Abstract
Human phospholipid scramblase 1 (SCR) catalyzes phospholipid transmembrane (flip-flop) motion. This protein is assumed to bind the membrane hydrophobic core through a transmembrane domain (TMD) as well as via covalently bound palmitoyl residues. Here, we explore the possible interaction of the SCR TMD with cholesterol by using a variety of experimental and computational biophysical approaches. Our findings indicate that SCR contains an amino acid segment at the C-terminal region that shows a remarkable affinity for cholesterol, although it lacks the CRAC sequence. Other 3-OH sterols, but not steroids lacking the 3-OH group, also bind this region of the protein. The newly identified cholesterol-binding region is located partly at the C-terminal portion of the TMD and partly in the first amino acid residues in the SCR C-terminal extracellular coil. This finding could be related to the previously described affinity of SCR for cholesterol-rich domains in membranes.
Introduction
Human phospholipid scramblase 1 (SCR) is an endofacial plasma transmembrane protein that has reported to function in the process of apoptosis as a nonspecific translocator of phosphatidylserine to the membrane outer monolayer (1–3). However, a number of studies (4–7) have suggested that the physiological role of this protein could in fact be unrelated to membranes, with SCR acting instead as a modulator of gene expression. SCR structure has not yet been resolved by crystallographic analysis, but experimental evidence suggests that scramblases present a structural dimorphism in the cell. SCR would exist in a palmitoylated form in plasma membrane caveolae-like, cholesterol (Chol)-rich lipid domains (8), where they would partition together with caveolin-1 and epidermal growth factor receptor (EGFR) (9), and in a nonacylated form in the nucleus, where they would play a role as a transcription factor with important functions in cellular proliferation and differentiation (4,5), including tumor suppression (6). Bateman et al. (7) proposed a structural model for scramblases according to which palmitoylation, rather than the transmembrane domain (TMD), would constitute the main membrane anchorage for these proteins. However, recent data from our laboratory and others have cast doubt on this assertion. Although mutant SCR lacking the TMD is still able to bind lipid bilayers (10), it has lost functional scramblase activity (11,12). Moreover, the isolated TMD binds and becomes inserted into lipid bilayers with great affinity (13).
In this study, we address the affinity of SCR for Chol in relation to the SCR trafficking to Chol-rich membrane domains, and the insertion of the TMD in these regions (Fig. 1 A). Chol recruitment amino acid consensus (CRAC) is a well-known Chol-binding domain in proteins, with the consensus pattern [L/V]-[X](1-5)-[Y]-[X](1-5)-[R/K]. This pattern is broadly conserved in many membrane proteins (14–16), although mutation of some of these amino acids does not necessarily entail the loss of Chol interaction. The human scramblase family contains a CRAC domain sequence at the C-terminus of the TMD, but in the first member of the family the consensus is not conserved (Fig. 1 B). Another consensus sequence, named CARC, constitutes exactly the reverse sequence of the CRAC domain, with an additional option for the central aromatic amino acid, which could be either Y or F (17). SCR contains the combination of basic arginine (at position X3), aromatic phenylalanine (X11), and aliphatic leucine (X12) residues in TMD found in the CARC motif, but the disposition of arginine does not fulfill the criterion of the CARC algorithm that five residues must separate the basic from the aromatic amino acid. In addition to the CRAC and CARC sequences, an important factor in protein-Chol binding is the three-dimensional orientation of the amino acid side chains with respect to the α/β faces of the sterol. Many tilted peptides, such as α-synuclein (18) and the TM glycoprotein gp41 of HIV-1 (19), bind Chol with a very exothermic energy of interaction, and these affinities are orientational in the space rather than sequence based.
Figure 1.
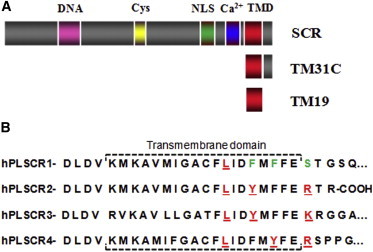
(A) SCR domains and sequence-derived peptides: DNA-binding domain (pink), Cys-rich domain (yellow), nuclear localization signal (green), calcium-binding domain (blue), and TMD (red). (B) Representation of the human phospholipid (hPL) SCR family putative TMD sequence. hPL SCRs 2, 3, and 4 contain a CRAC motif ([L/V]-[X](1-5)-[Y]-[X](1-5)-[R/K]) localized at the C-terminal end of the predicted transmembrane helix (amino acids in red), whereas SCR presumably is the result of several mutations (in green), such as the polar amino acid (serine) instead of a positively charged amino acid (arginine or lysine), and two phenylalanines in different positions, which appear as tyrosines in the rest of the family sequences. Thus, neither the CRAC nor the CRAC-like sequence ([L/V]-[X](1-5)-[F]-[X](1-5)-[R/K]) is completed in SCR TMD. Sequence information was taken from UNIPROT. Sequences were aligned according to Sahu et al. (3) and Bateman et al. (7).
Here, we conducted experimental and computational studies to examine the interaction of SCR and its TMD with lipid monolayers and bilayers of varying lipid composition, including different steroids. We focused on the recombinant, nonpalmitoylated protein and peptides derived from the C-terminus sequence: TM19, which comprises the TMD sequence, and TM31C, which comprises the TMD plus the extracellular coil sequence (Fig. 1 A). Our findings support the idea that a sequence in the C-terminal region of SCR, spanning part of the TMD and part of the adjacent extracellular coil, binds 3-OH sterols with high specificity. This may be related to the affinity of SCR for Chol-rich membrane domains.
Materials and Methods
Materials
TM31C (aa sequence: KMKAVMIGACFLIDFMFFESTGSQEQKSGVW) and TM19 (aa sequence: KMKAVMIGACFLIDFMFFE) were synthesized and purchased from PolyPeptide Group Laboratories (Strasbourg, France) and stored at −20°C in powder. For the different experiments, the peptides were dissolved in DMSO (Sigma) or HFIP (Fluka) immediately before use. All phospholipids and sterols were obtained from Avanti Polar Lipids (Birmingham, AL), and 8-aminonaphtalene-1,3,6-trisulfonic acid sodium salt (ANTS) and p-xylenebis(pyridinium) bromide (DPX) were obtained from Invitrogen (Life Technologies, Carlsbad, CA). The polyclonal C-terminal anti-scramblase antibody was obtained from Oncogene (Cambridge, UK) and the monoclonal N-terminal anti-scramblase antibody was obtained from Abcam (Cambridge, UK). Horseradish peroxidase (HRP)-linked anti-rabbit and anti-mouse antibodies were obtained from New England Biolabs (Ipswich, MA); 1-lauroyl-2-(1-pyrenebutyroyl)-sn-glycero-3-sphingomyelin (pyrene-SM) was synthesized as described in Muller et al. (20); and 1-oleoyl-2-{6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphoserine (C6-NBD-PS) was obtained from Avanti Polar Lipids (Birmingham, AL). The Chol oxidase/peroxidase kit was obtained from Biosystems (Barcelona, Spain). All other reagents were of analytical grade.
