD repeat region of tau (helices 1, 3, and 4). The spectra in solution with no lipid are in green and the spectra of lipid-bound protein are in black. The spectra in the left panel are from FL tau352 spin labeled at position 253 in R1/helix 1. The spectra in the middle and right panels are from the tau K19 construct, spin labeled at positions 315 and 353 in R3/helix 3 and R4/helix 4, respectively. A narrow peak marked by the asterisk in the right panel is from a Bruker standard.
Upon addition of liposomes composed of a 1:1 molar ratio of noncharged/charged lipids (POPC/POPS), we observed changes in the CW-ESR spectra for all studied tau variants that indicate restricted spin-label mobility and are characteristic of more constrained protein dynamics (Fig. 2, black). Thus, these spectral changes provide evidence for the interaction of tau K19 and tau352 with the POPC/POPS liposomes. The CW-ESR spectra for all spin-labeled liposome-bound tau variants are plotted in Fig. 3. As shown in the figure, under our experimental conditions, the tau MBD-lipid association is virtually complete. The contribution from unbound protein or unreacted spin label to the CW-ESR spectra is negligible (in some cases, it can be noticed as narrow lines of very low intensity superimposed on the spectrum from lipid-bound protein; Fig. 3).
Figure 3.
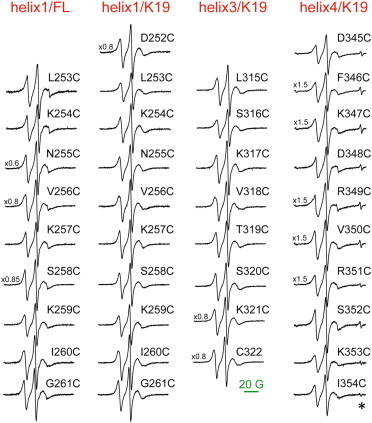
Complete set of CW-ESR spectra of lipid-bound tau. The spectra from spin-labeled residues in R1/helix 1 of FL tau are plotted in the leftmost column; all other spectra are from spin-labeled residues in the tau K19 construct (residues from R1/helix 1, R3/helix 3, and R4/helix 4 are plotted in the second, third, and fourth columns as indicated). The spectra were baseline corrected and normalized to the same number of spins. The line from a Bruker standard is marked by the asterisk. To see this figure in color, go online.
To assess secondary structure at the spin-labeled sites of membrane-bound tau, we conducted experiments to determine the accessibilities of the nitroxide spin label in each of the tau variants to the fast-relaxing colliders O2, which has high solubility in lipids (∼21% at equilibrium with air), and NiEDDA, which is polar and highly water soluble. In these experiments, we estimated the microwave power saturation effects on the central line in the nitroxide CW-ESR spectrum produced by either O2 or NiEDDA (Fig. S1). In principle, these data can be used to distinguish solvent exposed from buried residues as well as to determine the orientation of spin-labeled residues with respect to the membrane surface (23,24,53,61,62). The determined accessibilities to both O2 (Π(O2)) and NiEDDA (Π(NiEDDA)) are plotted in Fig. 4 a as a function of spin-label position (residue number). A careful examination of these data reveals an out-of-phase change in the accessibilities to O2 and NiEDDA with a periodicity of three or four residues that is characteristic of α-helical structure.
Figure 4.
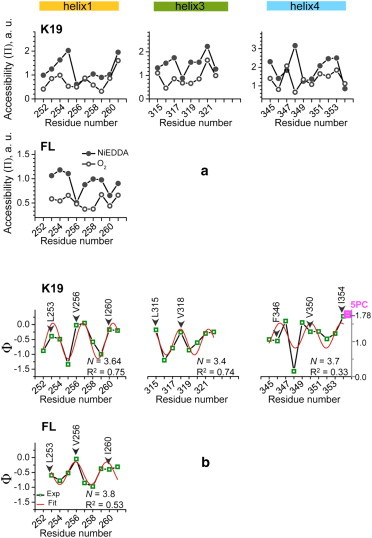
(a) Dependence of the accessibility parameter Π on the spin-label position for all studied sites in helices 1, 3, and 4, located in repeats R1, R3, and R4, respectively, of FL tau and tau K19. The data obtained in air (Π(O2)) and 5 mM NiEDDA (Π(NiEDDA)) are shown by solid and open circles, respectively, connected by black lines. (b) The dependence of the depth parameter, Φ, on spin-label position for all studied sites is shown in green squares connected by black lines. The experimental data were fitted to a cosine function to examine periodicity in Ф (see Materials and Methods); the fitted curves are shown in red and the resulting periodicity, N, and quality of fit, R2, are indicated. In both a and b, the data for helices 1, 3, and 4 in tau K19 are plotted in the upper plots and the data for helix 1/FL are shown in the lower plot. The positions of original hydrophobic residues are labeled and designated by arrows. The Ф parameter for spin-labeled lipid, 5PC, is shown in the upper-right plot in magenta (value of 1.78). Note that Ф scale for the 5PC sample, shown on a y axis on the right side of the graph, is shifted down relative to that for tau mutants.
The depth parameter Φ (Φ = ln[Π(O2)/Π(NiEDDA)]) can be used to better visualize the difference between Π(O2) and Π(NiEDDA), and consequently to assess protein topology. Spin-labels attached to more solvent-exposed residues, with smaller Π(O2) and larger Π(NiEDDA), have a smaller Φ parameter and vice versa. The calculated values of Φ for each of the studied residues in tau K19 and FL tau352 are plotted versus residue number in Fig. 4 b (green open squares connected by black lines). Within each of the studied amino acid segments (252–261, 315–322, and 345–354), Φ exhibits a clear periodic behavior. The data within each separate segment were fit to a periodic function, Φ(n) = Φ0 + Acos(2πn/N + b) (53,62) (Fig. 4 b, red line), and the resulting values of the periodicity N ranged between 3.4 and 3.8 amino acids per turn for the three individual segments. The average value of 3.63 ± 0.17 amino acids per turn agrees well with the theoretical α-helical periodicity of 3.6. A similar approach was used to analyze the periodicity of liposome-bound α-synuclein (44,53).
A close inspection of the data plotted in Fig. 4 b shows that spin labels positioned at sites of hydrophobic residues (L253, V256, and I260 in helix 1; L315 and V318 in helix 3; and F346, V351, and I354 in helix 4) have larger Φ-values (indicating lesser/greater accessibility to NiEDDA/O2), consistent with these positions being oriented toward the lipid membrane. Sites of hydrophilic residues (N255 and S258 in helix 1; S316, T319, and S320 in helix 3; and D348 and S352 in helix 4) are characterized by smaller Φ-values, consistent with greater solvent exposure. Interestingly, the helical arrangement of amino acids in helix 1 shows a shift of approximately one residue between the K19 fragment and the FL tau352 protein (Fig. 4 b, upper and lower leftmost panels). This might be a result of experimental uncertainty or might result from the influence of regions of the FL protein that are not present in the K19 fragment. In either case, the region in question clearly adopts an amphipathic helical structure in both contexts.
The helices in the tau MBD repeats are located at the membrane surface and do not penetrate deeply into the lipid bilayer
We noted that the entire set of CW-ESR nitroxide spectra possess nearly identical line shapes (Fig. 3). This observation points to similar dynamic properties among all spin-labeled residues. A coarse estimate of the dynamic properties of a spin-labeled site can be obtained from the inverse width of the central line in the CW-ESR nitroxide spectrum (ΔH0−1) (63). Values of ΔH0−1 for all recorded CW-ESR spectra in both K19 and FL tau fall into a narrow range of 0.3–0.37 Gauss−1, which is considerably larger than values typically observed for substantially immobilized spin-labeled residues in transmembrane helices (61). The observed values thus indicate a lack of significant spin-label immobilization and therefore are consistent with the helical segments being positioned on the membrane surface without penetrating deeply into the bilayer. Similar behavior was observed for spin-labeled residues in the membrane-bound region of human α-synuclein (53).
The Π(O2) data (Fig. 4 a) provide further support for the conclusion that the tau K19 helices are positioned at the membrane surface. The measured values for O2 accessibility exhibit a relatively flat pattern that is only weakly sensitive to the expected orientation of a given site toward the membrane or the solvent. This suggests a similar level of exposure to O2 for all sites, which is less consistent with complete burial of membrane-facing sites in the spin label in the O2-rich hydrocarbon region of the membrane, and instead is more consistent with these sites being positioned largely in the headgroup region of the membrane, which is expected to have an O2 content that is more similar to that present in the surrounding aqueous solvent (64).
To further assess the position of the K19 helices with respect to the membrane, we measured the O2 and NiEEDA accessibilities and determined the Φ parameter of a spin-labeled lipid 5PC (16:0-5 Doxyl PC) incorporated into liposomes of 1:1 POPC/POPS under the same conditions used for the spin-labeled tau mutants. We then compared the results with those obtained for spin-labeled sites in tau K19. 5PC is a well-characterized standard for estimating membrane immersion depth, with a paramagnetic nitroxide placed at the interface between the polar headgroup and hydrophobic acyl chain regions (65). Therefore, Φ-values that are either larger or smaller than those for 5PC should specify either deeper penetration in the lipid acyl chain region or a position closer to the membrane surface, respectively. Our results show that O2 had a pronounced effect on the microwave power saturation properties of 5PC spectrum, whereas the effect of 5 mM NiEDDA was marginal (Fig. S2 a). Thus, under our experimental conditions, 5PC was almost inaccessible to the hydrophilic relaxer, NiEDDA. This is dissimilar to our data for all spin-labeled tau mutants, since for each of them the microwave power saturation was affected by both O2 and NiEDDA, although the extent depended on the spin-label position (Fig. S2 c). Furthermore, for 5PC, we obtained a Φ parameter of 1.78, whereas the Φ-values for tau mutants were typically smaller, with some being close to zero (Fig. 4 b). These observations confirm that the helices in membrane-bound tau MBD are associated with the membrane periphery, in agreement with a previous analysis of micelle-bound K19 using NMR (50).
Long-range architecture of membrane-bound tau MBD: independent helices separated by flexible linkers
To evaluate the long-range architecture of tau K19 on the membrane surface and, in particular, the relative arrangement of the individual helices described above, we conducted DEER distance measurements on doubly spin-labeled cysteine mutants of the K19 fragment. We designed seven mutants with cysteines located either in the individual helices or in the intervening linker regions (linker 1 refers to the region between helix 1 and helix 3, and linker 2 refers to the region between helix 3 and helix 4). We used these seven pairs to evaluate distances between the following locations: helix 1/linker 1 (S258C/G273C), helix 1/helix 3 (S258C/S320C), helix 1/helix 4 (S258C/S352C), linker 1/helix 3 (G273C/S320C), helix 3/linker 2 (S320C/G335C), helix 3/helix 4 (S320C/S352C), and linker 2/helix 4 (G335C/S352C). After spin labeling with MTSSL, we subjected these K19 variants to PDS distance measurements both in the free state (no lipid present) and in the presence of 1:1 POPC/POPS liposomes as well as several membrane mimetics (spheroidal LPPG micelles (40 mM LPPG) and spheroidal and cylindrical (rod-like) SDS micelles (40 or 450 mM SDS, respectively)).
We evaluated whether any protein aggregation occurs in the absence or presence of membranes by comparing the experimental modulation depth (66) with a known value for the same experimental conditions (37,67). Under our experimental conditions, the previously determined modulation depth for doubly spin-labeled proteins is 0.23 (37,67). Smaller values most likely indicate low spin-labeling efficiency (37), whereas larger values point to a larger number of interacting spins (68) than two and consequently to protein aggregation. The modulation depths in our experimental DEER data for the various double spin-labeled K19 mutants were equal or very close to 0.23 (Fig. 5), strongly suggesting that the polypeptides remain monomeric. In addition, magnetic dilution experiments in which cysteine-free unlabeled protein was added to samples containing spin-labeled protein showed no alterations in the DEER signal and reconstructed distances, indicating that the distance distributions originate from intramolecular contributions, and largely excluding any possible presence of or contribution from protein aggregates in these measurements (Figs. S4 and S5).