Solid-phase binding assay
A screening dot-blot analysis was performed to assess the sterol analog affinities of TM31C and SCR (21). Lipids were dissolved in methanol/chloroform/water (2:1:0.8 v/v/v) at a final concentration of 800 μM, and 1 μl was spotted on a Hybond-C extra membrane and left for 1 h to let the lipid dry. After blocking with 0.75% skim milk in PBS for 1 h, the membrane was incubated for an additional hour with the peptide TM31C or with the recombinant protein SCR (500 nM final concentration), also in PBS. The membrane was then washed several times, followed by 1 h incubation of the anti-scramblase C-terminal and N-terminal antibody, respectively (1:1000). The blot was washed several times with PBS, pH 7.4, and incubated for 1 h with an HRP-linked anti-rabbit (for peptide) and anti-mouse antibody (for protein) (1:2000). After final washes to eliminate the nonbound secondary antibody, the blot was developed on a Curix 60 processor (AGFA, Belgium) using the Amersham Hyperfilm ECL (GE Healthcare, UK). The intensity of the sample signal was measured with a densitometer GS-800 (Bio-Rad, Stockholm, Sweden).
Langmuir balance measurements
TM31C-induced changes in surface pressure at the air/water interface and peptide-lipid monolayer interactions were studied at 25°C using a 1.25 ml multiwell Delta Pi-4 system (Kibron, Helsinki, Finland). Monolayers were formed by spreading a small amount of the lipid mixture made of POPC and increasing concentrations of Chol in a chloroform/methanol (2:1 v/v) solution on top of the assay buffer until the desired initial surface pressure was attained. Then, 3 μM peptide dissolved in DMSO (<0.5% of total volume) was individually injected with a micropipette through a hole connected to the subphase, and its surface activity was followed by means of surface pressure changes with constant stirring.
Vesicle content efflux measurements
Large unilamellar vesicles (LUVs) were prepared in assay buffer in the presence of ANTS and DPX in a 1:3.6 ratio. LUVs were prepared by extruding multilamellar vesicle (MLV) suspensions through polycarbonate filters (0.1 μm pore diameter). The lipid suspension was then passed through a Sephadex PD-10 column to discard nonencapsulated dye. Vesicle content efflux or leakage was measured at 23°C in a Fluoromax luminescence spectrometer by following ANTS externalization after peptide (dissolved in DMSO) addition in a 1:50 mol ratio. ANTS was excited at 355 nm and emission was recovered at 530 nm using a cutoff filter at 470 nm. Initial vesicle suspension was used as the 0% fluorescence signal, and 100% fluorescence was obtained after breakdown of the vesicles by addition of 1 mM Triton X-100 (22,23).
Differential scanning calorimetry
Differential scanning calorimetry (DSC) measurements were performed using a VP-DSC high-sensitivity scanning microcalorimeter (Microcal, Northampton, MA). For preparation of peptide-MLVs, the appropriate amounts of either pure SOPC (0.5 mM final concentration) or SOPC/Chol 1:1 (2.5 mg/ml final concentration) (16) in chloroform/methanol (2:1 v/v) and peptide in HFIP were mixed and the solvent was evaporated exhaustively. The MLVs were then prepared by slowly hydrating the peptide-containing lipid film with assay buffer at a temperature above the lipid-phase transition temperature, with continuous stirring with a glass rod followed by vigorous vortexing of the sample at 50°C. The samples were then carefully degassed before measurements were conducted. The assay buffer was scanned as a background. The scan rate was 90°C/h. Samples were scanned several times to ensure the reproducibility of the endotherms. Data were analyzed using ORIGIN software provided by MicroCal. The final volume of the cell was 0.5 ml. The SOPC concentration was measured with a molybdate assay, and the Chol concentration was measured with the use of a Chol oxidase/peroxidase kit.
In silico studies of Chol-SCR interactions
Molecular modeling studies were performed using the Hyperchem 8 program (ChemCAD, Obernay, France). A model of the FLIDFMFFESTGSQE peptide was first obtained in vacuo and then merged with Chol. Various starting conditions were assayed and only the one that led to the highest energy of interaction was selected. Geometry optimization of the Chol/SCR complex was achieved using the unconstrained optimization rendered by the Polak-Ribière conjugate gradient algorithm (24). Molecular-dynamics simulations were then performed for iterative periods of 1 ns in vacuo with the Bio+ (CHARMM) force field (25). The energy of interaction was determined with the Molegro Molecular Viewer (26).
SCR purification and reconstitution into lo-containing LUVs
SCR was overexpressed as an MBP-fusion protein and purification was performed essentially as described previously by Zhou et al. (2). For the reconstitution assay, a mixture of DOPC/eSM/Chol (2:1:1 or 1:1:1 mol ratio), POPC/PS/Chol (9:1:3.3 mol ratio), or PC/PS (9:1 mol ratio) was dried under a stream of nitrogen and resuspended in 20 mM Tris (pH 7.4), 100 mM KCl, and 0.1 mM EGTA. The protein samples to be reconstituted were added to the liposomes at a 5 mM final lipid concentration in the presence of 30 mM OG and dialyzed overnight at 4°C in a buffer containing Bio-Beads SM-2 (1 g/l). A sucrose gradient was performed afterward to isolate reconstituted from nonreconstituted proteins (12,27).
Flip-flop assay
Proteoliposomes (100 μM final lipid concentration) were transferred to a stirred fluorescence cuvette at 37°C, and 5 mol % pyrene-SM was added to label the vesicles externally (12,20). Initial fluorescence was recorded and 5 mM calcium was added. Fluorescence was monitored for 80 min (λex= 344 nm, λem= 380–520 nm) in an Aminco Bowman Series 2 spectrofluorometer (Rochester, NY) and the decay of the Iex/Imon ratio was attributed to a calcium-induced change of the py-SM position from the outer to the inner monolayer, promoted by the reconstituted proteins (20).
Alternatively, proteoliposomes were labeled with externally added 0.1% C6-NBD-PS and the sample was incubated for 30 min on ice to ensure complete incorporation of the probe into the liposomes. Phospholipid flip-flop was initiated by adding 5 mM Ca2+ followed by incubation at 37°C. At different time intervals, an aliquot was taken and immediately added to a stirred cuvette (5-fold dilution) at 23°C in an Aminco Bowman Series 2 spectrofluorometer, and the reaction was stopped by addition of 5 mM sodium dithionite (28). Flip-flop was quantified by comparing NBD fluorescence before (F0) and after (Fx) bleaching by the addition of dithionite, according to the equation F (%) = (Fx/F0) × 100.
Results
Lipid overlay assay
Both the SCR protein and the TM31C peptide were studied using a solid-phase binding assay, which enables rapid identification of the lipid ligands with which proteins interact. In this case, Chol and six steroid analogs were spotted onto a nitrocellulose membrane and incubated with the protein/peptide samples. The immunoblotted membranes are shown in Fig. 2, A and B. The protein and peptide exhibit a remarkable preferential binding for sterols containing the polar alcohol group in ring A, which seems to be a major structural requirement for interaction with the scramblase TMD.
Figure 2.
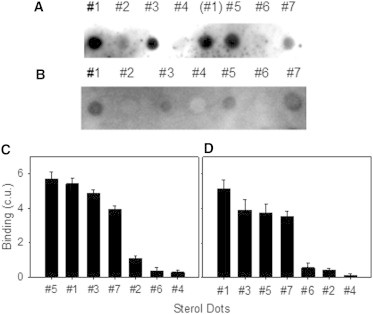
Solid-phase binding assay. Dot blots of TM31C (A) and SCR (B) binding to Chol (#1) and different analogs: cholestanone (#2), ergosterol (#3), 5α-cholestane (#4), 7-dehydrocholesterol (#5), Chol-one (#6), and coprostanol (#7). (C and D) The results are presented in decreasing order of peptide (C) and SCR (D) binding to sterol analogs. The binding indicates the importance of the alcohol group in ring A for interaction with the scramblase TMD.
Binding quantitation is displayed in Fig. 2, C and D, for TM31C and SCR, respectively. In both cases, Chol (#1), ergosterol (#3), and 7-dehydrocholesterol (#5) are the sterols for which SCR and TM31C exhibit the strongest interaction, and both show a lower binding for coprostanol (#7), which includes the alcohol group in ring A but lacks the double bond in ring B of Chol. Apparently this absence of a double bond lowers the scramblase TMD’s affinity for the sterol. Finally, in both cases, the steroids that exhibited the lowest protein/peptide binding were cholestanone (#2) and cholestenone (Chol-one) (#6), which include a ketone group instead of an alcohol group in ring A, followed by 5α-cholestane (#4), which lacks any oxygenated chemical group in ring A and the double bonds in ring B. Hence, the solid-phase binding assay indicates a strong affinity between SCR TMD and Chol or similar C3-OH sterols.
Peptide interaction with mono- and bilayers containing Chol
To substantiate the lipid overlay assays that suggested a scramblase TMD affinity for Chol, we carried Langmuir balance measurements out to check for TM31C insertion into pure POPC and POPC/Chol lipid films. TM31C peptide is a powerful tool for exploring SCR TMD affinity for Chol in model and cell membranes, and is more manageable than the native protein. Insertion of TM31C into PC monolayers was previously reported (13), with a critical surface pressure πc (the maximum surface pressure beyond which no peptide insertion occurs) at 33 mN/m. At π0 = 24 ± 0.5 mN/m (the initial monolayer pressure), a Δπ (surface pressure increase) of 3.9 ± 0.17 mN/m was reported. With the addition of increasing concentrations of Chol to PC monolayers, a dual effect is detected (Fig. 3 A): 1) TM31C shows greater affinity for monolayers containing up to 20 mol % Chol than for pure PC, even if the mean molecular area decreases under these conditions (29). Due to the increasing presence of Chol, monolayers become more compressed at the same π0, impeding peptide insertion and diminishing Δπ, but the affinity for Chol nonetheless increases. At 20 mol % Chol, Δπ rises to 5.9 ± 0.18 mN/m with a mean molecular area of 56 Å2, which is 11 Å2 lower than observed for pure PC monolayers. 2) From this point on, at 30–50 mol % Chol, peptide insertion into monolayers decays, probably owing to the high packing of the lipid molecules, which hampers the peptide affinity for Chol. The slope of the time courses of peptide-induced π changes correlates with the observed equilibrium Δπ (Fig. 3 A). Thus, the Langmuir balance data are in agreement with the above binding data obtained with both the peptide and the protein, showing that scramblase TMD has a large and measurable affinity for Chol when it is in solid phase or included in a monolayer lipid film.
Figure 3.
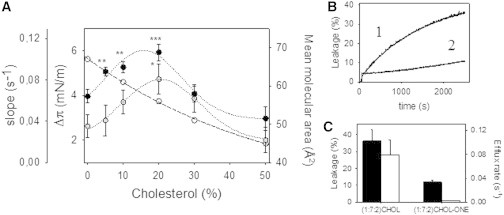
(A) Surface pressure measurements of TM31C in PC/Chol monolayers. The increase in surface pressure (●) is shown after insertion of TM31C into lipid monolayers (π0 = 24 ± 0.5 mN/m) made of PC and increasing concentrations of Chol; the calculated slopes from the time traces are represented by ○, dotted line. Experimental data are fitted to a polynomial function (degree = 2). Symbols correspond to the mean ± SDs of four to six independent measurements. The mean molecular areas (○, dashed line) were taken from Li et al. (29) to show the increased monolayer packing. (B and C) ANTS-DPX vesicle content efflux. (B) Time course of TMC31-induced release of intravesicular aqueous contents for POPC/PSM/Chol (1) and POPC/PSM/Chol-one (2) 1:7:2 composition. (C) TM31C-induced total release (black bars) and corresponding initial rates (white bars).