Figure 5.
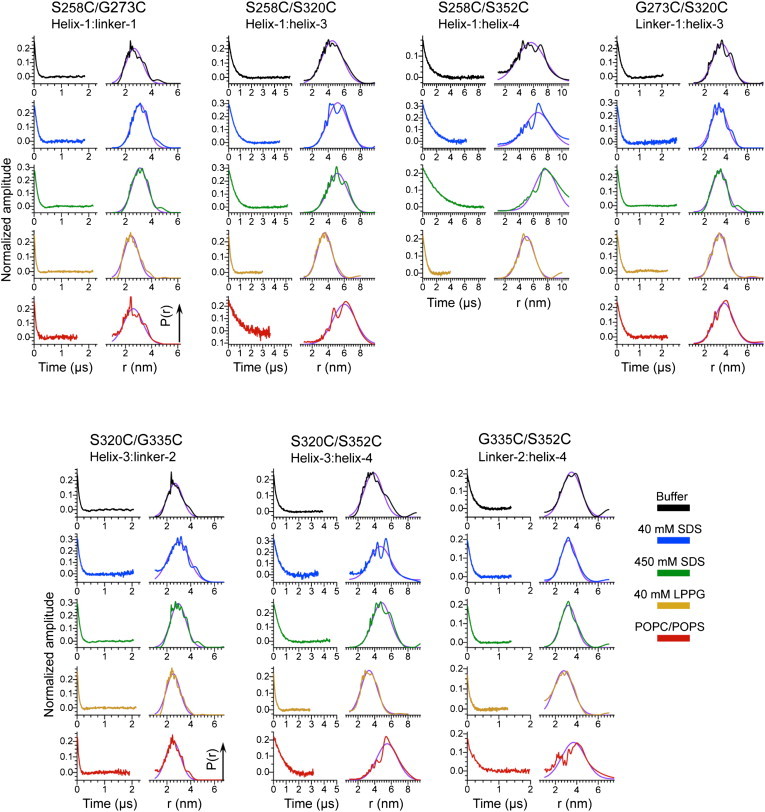
Experimental time-domain DEER data (left side of each panel) and reconstructed distance distributions for doubly spin-labeled cysteine mutants in seven K19 tau constructs (S258C/G273C, S258C/S320C, S258C/S352C, G273C/S320C, S320C/G335C, S320C/S352C, and G335C/S352C). The data for buffer, micelles of 40 mM SDS, micelles of 450 mM SDS, micelles of 40 mM LPPG, and equimolar POPC/POPS liposomes are shown in black, blue, green, orange, and red, respectively. The experimentally obtained distance distributions were modeled by fitting them to Gaussian functions, with the fits shown in violet. Distances obtained from the fits are listed in Table 1.
We fit the experimental DEER-derived distance distributions by using Gaussian functions to estimate both the average distance and distance distribution width (Fig. 5). In all cases, a single Gaussian provided a satisfactory representation of the experimental data. In the case of the S258C/S352C (helix 1/helix 4) double mutant in the presence of lipid vesicles, we were unable to record a distance due to an apparently very long (exceeding 6 nm) interspin distance, since short phase relaxation times precluded measurement of the DEER signal out to the long dipole evolution times required for such distance. The averaged distance and distance distribution, characterized by Gaussian mean and full width at half-maximum (FWHM, Δr), varied depending on the spin-label location (Table 1). Generally, the distributions were considerably broader (in some cases as wide as 3 nm) than those observed in well-structured proteins in solution (43,67,69) and instead were consistent with the distributions typically observed for highly unstructured proteins (27).
Table 1.
Mean distances, r, and widths, Δr, derived from Gaussian function fits of the experimentally obtained interspin distance distributions for doubly spin-labeled cysteine mutants in tau K19
| Conditions |
r ± Δr/2 |
||||||
|---|---|---|---|---|---|---|---|
| S258C/G273C |
S258C/S320C |
S258C/S352C |
G273C/S320C |
S320C/G335C |
S320C/S352C |
G335C/S352C |
|
| helix 1/linker 1 | helix 1/helix 3 | helix 1/helix 4 | linker 1/helix 3 | helix 3/linker 2 | helix 3/helix 4 | linker 2/helix 4 | |
| Buffer | 2.7 ± 0.7 | 4.5 ± 1.5 | 5.7 ± 2.2 | 3.6 ± 1.0 | 2.7 ± 0.7 | 3.8 ± 1.3 | 3.5 ± 1.1 |
| 40 mM SDS | 3.1 ± 0.7 | 5.2 ± 1.5 | 6.6 ± 2.1 | 3.4 ± 0.9 | 3.0 ± 1.1 | 4.7 ± 1.5 | 3.2 ± 0.9 |
| 450 mM SDS | 3.1 ± 0.7 | 5.2 ± 1.4 | 7.7 ± 1.8 | 3.4 ± 0.8 | 2.9 ± 0.8 | 4.8 ± 1.3 | 3.2 ± 0.9 |
| 40 mM LPPG | 2.4 ± 0.7 | 3.5 ± 1.1 | 5.0 ± 1.3 | 3.4 ± 0.8 | 2.5 ± 0.7 | 3.3 ± 1.2 | 2.8 ± 1.1 |
| POPC/POPS | 2.6 ± 0.9 | 6.0 ± 1.7 | N/A | 3.8 ± 1.1 | 2.5 ± 0.7 | 5.5 ± 1.5 | 3.7 ± 1.3 |
Here, Δr is defined as FWHM. All distances are in nanometers. N/A, not available.
The dependence of the measured distances on the different conditions examined generally falls into two categories depending on whether both of the spin labels were located in a helical segment or one of the labels was located in a linker region. Helix-to-linker distances were found to be relatively independent of whether the sample contained lipid vesicles or membrane mimetics, with a value of ∼3 nm observed in all cases (average difference between LPPG micelle and POPC/POPS vesicle data of 0.4 nm with a standard deviation (SD) of 0.4, paired t-test, p = 0.15). In contrast, helix-to-helix distances were found to be dependent on the presence and type of membrane or membrane mimetic (average difference between LPPG micelle and POPC/POPS vesicle data, using an estimate of 8 nm for the helix 1/helix 4 distance on vesicles, of 2.5 nm with an SD of 0.4, paired t-test, p = 0.009). In general, the shortest helix-to-helix distances were observed in the presence of 40 mM LPPG, with longer distances observed in the absence of any lipid or membrane mimetic, even longer distances observed in the presence of either 40 or 450 mM SDS, and the longest distances observed in the presence of POPC/POPS vesicles. This dependence suggests that the confinement of the membrane-binding helices to the surface of small spheroidal micelles decreases the average distance between them compared with the distance sampled in the membrane-free state, but when a larger binding surface is available, such as that of a cylindrical micelle or a lipid vesicle, the helices are free to move farther away from each other. Thus, the data support a model in which the regions between the helices are relatively flexible, and the helices are neither directly associated with each other nor arranged in any specific way with respect to each other by the intervening regions.
Under our conditions, it is expected that 40 mM SDS solutions will form spheroidal micelles with a diameter of ∼3.5 nm (70), whereas 450 mM SDS solutions will form cylindrical or rod-like micelles with a considerably larger size and surface area (27). Therefore, it was unexpected that the distance distributions measured in 40 mM SDS were nearly identical to those measured in 450 mM SDS, and were associated with significantly larger interhelix distances (as well as somewhat broader distributions) than those observed in 40 mM LPPG (which forms spheroidal micelles with a diameter of ∼4 nm (71)). This observation suggests that protein molecules at both SDS concentrations are bound to rod-like micelles. We previously found that the membrane-binding protein α-synuclein is able to influence the topology of SDS micelles and cause the formation of rod-like micelles at SDS concentrations where spheroidal micelles are expected in the absence of protein (27). Although we postulated that this activity of synuclein may reflect a physiologically relevant capacity to remodel membranes, our current data suggest that the tau MBD may also possess the ability to remodel detergent micelle topology. Synuclein has been reported to bridge individual detergent micelles under certain conditions (72), and it may be that binding of individual tau MBD helices to different spheroidal micelles can also cluster them together and thereby promote their fusion into cylindrical micelles. Whether such a capacity is in any way related to tau function or pathology remains to be investigated.
Discussion
Tau-membrane interactions can enhance tau aggregation in vitro (12,13,15), consistent with a possible role for membranes in inducing tau aggregation in vivo (21,22). Tau can be localized to membranes indirectly via protein-protein interactions with membrane-associated proteins (16–18), but the MBD of tau also binds directly to membrane surfaces (19,20,50,57). It is possible that protein-protein interactions that localize tau at the membrane also promote direct membrane binding by the MBD, which can then lead to tau aggregation. Despite growing interest in tau-membrane interactions, relatively little is known about the structural changes in tau that accompany membrane binding. An analysis based on x-ray and neutron scattering measurements indicated that tau becomes more compact upon membrane binding before membrane-induced aggregation (15). Detailed NMR studies of MBD fragments associated with detergent or lysophospholipid micelles have shown that upon binding to phospholipids or detergents, regions within each of the repeats of the MBD adopt a highly helical structure (50,57). Interestingly, these same regions exhibit a much weaker propensity for helical structure even in the free state of the protein (56,57). Studies of individual tau repeats have shown that helical structure can also be induced in similar regions by fluorinated alcohols, and that the formation of helical structure promotes tau aggregation (73–76). CD data show that the MBD also accrues helical structure upon binding to lipid vesicles (50,57), but no detailed structural information is available for the vesicle-bound form of the protein. In this study, we set out to determine whether regions that become helical when the tau MBD binds to micelles are also helical in the membrane-bound of the protein, and to assess the relative arrangements of any such helices on micelle and vesicle surfaces.
By applying an approach combining CW- and pulse-ESR experiments, we were able to access and characterize at different levels the structure of membrane-bound tau K19, a fragment that corresponds to the MBD repeats of 3R isoforms of human tau protein. Our results show that helical segments previously identified in micelle-bound tau K19 also adopt helical structure when tau K19 binds to lipid vesicles. This result is consistent with the weak but detectable helical propensity of these regions in the lipid-free form of tau (56,57), and also with the induced helicity observed in CD spectra of membrane-bound tau MBD fragments (50,57). Furthermore, we show that the helical segment within R1 of vesicle-bound K19 is also helical in vesicle-bound tau352, the shortest FL human tau isoform. A phase shift of approximately one residue in the helical structure of R1 is observed for membrane-bound tau352 relative to K19, suggesting that the presence of protein regions outside the MBD may influence MBD structure in the membrane-bound state. This is consistent with indications for long-range structure in FL lipid-free tau (58), as well with indications that the membrane-bound form of tau is compact (15). Nevertheless, a very clear helical periodicity is evident for the helical region of R1 in membrane-bound tau352, confirming that this region becomes helical in the context of the FL protein as well.
We also obtained clear evidence that the helical structure in the MBD repeats of membrane-bound tau is surface associated and does not assume a transmembrane configuration. Indeed, we find that the amphipathic helical structure is largely restricted to the lipid headgroup region of the bilayer and does not penetrate deeply into the acyl chain region of the membrane. This is also consistent with the previous observation that the micelle-bound MBD helices do not penetrate deeply into the hydrophobic micelle interior (50). As the short length of the MBD helical segments (approximately nine residues) limits the number of hydrophobic side chains on the apolar side of the helix (three hydrophobes per helix), a shallow penetration into the acyl chain region of the membrane is perhaps not surprising. In addition, the lysine side chains that decorate the boundary between the polar and apolar sides of the individual helices would favor interactions with negatively charged lipid headgroups, and may also act to prevent deep penetration beyond the headgroup region.