As previously reported, TM31C in Chol-containing LUV bilayers can promote a large peptide-induced release of vesicular aqueous contents (see, e.g., the POPC/pSM/Chol 1:7:2 (lo + so) composition (13)). When Chol was replaced with Chol-one, an abrupt decrease was observed in both the extent of release and the initial efflux rate (Fig. 3, B and C). Thus, the SCR TMD’s affinity for Chol in solid phase and mono- or bilayers is confirmed. In the latter case, it is important to remember that Chol-one cannot form lo domains in vesicles as does Chol, suggesting that the presence of these domains or the presence of a phase boundary is also important. Domain boundaries would favor peptide insertion and accommodation in the bilayer and promote leakage.
TM31C-induced Chol phase separation detected by differential scanning calorimetry
Calorimetric studies were performed to observe scramblase TMD behavior in SOPC/Chol 1:1 (mol ratio) bilayers according to a previously described method (16,30). CRAC-motif-containing peptides, such as the caveolin-1 peptide, can alter membrane properties, forming Chol-rich domains by forcing the sterol beyond its solubility limit and thus causing formation of Chol crystals (anhydrous Chol). Due to single-residue mutations, scramblase TMD lacks the CRAC sequence (Fig. 1 B), but nonetheless displays a tendency to interact with Chol. We therefore developed an experiment to quantify the interaction thermodynamically. In addition to TM31C, we used TM19, a peptide that comprises only the putative TMD sequence (Fig. 1 A).
Fig. 4 A shows the heating endothermic phase transitions for the SOPC/Chol lipid mixture and mixtures with increasing concentrations of TM31C. In the pure lipid system, there is no detectable endotherm near 36°C (thermogram 1), but a broad endotherm starts to appear in the presence of 2.5 mol % TM31C (thermogram 2). Increasing the peptide concentrations up to 7.5 mol % results in the appearance of an even more prominent and cooperative endotherm at 36°C (thermograms 3 and 4), with the consequent exothermic peak at 24°C observed in the cooling scans (not shown). These signals can be attributed to the polymorphic transition of anhydrous Chol crystals extracted from the lipid mixture by the peptide. In contrast, TM19 seems to initially promote a modest anhydrous Chol formation in vesicles (Fig. 4 B, thermogram 2), but from that point on (Fig. 4 B, thermograms 3 and 4), at 5 and 7.5 mol %, crystal formation does not occur and the endotherms do not show any further changes (see Table 1).
Figure 4.
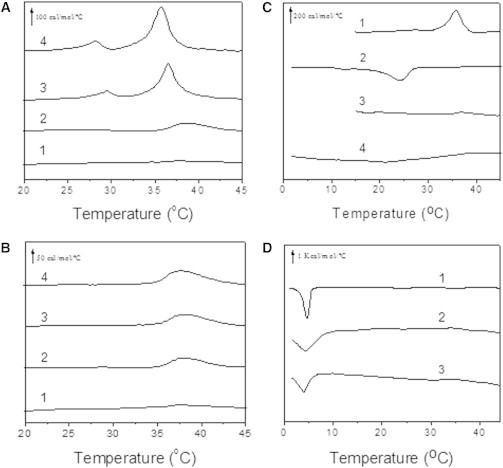
(A and B) DSC of pure SOPC/Chol 1:1 (mol:mol) (thermogram 1) and the same lipid mixtures with 2.5 mol % (2A or 2B), 5 mol % (3A or 3B), and 7.5 mol % (4A or 4B) TM31C or TM19, respectively. Scans are offset along the y axis for clarity of presentation. The scan rate was 1.5°C/min from 10°C to 50°C. The lipid concentration was 4 mM in 20 mM PIPES (pH 7.4), 150 mM NaCl, 1 mM EDTA. (C) Heating-cooling scans of 5 mol % TM31C- (thermograms 1 and 2) or TM19- (thermograms 3 and 4) induced changes when inserted into SOPC/Chol 1:1 (mol ratio). (D) DSC measurements of pure SOPC (thermogram 1) and mixtures with 7.5 mol % TM31C (thermogram 2) or TM19 (thermogram 3). Only cooling scans are shown in D.
Table 1.
Peptide induction of anhydrous cholesterol crystallite formation
| TM31C (long peptide) | TM19 (short peptide) | |||||
|---|---|---|---|---|---|---|
| Parameters | 2.5 mol % | 5 mol % | 7.5 mol % | 2.5 mol % | 5 mol % | 7.5 mol % |
| ΔH (cal/mol) | 290 | 623 | 864 | 131 | 149 | 189 |
| Tm (°C) | 38.7 | 36.4 | 35.6 | 37.6 | 38 | 37.8 |
| T1/2 (°C) | 4.4 | 1.9 | 1.8 | 4.2 | 4.1 | 4.2 |
The thermodynamic correspond to the anhydrous cholesterol endothermic phase transition near 36°C.
The behavior of short and long peptides with other lipid compositions, such as pure pSM and pure DPPC, was checked previously (13) and the obtained endotherms were virtually the same for both peptides. Thus, the endothermic differences obtained with SOPC/Chol membranes in this case are apparently the result of the lack of the C-ter extracellular coil in TM19 peptide, suggesting that this segment is a crucial element for the interaction with Chol.
Peptides that preferentially interact with Chol-rich domains sequester Chol, leaving other parts of the membrane depleted of Chol. As a consequence, the Chol-poor regions exhibit a higher enthalpic and cooperative transition of the phospholipid partner. In contrast, when peptides interact preferentially with Chol-depleted regions, the cooperativity of the phospholipid phase transition decreases, resulting in a broader and lower transition enthalpy. We performed serial cooling scans from 50°C to 1°C to discern the peptide location in Chol-depleted or Chol-rich domains (Fig. 4 C). The lack of an SOPC transition at 6°C indicates that TM19 partitions mainly into Chol-depleted domains and is therefore unable to accomplish the large formation of Chol crystals that TM31C elicits. However, even the long peptide does not give rise to an increase in the SOPC transition enthalpy, possibly because this peptide partitions, to some extent, into both Chol-rich and Chol-depleted domains. Traditionally, SCR has been reconstituted in Chol-free vesicles, and the possibility of simultaneous partitioning was evidenced by the TM31C peptide-induced widening of the transition of pure SOPC, just as observed for TM19 (Fig. 4 D).