Because amphipathic helices can often pack together into globular folds, we investigated the relative arrangements of the individual tau MBD helices on membrane and micelle surfaces. Our results, based on ESR distance measurements, clearly demonstrate that the helices within the tau MBD are widely dispersed on the surface of lipid vesicles: neighboring helices (R1-R3 and R3-R4) are separated by an average distance of ∼6 nm, whereas the distance separating R1 from R4 is beyond the reach of the current measurements. These large interhelix distances suggest that the connecting linker regions adopt highly extended conformations. When combined with the observation that the width of the distance distributions is typical of that found between sites in highly disordered proteins, our data suggest that the individual helices are linked by largely disordered polypeptide segments on the membrane surface. This assertion is further supported by the observation that when tau is confined to smaller surfaces, such as those of cylindrical or spheroidal micelles, the distances between the helices decrease with the binding surface area, indicating that the linker regions are able to adapt to different spatial constraints. Although for the sake of simplicity only 3R versions of tau were used in this study, there is no reason to believe that the presence of R2 would qualitatively alter the observed behavior of the MBD in the context of lipid vesicles, given that R2 behaves similarly to the other repeats in the context of micelles (57). Our results are summarized in a schematic illustration in Fig. 6, which shows the location of the individual helices within each MBD repeat, and the variation of the interhelix distances in going from vesicles to micelles.
Figure 6.
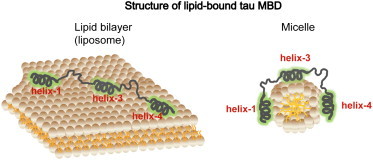
Schematic illustration of tau MBD-membrane interactions. Based on our CW-ESR microwave power saturation experiments, short surface-associated helices form within each MBD repeat upon membrane binding, located at residues 253–261 (helix 1), 315–323 (helix 3), and 346–355 (helix 4). Based on our long-range DEER distance measurements, these helices are connected by flexible linkers and the MBD can adopt either more extended conformations in the case of more expansive membrane surfaces, such as those of ∼75 nm diameter liposomes (left), or more compact conformations in the case of smaller surfaces, such as those of ∼4 nm diameter micelles (right). Although helix-to-helix distances change considerably between vesicles and micelles, helix linker distances are relatively unaffected.
It is interesting to consider the potential implications of our findings for tau function and aggregation. The induction of helical structure upon membrane binding by disordered amyloidogenic proteins is a somewhat common theme that has now been observed for tau, α-synuclein, the Aβ peptide, and amylin. In the latter three cases, models have emerged that posit that amphipathic helical structure may serve to bring protein molecules into close contact with one another, and in particular to cluster and align nearby amyloidogenic nonhelical regions in a way that would facilitate the nucleation of intermolecular β-sheet formation (77–79). In light of this, it is interesting to consider that three of the four helical regions of the tau MBD immediately follow a region that has been demonstrated to effectively nucleate tau filament formation (the PHF6 and PHF6∗ regions in R2 and R3) (80,81) or to potently modulate tau filament formation (the Module B motif in R4) (82) (the helical region in R1 is an exception because it follows a segment that contains multiple proline residues and therefore is unlikely to nucleate β-sheet formation). Thus, it seems plausible that also in the case of tau, membrane binding may facilitate aggregation and filament formation via the clustering of membrane-bound amphipathic helices, which would then bring into close proximity and alignment highly amyloidogenic nonhelical regions that can participate in the earliest steps of intermolecular β-sheet formation (83).
With respect to normal tau function, we previously postulated that the amphipathic helices formed by the MBD upon micelle binding could also serve to mediate tau-microtubule interactions. Independent binding of each helical segment to a tubulin heterodimer site within the microtubule structure would allow tau to bridge between tubulin dimers both within the same protofilament and across different protofilaments. Although this hypothesis remains to be proved or disproved, our observation that these helices also form on membrane surfaces strengthens the argument that these regions of the protein are disposed to form helical structure when tau forms complexes with a variety of binding partners.
Acknowledgments
This study was made possible by NPRP grant 4-1371-1-223 from Qatar National Research Fund (a member of the Qatar Foundation). This work was also supported by NIH/NIA grant R37AG019391 to D.E. and NIH/NIBIB grant R01EB003150 and NIH/NIGMS grant P41GM103521 to J.F.
Contributor Information
Jack H. Freed, Email: jhf3@cornell.edu.
David Eliezer, Email: dae2005@med.cornell.edu.
Supporting Material
References
- 1.Cleveland D.W., Hwo S.Y., Kirschner M.W. Purification of tau, a microtubule-associated protein that induces assembly of microtubules from purified tubulin. J. Mol. Biol. 1977;116:207–225. doi: 10.1016/0022-2836(77)90213-3. [DOI] [PubMed] [Google Scholar]
- 2.Cleveland D.W., Hwo S.Y., Kirschner M.W. Physical and chemical properties of purified tau factor and the role of tau in microtubule assembly. J. Mol. Biol. 1977;116:227–247. doi: 10.1016/0022-2836(77)90214-5. [DOI] [PubMed] [Google Scholar]
- 3.Goedert M., Spillantini M.G., Crowther R.A. Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron. 1989;3:519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 4.Andreadis A., Brown W.M., Kosik K.S. Structure and novel exons of the human tau gene. Biochemistry. 1992;31:10626–10633. doi: 10.1021/bi00158a027. [DOI] [PubMed] [Google Scholar]
- 5.Landau S.M., Frosch M.P. Tracking the earliest pathologic changes in Alzheimer disease. Neurology. 2014;82:1576–1577. doi: 10.1212/WNL.0000000000000392. [DOI] [PubMed] [Google Scholar]
- 6.Overk C.R., Masliah E. Pathogenesis of synaptic degeneration in Alzheimer’s disease and Lewy body disease. Biochem. Pharmacol. 2014;88:508–516. doi: 10.1016/j.bcp.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crowther T., Goedert M., Wischik C.M. The repeat region of microtubule-associated protein tau forms part of the core of the paired helical filament of Alzheimer’s disease. Ann. Med. 1989;21:127–132. doi: 10.3109/07853898909149199. [DOI] [PubMed] [Google Scholar]
- 8.Lee V.M., Goedert M., Trojanowski J.Q. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 9.Goedert M., Jakes R. Mutations causing neurodegenerative tauopathies. Biochim. Biophys. Acta. 2005;1739:240–250. doi: 10.1016/j.bbadis.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Wolfe M.S. Tau mutations in neurodegenerative diseases. J. Biol. Chem. 2009;284:6021–6025. doi: 10.1074/jbc.R800013200. [DOI] [PubMed] [Google Scholar]
- 11.Spillantini M.G., Goedert M. Tau pathology and neurodegeneration. Lancet Neurol. 2013;12:609–622. doi: 10.1016/S1474-4422(13)70090-5. [DOI] [PubMed] [Google Scholar]
- 12.Chirita C.N., Necula M., Kuret J. Anionic micelles and vesicles induce tau fibrillization in vitro. J. Biol. Chem. 2003;278:25644–25650. doi: 10.1074/jbc.M301663200. [DOI] [PubMed] [Google Scholar]
- 13.Elbaum-Garfinkle S., Ramlall T., Rhoades E. The role of the lipid bilayer in tau aggregation. Biophys. J. 2010;98:2722–2730. doi: 10.1016/j.bpj.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Künze G., Barré P., Huster D. Binding of the three-repeat domain of tau to phospholipid membranes induces an aggregated-like state of the protein. Biochim. Biophys. Acta. 2012;1818:2302–2313. doi: 10.1016/j.bbamem.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones E.M., Dubey M., Chi E.Y. Interaction of tau protein with model lipid membranes induces tau structural compaction and membrane disruption. Biochemistry. 2012;51:2539–2550. doi: 10.1021/bi201857v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brandt R., Léger J., Lee G. Interaction of tau with the neural plasma membrane mediated by tau’s amino-terminal projection domain. J. Cell Biol. 1995;131:1327–1340. doi: 10.1083/jcb.131.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein C., Kramer E.M., Trotter J. Process outgrowth of oligodendrocytes is promoted by interaction of fyn kinase with the cytoskeletal protein tau. J. Neurosci. 2002;22:698–707. doi: 10.1523/JNEUROSCI.22-03-00698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gauthier-Kemper A., Weissmann C., Brandt R. The frontotemporal dementia mutation R406W blocks tau’s interaction with the membrane in an annexin A2-dependent manner. J. Cell Biol. 2011;192:647–661. doi: 10.1083/jcb.201007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Surridge C.D., Burns R.G. The difference in the binding of phosphatidylinositol distinguishes MAP2 from MAP2C and Tau. Biochemistry. 1994;33:8051–8057. doi: 10.1021/bi00192a009. [DOI] [PubMed] [Google Scholar]
- 20.Shea T.B. Phospholipids alter tau conformation, phosphorylation, proteolysis, and association with microtubules: implication for tau function under normal and degenerative conditions. J. Neurosci. Res. 1997;50:114–122. doi: 10.1002/(SICI)1097-4547(19971001)50:1<114::AID-JNR12>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 21.Gray E.G., Paula-Barbosa M., Roher A. Alzheimer’s disease: paired helical filaments and cytomembranes. Neuropathol. Appl. Neurobiol. 1987;13:91–110. doi: 10.1111/j.1365-2990.1987.tb00174.x. [DOI] [PubMed] [Google Scholar]
- 22.Mena R., Edwards P.C., Wischik C.M. Staging the pathological assembly of truncated tau protein into paired helical filaments in Alzheimer’s disease. Acta Neuropathol. 1996;91:633–641. doi: 10.1007/s004010050477. [DOI] [PubMed] [Google Scholar]
- 23.Altenbach C., Greenhalgh D.A., Hubbell W.L. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 1994;91:1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klare J.P., Steinhoff H.-J. Structural information from spin-labelling membrane-bound proteins. In: Harmer J.R., Timmel C.R., editors. Vol. 152. Springer; Heidelberg: 2014. pp. 205–248. (Structure and Bonding). [Google Scholar]
- 25.Zou P., Mchaourab H.S. Alternating access of the putative substrate-binding chamber in the ABC transporter MsbA. J. Mol. Biol. 2009;393:574–585. doi: 10.1016/j.jmb.2009.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Georgieva E.R., Ramlall T.F., Eliezer D. Membrane-bound alpha-synuclein forms an extended helix: long-distance pulsed ESR measurements using vesicles, bicelles, and rodlike micelles. J. Am. Chem. Soc. 2008;130:12856–12857. doi: 10.1021/ja804517m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Georgieva E.R., Ramlall T.F., Eliezer D. The lipid-binding domain of wild type and mutant alpha-synuclein: compactness and interconversion between the broken and extended helix forms. J. Biol. Chem. 2010;285:28261–28274. doi: 10.1074/jbc.M110.157214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borbat P.P., Crepeau R.H., Freed J.H. Multifrequency two-dimensional Fourier transform ESR: an X/Ku-band spectrometer. J. Magn. Reson. 1997;127:155–167. doi: 10.1006/jmre.1997.1201. [DOI] [PubMed] [Google Scholar]
- 29.Pannier M., Veit S., Spiess H.W. Dead-time free measurement of dipole-dipole interactions between electron spins. 2000. J. Magn. Reson. 2011;213:316–325. doi: 10.1016/j.jmr.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 30.Chiang Y.W., Borbat P.P., Freed J.H. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J. Magn. Reson. 2005;172:279–295. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 31.Chiang Y.W., Borbat P.P., Freed J.H. Maximum entropy: a complement to Tikhonov regularization for determination of pair distance distributions by pulsed ESR. J. Magn. Reson. 2005;177:184–196. doi: 10.1016/j.jmr.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 32.Hubbell W.L., Cafiso D.S., Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 33.Jeschke G. DEER distance measurements on proteins. Annu. Rev. Phys. Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 34.Bordignon E. Site-directed spin labeling of membrane proteins. Top. Curr. Chem. 2012;321:121–157. doi: 10.1007/128_2011_243. [DOI] [PubMed] [Google Scholar]