From this series of DSC experiments, we conclude that even in the absence of a CRAC sequence, TM31C is more effective at promoting the formation of Chol-rich domains than the shorter peptide TM19, which comprises only the putative TMD domain. These data suggest that the C-terminal extracellular amino acids contribute to the interaction with Chol. Long TMDs favor insertion into lo domains, avoiding the mismatch that occurs upon insertion into ld domains (31,32). For instance, protein TMDs anchored to Golgi membranes have an average length of 15 aa, whereas TMDs of proteins in the plasma membrane, with 3-fold more Chol, exhibit an average length of ∼22 aa (33). In the case of SCR, amino acids such as Ser, Thr, and Gly can be found in the membrane interface in contact with lipid headgroups (34). Additionally, the two snorkeling Lys found at the beginning of the TMD sequence (35) could also favor the protein partitioning into Chol-rich domains. The 19 aa length of SCR TMD does not fulfill the length requirement for avoiding the abovementioned mismatch, and thus the SCR C-terminal juxtamembrane segment’s interaction with Chol could favor protein insertion into lo domains.
In silico studies of Chol-SCR interactions
We used several starting conditions and conducted a series of molecular modeling simulations on a short sequence [298FLIDFMFFESTGSQE311] that includes the last C-terminal amino acids of the putative TMD and the first amino acids of the protein C-terminal extracellular coil, which appear to be involved in the interaction with Chol. A detailed analysis of the energy of interaction of Chol and this sequence showed that the interaction complex involves essentially eight residues—five in the SCR TMD (F298, L299, I300, M303, and E306) and three in the protein C-terminal juxtamembrane positions (S307, S310, and Q311)—for a total energy of interaction of −64.39 KJ.mol−1 (Fig. 5 A).
Figure 5.
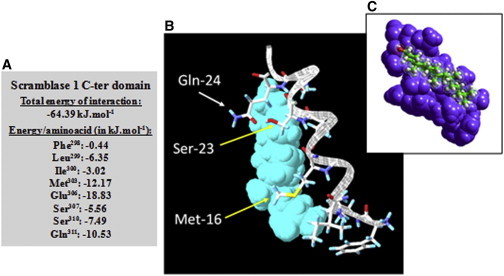
Docking of Chol on hPL SCR. A short part of TM31C, [298FLIDFMFFESTGSQ311], contains the highest enthalpic interaction with Chol. (A) Detailed analysis of the energetics of Chol interaction with the protein TMD and other surrounding amino acids that presumably are involved in the interaction. (B) Surface view of the complex, with important amino acid side chains presented as interacting with the sterol. The TMD amino acid M303 is represented as Met-16, which interacts with the Chol ring structure, and the C-terminal coil amino acids S309 and Q311, which interact with the small polar alcohol headgroup, are represented as Ser-23 and Gln-24 respectively. E306, at the end of TMD (which is not represented), contains the highest enthalpic interaction with Chol. (C) Overall illustration of the interaction of scramblase TMD C-ter coil (in blue) and Chol.
Surprisingly, the last three Fs of the TMDs, which theoretically might interact through their aromatic lateral chains with the lipid rings, do not interact with Chol. Molecular modeling studies have shown that the remaining sequence can have a good fit for Chol, as illustrated in Fig. 5, B and C. The synthetic peptide TM19 ending in Glu306 would lose the energies of interaction of S307, S310, and Q311. As a result, Chol could not interact properly with Glu as a terminal residue, as experimentally observed (Fig. 4 B).
SCR reconstitution in ld- and lo-domain-containing membranes
The recombinant purified protein could be reconstituted in ld membranes (e.g., PC/PS 9:1 (mol/mol) (12)) or in lo-containing membranes (e.g., DOPC/PSM/Chol 2:1:1 or 1:1:1 (mol ratio); Fig. 6, A and B). After reconstitution into lo-containing LUVs made of PC/SM/Chol 2:1:1 (mol ratio), flip-flop activity was assayed according to the pyrene excimer method (20) and the NBD-dithionite method (28). Freshly synthesized pyrene-sphingomyelin (py-SM) was used, and protein reconstituted in ld PC/PS 9:1 LUVs was used as a positive control for lipid scrambling.
Figure 6.
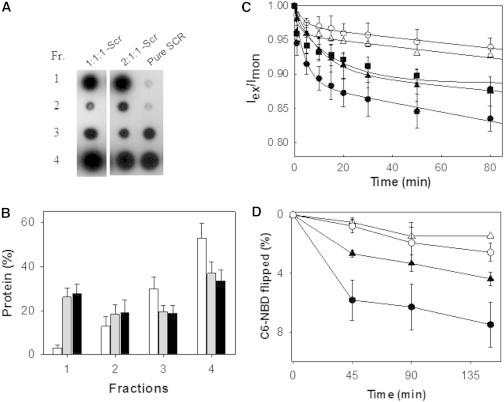
(A) Dot blots of the fractions recovered from the sucrose gradient (27) (fractions 1–4, samples recovered from top to bottom of the sucrose gradient). The yield of protein reconstitution into PC/SM/Chol 2:1:1 and 1:1:1 vesicles was virtually the same (∼50% of SCR successfully reconstituted). (B) SCR quantification in each recovered fraction when reconstituted in PC/SM/Chol 2:1:1 vesicles (gray bars) or PC/PS 9:1 vesicles (black bars). The recovered top fraction was used for the flip-flop assay. White bars correspond to pure ultracentrifuged protein. (C and D) Influence of Chol on py-SM (C) and NBD-PS (D) transbilayer movement promoted by SCR. All measurements were made in the presence of 5 mM calcium. (Δ) Liposomes. (▲) SCR proteoliposomes of DOPC/SM/Chol 2:1:1. (▪) SCR proteoliposomes of POPC/PS/Chol 9:1:3.3. (●) SCR proteoliposomes of PC/PS 9:1. (○) SCR proteoliposomes in the absence of Ca2+. Average values ± SEM (n ≥ 3).
The data in Fig. 6 C show that for PC/PS 9:1 and PC/SM/Chol 2:1:1, in the presence of SCR, the proportion of pyrene excimer/monomer decreased, particularly in the first 30 min after 5 mM Ca2+ addition, which fully activated the protein. This is taken as an indication of transmembrane lipid motion, or flip-flop (12). The protein reconstituted in lo-containing membranes was expected to promote faster or more extensive py-SM translocation due to the presence of Chol, but in fact the scrambling motion was somewhat lower than that observed for the protein reconstituted in pure ld phase. The Chol-ordering effect in the vicinity of the SCR might inhibit the translocation mechanism. SCR was reconstituted in the POPC/PS/Chol 9:1:3.3 (mol ratio) composition as well, to exclude the possibility that the lower translocation rate recorded for the 2.1:1 composition could be due to the lack of a negative charge in the membrane, and the scrambling rate was found to be the same as in the 2:1:1 composition (Fig. 6 C).
Similar results were obtained for SCR reconstituted in PC/PS 9:1 and PC/SM/Chol 2:1:1 compositions with the NBD-PS probe (Fig. 6 D). Zhao et al. (36) suggested that cysteines in the SCR N-terminal structure could be responsible for the decreased activity of the recombinant protein as compared with the native form. Thus, it appears that Chol has a dual effect of helping in membrane insertion and inhibiting translocation.
Discussion
SCR, a 35 kDa protein, is an integral membrane protein that is located in plasma membrane liquid-ordered domains when it is palmitoylated. One of the major lipid components in these domains is Chol, which increases the membrane thickness of the plasma membrane to accommodate the TMDs of many proteins. However, Chol can also interact directly with these protein domains via hydrophobic or van der Waals forces, as described for caveolin-1 and human acetylcholine receptor TMDs (37,38). In this work, we provide evidence of a close relationship between Chol and some TMD and extracellular coil amino acids of SCR (Fig. 7) that would facilitate SCR accommodation and stabilization in lo domains.
Figure 7.
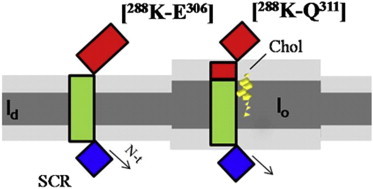
Suggested outline of the SCR C-terminal domain disposition in membranes when inserted into Chol-depleted domains (left) and Chol-rich domains (right). Red, extracellular segment; green, the putative TMD; blue, the cytoplasmic portion.
Solid-phase binding and mono-/bilayer insertion measurements
We performed a series of lipid interaction experiments with the aim of studying peptide-induced membrane perturbation and insertion. Similar results were obtained for SCR and TM31C: the highest binding was observed for the sterols that, like Chol, contain an alcohol group in ring A. The Langmuir balance observation of increased TM31C affinity for Chol deserves some comment (Fig. 3 A). The peptide presents a preferential interaction with Chol-containing membranes in comparison with pure PC. Even when lipid packing (at 20 mol % Chol) is high due to the presence of Chol, the peptide achieves insertion, and Δπ rises from 4 to 6 mN/m. Thereafter, at very high Chol concentrations (30–50 mol %), the lipid packing is too high for the peptide to be included in the lipid film. The increase in surface pressure is significantly smaller when the lipid film has no Chol or when it is highly enriched in it, an effect that has been observed with other proteins such as FraC and equinatoxin II in PC/SM host lipid mixtures (39,40).
Addition of TM31C to liposomes made of PC/SM/Chol (1:7:2) resulted in the gradual release of up to 40% of the encapsulated solute (Fig. 3, B and C), with a fairly high efflux rate. In comparison, with the same lipid composition, but with 20 mol % Chol-one instead of Chol, the total ANTS leakage was several times lower, with a very poor efflux rate. Thus, the phase boundary between the ld and lo domains appears to be important for SCR peptide insertion and probably also for protein docking.
Contribution of the SCR extracellular segment to the interaction with Chol
DSC has been used to identify Chol-rich domains induced by peptides or proteins containing a CRAC domain. The formation of crystals of anhydrous Chol in the presence of NAP-22 (41), caveolin-1 (16), or other peptides has been detected by this technique. Two criteria must be taken into account as an indication of Chol recruitment into pools: 1), the appearance of Chol crystals in a membrane in which Chol is in principle miscible in the absence of peptides/proteins; and 2), the recovery of the phase transition cooperativity of the phospholipid partner.
It is rare for a protein not to discriminate between these domains, but this does occur with caveolin-1 peptide (16), which promotes Chol crystal formation but also stays in the Chol-depleted domains, thus blurring the phospholipid phase transition signal. This is also the case for the scramblase peptide TM31C (Fig. 4 A), which in contrast to TM19 (Fig. 4 B) promotes Chol crystallite formation. However, both TM19 and the long peptide can be detected in Chol-depleted pools abolishing the SOPC phase transition at 6°C (Fig. 4 C). This is evidenced by the fact that both peptides lower the transition enthalpy of pure SOPC (Fig. 4 D), just as they do that of DPPC and pSM (13).
Traditionally, SCR has been reconstituted in Chol-free membranes, such that the C-terminal part of the protein can be easily inserted. However, assuming that the amino acid sequence [288K-E306] is the SCR TMD, the TMD represented by TM19 is too short to insert into Chol-rich membranes, suggesting an important role for the extracellular juxtamembrane amino acids in the interaction with Chol. This phenomenon is not exceptional. A juxtamembrane segment found in the gp41 protein of HIV is believed to play a role in sequestering the whole protein to a Chol-rich domain (42). In addition, certain amino acids involved in CRAC or CARC domains are found outside or shared between the TMD and the extracellular portions, as is the case with the nicotinic acetylcholine receptor protein TMDs (38) and the translocator protein 18 kDa of the mitochondrial outer membrane (43), thus supporting the involvement of extramembranous amino acid residues in Chol interactions.
Using molecular modeling, we predicted that Chol would be docked onto the sequence 298[FLIDFMFFESTGSQ]311, with a favorable total energy of interaction of −64.39 KJ.mol−1 (Fig. 5 A). Most of the interaction involves amino acid residues that traditionally have not been predicted to be involved in this binding. On the contrary, the modeling excludes other attractive amino acids. The strongest predicted interaction with Chol cycles relies on TMD Leu299 and Met303 via hydrophobic interactions with an energy of interaction of −5 KJ/mol per residue, which is fully consistent with this type of interaction. Additionally, Glu306 and the juxtamembrane polar amino acids Ser307, Ser310, and Gln311, which would be located at the membrane interface (34), also appear to interact strongly with Chol.
The Chol ordering effect decreases SCR activity
Some lipid compositions that give rise to Chol-rich domains were tested for protein reconstitution. The scrambling activity was lower when Chol was present in the membrane (Fig. 6, C and D), showing that the Chol ordering effect hampers the transbilayer distribution of lipids, in agreement with other studies (44–46).
Zhao et al. (36) hypothesized that palmitoylation, apart from anchoring the protein to the membrane, assists the protein calcium-binding domain in forming the loop configuration required to complex the metal ion. We propose that when the protein properly binds calcium, the SCR TMD backbone dynamics could provide a hydrophilic pathway along which a polar lipid headgroup could transverse the bilayer (47). Alternatively, it could disorder the bilayer at the helix/lipid interface for a more favorable translocation (48).