- 35.Borbat P.P., Freed J.H. Measuring distances by pulsed dipolar ESR spectroscopy: spin-labeled histidine kinases. Methods Enzymol. 2007;423:52–116. doi: 10.1016/S0076-6879(07)23003-4. [DOI] [PubMed] [Google Scholar]
- 36.Mchaourab H.S., Steed P.R., Kazmier K. Toward the fourth dimension of membrane protein structure: insight into dynamics from spin-labeling EPR spectroscopy. Structure. 2011;19:1549–1561. doi: 10.1016/j.str.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Georgieva E.R., Borbat P.P., Boudker O. Conformational ensemble of the sodium-coupled aspartate transporter. Nat. Struct. Mol. Biol. 2013;20:215–221. doi: 10.1038/nsmb.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z., Fleissner M.R., Freed J.H. Multifrequency electron spin resonance study of the dynamics of spin labeled T4 lysozyme. J. Phys. Chem. B. 2010;114:5503–5521. doi: 10.1021/jp910606h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhatnagar J., Borbat P.P., Crane B.R. Structure of the ternary complex formed by a chemotaxis receptor signaling domain, the CheA histidine kinase, and the coupling protein CheW as determined by pulsed dipolar ESR spectroscopy. Biochemistry. 2010;49:3824–3841. doi: 10.1021/bi100055m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward R., Pliotas C., Schiemann O. Probing the structure of the mechanosensitive channel of small conductance in lipid bilayers with pulsed electron-electron double resonance. Biophys. J. 2014;106:834–842. doi: 10.1016/j.bpj.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borbat P., Ramlall T.F., Eliezer D. Inter-helix distances in lysophospholipid micelle-bound alpha-synuclein from pulsed ESR measurements. J. Am. Chem. Soc. 2006;128:10004–10005. doi: 10.1021/ja063122l. [DOI] [PubMed] [Google Scholar]
- 42.Borbat P.P., Freed J.H. Pulse dipolar ESR: distance measurements. In: Harmer J.R., Timmel C.R., editors. Vol. 152. Springer; Heidelberg: 2014. pp. 1–82. (Structure and Bonding). [Google Scholar]
- 43.Borbat P.P., McHaourab H.S., Freed J.H. Protein structure determination using long-distance constraints from double-quantum coherence ESR: study of T4 lysozyme. J. Am. Chem. Soc. 2002;124:5304–5314. doi: 10.1021/ja020040y. [DOI] [PubMed] [Google Scholar]
- 44.Jao C.C., Hegde B.G., Langen R. Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement. Proc. Natl. Acad. Sci. USA. 2008;105:19666–19671. doi: 10.1073/pnas.0807826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siddiqua A., Luo Y., Margittai M. Conformational basis for asymmetric seeding barrier in filaments of three- and four-repeat tau. J. Am. Chem. Soc. 2012;134:10271–10278. doi: 10.1021/ja303498q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pornsuwan S., Giller K., Bennati M. Long-range distances in amyloid fibrils of α-synuclein from PELDOR spectroscopy. Angew. Chem. Int. Ed. Engl. 2013;52:10290–10294. doi: 10.1002/anie.201304747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borbat P.P., Freed J.H. Pros and cons of pulse dipolar ESR: DQC and DEER. EPR Newsl. 2007;17:21–33. [Google Scholar]
- 48.Caron J.M., Berlin R.D. Interaction of microtubule proteins with phospholipid vesicles. J. Cell Biol. 1979;81:665–671. doi: 10.1083/jcb.81.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.LoPresti P., Szuchet S., Binder L.I. Functional implications for the microtubule-associated protein tau: localization in oligodendrocytes. Proc. Natl. Acad. Sci. USA. 1995;92:10369–10373. doi: 10.1073/pnas.92.22.10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barré P., Eliezer D. Folding of the repeat domain of tau upon binding to lipid surfaces. J. Mol. Biol. 2006;362:312–326. doi: 10.1016/j.jmb.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 51.Elbaum-Garfinkle S., Cobb G., Rhoades E. Tau mutants bind tubulin heterodimers with enhanced affinity. Proc. Natl. Acad. Sci. USA. 2014;111:6311–6316. doi: 10.1073/pnas.1315983111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wells K., Farooqui A.A., Horrocks L.A. Neural membrane phospholipids in Alzheimer disease. Neurochem. Res. 1995;20:1329–1333. doi: 10.1007/BF00992508. [DOI] [PubMed] [Google Scholar]
- 53.Jao C.C., Der-Sarkissian A., Langen R. Structure of membrane-bound alpha-synuclein studied by site-directed spin labeling. Proc. Natl. Acad. Sci. USA. 2004;101:8331–8336. doi: 10.1073/pnas.0400553101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Der-Sarkissian A., Jao C.C., Langen R. Structural organization of alpha-synuclein fibrils studied by site-directed spin labeling. J. Biol. Chem. 2003;278:37530–37535. doi: 10.1074/jbc.M305266200. [DOI] [PubMed] [Google Scholar]
- 55.Longhi S., Belle V., Carrière F. Probing structural transitions in both structured and disordered proteins using site-directed spin-labeling EPR spectroscopy. J. Pept. Sci. 2011;17:315–328. doi: 10.1002/psc.1344. [DOI] [PubMed] [Google Scholar]
- 56.Eliezer D., Barré P., Heend L. Residual structure in the repeat domain of tau: echoes of microtubule binding and paired helical filament formation. Biochemistry. 2005;44:1026–1036. doi: 10.1021/bi048953n. [DOI] [PubMed] [Google Scholar]
- 57.Barré P., Eliezer D. Structural transitions in tau k18 on micelle binding suggest a hierarchy in the efficacy of individual microtubule-binding repeats in filament nucleation. Protein Sci. 2013;22:1037–1048. doi: 10.1002/pro.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mukrasch M.D., Bibow S., Zweckstetter M. Structural polymorphism of 441-residue tau at single residue resolution. PLoS Biol. 2009;7:e34. doi: 10.1371/journal.pbio.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smet C., Leroy A., Lippens G. Accepting its random coil nature allows a partial NMR assignment of the neuronal Tau protein. ChemBioChem. 2004;5:1639–1646. doi: 10.1002/cbic.200400145. [DOI] [PubMed] [Google Scholar]