How can SCR perform its scrambling role when it is embedded in liquid-ordered domains? Although many proteins and receptors are localized to lo domains, the effect of ligand on this association is highly variable. In the case of EGFR, which colocalizes with SCR in rafts (9), the protein rapidly moves out from lipid rafts upon activation by ligand (49). For this receptor, events such as ligand binding, dimerization, and autophosphorylation are all enhanced after raft disruption occurs (50–52). Thus, apart from serving in signal initiation, rafts may also be important platforms for suppressing false signaling coming from proteins such as EGFR and perhaps SCR. SCR anchored to lipid rafts serves as an important effector for cell proliferation and maturation (53) by promoting, among other effects, the kinase activity of cellular c-Src. It is therefore conceivable that SCR could modulate its activities by its localization in the cell, including its role as a transcription factor in the nucleus (4,54) and its possible migration from rafts to the bulk of the plasma membrane or to the lo/ld interface to promote lipid scrambling.
SCR palmitoylated and anchored to Chol-rich membranes
The notion of scramblase anchoring to the membrane was challenged by Bateman et al. (7), who proposed that the highly hydrophobic α-helical domain would remain buried in the protein core, with the palmitoyl residues serving as the only tether to the bilayer. Our previous experimental work (13) provided strong evidence that the 288–306 peptide SCR could be membrane inserted. We also detected an affinity of TM31C for liquid-ordered (lo) domains, and more specifically for phase boundaries limiting lo domains. Palmitoylation is important for protein sorting into membrane rafts, as demonstrated for members of the Src family in cells (55) and for G-proteins in model membranes (56). The packing order conferred by the structure of the lipid modification is considered to be an important determinant for a protein to be targeted to a Chol-rich domain, rather than the TMD amino acid composition or hydrophobic mismatch prevention (31).
In the case of SCR, palmitoylation functions primarily as a sorting signal (8) that is required for its plasma membrane transport and for subsequent anchoring to Chol-enriched domains. However, protein TMD might also be involved in anchoring SCR to Chol-rich membranes. Our data are consistent with the hypothesis of direct lipid-protein contact, with all of the tested methods showing large, measurable SCR-Chol interactions. We propose that both palmitoylation and the protein amino acid segment [298F-Q311] anchor SCR to Chol-rich membranes (Fig. 7). Additionally, Chol inhibits protein scrambling activity by ordering the membrane around the TMD. Calcium binding could stimulate the proper conformation of TMD to promote lipid flip-flop.
Acknowledgments
This work was supported in part by grants from the Spanish Ministerio de Economía (BFU 2012-36241 to F.M.G. and BFU 2011-28566 to A.A.) and the Basque Government (IT849-13 to F.M.G. and IT838-13 to A.A.). I.M.D.P. was a predoctoral student supported by the Basque Government.
References
- 1.Bassé F., Stout J.G., Wiedmer T. Isolation of an erythrocyte membrane protein that mediates Ca2+-dependent transbilayer movement of phospholipid. J. Biol. Chem. 1996;271:17205–17210. doi: 10.1074/jbc.271.29.17205. [DOI] [PubMed] [Google Scholar]
- 2.Zhou Q., Zhao J., Sims P.J. Molecular cloning of human plasma membrane phospholipid scramblase. A protein mediating transbilayer movement of plasma membrane phospholipids. J. Biol. Chem. 1997;272:18240–18244. doi: 10.1074/jbc.272.29.18240. [DOI] [PubMed] [Google Scholar]
- 3.Sahu S.K., Gummadi S.N., Aradhyam G.K. Phospholipid scramblases: an overview. Arch. Biochem. Biophys. 2007;462:103–114. doi: 10.1016/j.abb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Efraim I., Zhou Q., Sims P.J. Phospholipid scramblase 1 is imported into the nucleus by a receptor-mediated pathway and interacts with DNA. Biochemistry. 2004;43:3518–3526. doi: 10.1021/bi0356911. [DOI] [PubMed] [Google Scholar]
- 5.Wyles J.P., Wu Z., Cole S.P. Nuclear interactions of topoisomerase II alpha and beta with phospholipid scramblase 1. Nucleic Acids Res. 2007;35:4076–4085. doi: 10.1093/nar/gkm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silverman R.H., Halloum A., Sims P.J. Suppression of ovarian carcinoma cell growth in vivo by the interferon-inducible plasma membrane protein, phospholipid scramblase 1. Cancer Res. 2002;62:397–402. [PubMed] [Google Scholar]
- 7.Bateman A., Finn R.D., Söding J. Phospholipid scramblases and Tubby-like proteins belong to a new superfamily of membrane tethered transcription factors. Bioinformatics. 2009;25:159–162. doi: 10.1093/bioinformatics/btn595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiedmer T., Zhao J., Sims P.J. Palmitoylation of phospholipid scramblase 1 controls its distribution between nucleus and plasma membrane. Biochemistry. 2003;42:1227–1233. doi: 10.1021/bi026679w. [DOI] [PubMed] [Google Scholar]
- 9.Sun J., Nanjundan M., Sims P.J. Plasma membrane phospholipid scramblase 1 is enriched in lipid rafts and interacts with the epidermal growth factor receptor. Biochemistry. 2002;41:6338–6345. doi: 10.1021/bi025610l. [DOI] [PubMed] [Google Scholar]
- 10.Posada I.M.D., Sánchez-Magraner L., Goñi F.M. Membrane binding of human phospholipid scramblase 1 cytoplasmic domain. Biochim. Biophys. Acta. 2014;1838:1785–1792. doi: 10.1016/j.bbamem.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Francis V.G., Mohammed A.M., Gummadi S.N. The single C-terminal helix of human phospholipid scramblase 1 is required for membrane insertion and scrambling activity. FEBS J. 2013;280:2855–2869. doi: 10.1111/febs.12289. [DOI] [PubMed] [Google Scholar]