- 60.Harbison N.W., Bhattacharya S., Eliezer D. Assigning backbone NMR resonances for full length tau isoforms: efficient compromise between manual assignments and reduced dimensionality. PLoS ONE. 2012;7:e34679. doi: 10.1371/journal.pone.0034679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perozo E., Cortes D.M., Cuello L.G. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat. Struct. Biol. 1998;5:459–469. doi: 10.1038/nsb0698-459. [DOI] [PubMed] [Google Scholar]
- 62.Jao C.C., Hegde B.G., Langen R. Roles of amphipathic helices and the bin/amphiphysin/rvs (BAR) domain of endophilin in membrane curvature generation. J. Biol. Chem. 2010;285:20164–20170. doi: 10.1074/jbc.M110.127811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mchaourab H.S., Lietzow M.A., Hubbell W.L. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 64.Subczynski W.K., Hyde J.S. Concentration of oxygen in lipid bilayers using a spin-label method. Biophys. J. 1983;41:283–286. doi: 10.1016/S0006-3495(83)84439-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Griffith O.H., Dehlinger P.J., Van S.P. Shape of the hydrophobic barrier of phospholipid bilayers (evidence for water penetration in biological membranes) J. Membr. Biol. 1974;15:159–192. doi: 10.1007/BF01870086. [DOI] [PubMed] [Google Scholar]
- 66.Jeschke G., Sajid M., Godt A. Three-spin correlations in double electron-electron resonance. Phys. Chem. Chem. Phys. 2009;11:6580–6591. doi: 10.1039/b905724b. [DOI] [PubMed] [Google Scholar]
- 67.Georgieva E.R., Roy A.S., Freed J.H. Effect of freezing conditions on distances and their distributions derived from double electron electron resonance (DEER): a study of doubly-spin-labeled T4 lysozyme. J. Magn. Reson. 2012;216:69–77. doi: 10.1016/j.jmr.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Milov A.D., Ponomarev A.B., Tsvetkov Y.D. Modulation beats of signal of double electron-electron resonance in spin echo for biradical systems. J. Struct. Chem. 1984;25:710–713. [Google Scholar]
- 69.Kazmier K., Alexander N.S., McHaourab H.S. Algorithm for selection of optimized EPR distance restraints for de novo protein structure determination. J. Struct. Biol. 2011;173:549–557. doi: 10.1016/j.jsb.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duplatre G., Marques M.F.F., Miguel M.G. Size of sodium dodecyl sulfate micelles in aqueous solutions as studied by positron annihilation lifetime spectroscopy. J. Phys. Chem. 1996;100:16608–16612. [Google Scholar]
- 71.Oliver R.C., Lipfert J., Columbus L. Dependence of micelle size and shape on detergent alkyl chain length and headgroup. PLoS ONE. 2013;8:e62488. doi: 10.1371/journal.pone.0062488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Giehm L., Oliveira C.L., Otzen D.E. SDS-induced fibrillation of alpha-synuclein: an alternative fibrillation pathway. J. Mol. Biol. 2010;401:115–133. doi: 10.1016/j.jmb.2010.05.060. [DOI] [PubMed] [Google Scholar]
- 73.Minoura K., Tomoo K., Taniguchi T. Amphipathic helical behavior of the third repeat fragment in the tau microtubule-binding domain, studied by (1)H NMR spectroscopy. Biochem. Biophys. Res. Commun. 2002;294:210–214. doi: 10.1016/S0006-291X(02)00457-6. [DOI] [PubMed] [Google Scholar]
- 74.Minoura K., Yao T.M., Ishida T. Different associational and conformational behaviors between the second and third repeat fragments in the tau microtubule-binding domain. Eur. J. Biochem. 2004;271:545–552. doi: 10.1046/j.1432-1033.2003.03956.x. [DOI] [PubMed] [Google Scholar]
- 75.Tomoo K., Yao T.M., Ishida T. Possible role of each repeat structure of the microtubule-binding domain of the tau protein in in vitro aggregation. J. Biochem. 2005;138:413–423. doi: 10.1093/jb/mvi142. [DOI] [PubMed] [Google Scholar]
- 76.Hiraoka S., Yao T.M., Ishida T. Conformational transition state is responsible for assembly of microtubule-binding domain of tau protein. Biochem. Biophys. Res. Commun. 2004;315:659–663. doi: 10.1016/j.bbrc.2004.01.107. [DOI] [PubMed] [Google Scholar]
- 77.Anderson V.L., Ramlall T.F., Eliezer D. Identification of a helical intermediate in trifluoroethanol-induced alpha-synuclein aggregation. Proc. Natl. Acad. Sci. USA. 2010;107:18850–18855. doi: 10.1073/pnas.1012336107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kirkitadze M.D., Condron M.M., Teplow D.B. Identification and characterization of key kinetic intermediates in amyloid beta-protein fibrillogenesis. J. Mol. Biol. 2001;312:1103–1119. doi: 10.1006/jmbi.2001.4970. [DOI] [PubMed] [Google Scholar]
- 79.Apostolidou M., Jayasinghe S.A., Langen R. Structure of alpha-helical membrane-bound human islet amyloid polypeptide and its implications for membrane-mediated misfolding. J. Biol. Chem. 2008;283:17205–17210. doi: 10.1074/jbc.M801383200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.von Bergen M., Friedhoff P., Mandelkow E. Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming beta structure. Proc. Natl. Acad. Sci. USA. 2000;97:5129–5134. doi: 10.1073/pnas.97.10.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.von Bergen M., Barghorn S., Mandelkow E. Mutations of tau protein in frontotemporal dementia promote aggregation of paired helical filaments by enhancing local beta-structure. J. Biol. Chem. 2001;276:48165–48174. doi: 10.1074/jbc.M105196200. [DOI] [PubMed] [Google Scholar]
- 82.DeTure M.A., Di Noto L., Purich D.L. In vitro assembly of Alzheimer-like filaments. How a small cluster of charged residues in Tau and MAP2 controls filament morphology. J. Biol. Chem. 2002;277:34755–34759. doi: 10.1074/jbc.M201201200. [DOI] [PubMed] [Google Scholar]
- 83.Abedini A., Raleigh D.P. A role for helical intermediates in amyloid formation by natively unfolded polypeptides? Phys. Biol. 2009;6:015005. doi: 10.1088/1478-3975/6/1/015005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