- 12.Sánchez-Magraner L., Posada I.M., Goñi F.M. The C-terminal transmembrane domain of human phospholipid scramblase 1 is essential for the protein flip-flop activity and Ca2+-binding. J. Membr. Biol. 2014;247:155–165. doi: 10.1007/s00232-013-9619-7. [DOI] [PubMed] [Google Scholar]
- 13.Posada I.M.D., Busto J.V., Alonso A. Membrane binding and insertion of the predicted transmembrane domain of human scramblase 1. Biochim. Biophys. Acta. 2014;1838(1 Pt B):388–397. doi: 10.1016/j.bbamem.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 14.Li H., Yao Z., Papadopoulos V. Cholesterol binding at the cholesterol recognition/interaction amino acid consensus (CRAC) of the peripheral-type benzodiazepine receptor and inhibition of steroidogenesis by an HIV TAT-CRAC peptide. Proc. Natl. Acad. Sci. USA. 2001;98:1267–1272. doi: 10.1073/pnas.031461598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jamin N., Neumann J.M., Lacapère J.J. Characterization of the cholesterol recognition amino acid consensus sequence of the peripheral-type benzodiazepine receptor. Mol. Endocrinol. 2005;19:588–594. doi: 10.1210/me.2004-0308. [DOI] [PubMed] [Google Scholar]
- 16.Epand R.M., Sayer B.G., Epand R.F. Caveolin scaffolding region and cholesterol-rich domains in membranes. J. Mol. Biol. 2005;345:339–350. doi: 10.1016/j.jmb.2004.10.064. [DOI] [PubMed] [Google Scholar]
- 17.Fantini J., Barrantes F.J. How cholesterol interacts with membrane proteins: an exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front. Physiol. 2013;4:31. doi: 10.3389/fphys.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crowet J.M., Lins L., Brasseur R. Tilted properties of the 67-78 fragment of alpha-synuclein are responsible for membrane destabilization and neurotoxicity. Proteins. 2007;68:936–947. doi: 10.1002/prot.21483. [DOI] [PubMed] [Google Scholar]
- 19.Charloteaux B., Lorin A., Brasseur R. The N-terminal 12 residue long peptide of HIV gp41 is the minimal peptide sufficient to induce significant T-cell-like membrane destabilization in vitro. J. Mol. Biol. 2006;359:597–609. doi: 10.1016/j.jmb.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 20.Müller P., Schiller S., Herrmann A. Continuous measurement of rapid transbilayer movement of a pyrene-labeled phospholipid analogue. Chem. Phys. Lipids. 2000;106:89–99. doi: 10.1016/s0009-3084(00)00134-1. [DOI] [PubMed] [Google Scholar]
- 21.Dowler S., Kular G., Alessi D.R. Protein lipid overlay assay. Sci. STKE. 2002;2002:pl6. doi: 10.1126/stke.2002.129.pl6. [DOI] [PubMed] [Google Scholar]
- 22.Ellens H., Bentz J., Szoka F.C. H+- and Ca2+-induced fusion and destabilization of liposomes. Biochemistry. 1985;24:3099–3106. doi: 10.1021/bi00334a005. [DOI] [PubMed] [Google Scholar]
- 23.Goñi F.M., Villar A.V., Alonso A. Interaction of phospholipases C and sphingomyelinase with liposomes. Methods Enzymol. 2003;372:3–19. doi: 10.1016/S0076-6879(03)72001-1. [DOI] [PubMed] [Google Scholar]
- 24.Fantini J., Carlus D., Yahi N. The fusogenic tilted peptide (67-78) of α-synuclein is a cholesterol binding domain. Biochim. Biophys. Acta. 2011;1808:2343–2351. doi: 10.1016/j.bbamem.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 25.Singh R.P., Brooks B.R., Klauda J.B. Binding and release of cholesterol in the Osh4 protein of yeast. Proteins. 2009;75:468–477. doi: 10.1002/prot.22263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomsen R., Christensen M.H. MolDock: a new technique for high-accuracy molecular docking. J. Med. Chem. 2006;49:3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 27.Yethon J.A., Epand R.F., Andrews D.W. Interaction with a membrane surface triggers a reversible conformational change in Bax normally associated with induction of apoptosis. J. Biol. Chem. 2003;278:48935–48941. doi: 10.1074/jbc.M306289200. [DOI] [PubMed] [Google Scholar]
- 28.McIntyre J.C., Sleight R.G. Fluorescence assay for phospholipid membrane asymmetry. Biochemistry. 1991;30:11819–11827. doi: 10.1021/bi00115a012. [DOI] [PubMed] [Google Scholar]
- 29.Li X.M., Momsen M.M., Brown R.E. Cholesterol decreases the interfacial elasticity and detergent solubility of sphingomyelins. Biochemistry. 2001;40:5954–5963. doi: 10.1021/bi002791n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Epand R.M. Do proteins facilitate the formation of cholesterol-rich domains? Biochim. Biophys. Acta. 2004;1666:227–238. doi: 10.1016/j.bbamem.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 31.Schäfer L.V., de Jong D.H., Marrink S.J. Lipid packing drives the segregation of transmembrane helices into disordered lipid domains in model membranes. Proc. Natl. Acad. Sci. USA. 2011;108:1343–1348. doi: 10.1073/pnas.1009362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sprong H., van der Sluijs P., van Meer G. How proteins move lipids and lipids move proteins. Nat. Rev. Mol. Cell Biol. 2001;2:504–513. doi: 10.1038/35080071. [DOI] [PubMed] [Google Scholar]
- 33.Bretscher M.S., Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 34.MacCallum J.L., Bennett W.F., Tieleman D.P. Distribution of amino acids in a lipid bilayer from computer simulations. Biophys. J. 2008;94:3393–3404. doi: 10.1529/biophysj.107.112805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strandberg E., Killian J.A. Snorkeling of lysine side chains in transmembrane helices: how easy can it get? FEBS Lett. 2003;544:69–73. doi: 10.1016/s0014-5793(03)00475-7. [DOI] [PubMed] [Google Scholar]
- 36.Zhao J., Zhou Q., Sims P.J. Palmitoylation of phospholipid scramblase is required for normal function in promoting Ca2+-activated transbilayer movement of membrane phospholipids. Biochemistry. 1998;37:6361–6366. doi: 10.1021/bi980218m. [DOI] [PubMed] [Google Scholar]
- 37.Aoki S., Epand R.M. Caveolin-1 hydrophobic segment peptides insertion into membrane mimetic systems: role of proline residue. Biochim. Biophys. Acta. 2012;1818:12–18. doi: 10.1016/j.bbamem.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Baier C.J., Fantini J., Barrantes F.J. Disclosure of cholesterol recognition motifs in transmembrane domains of the human nicotinic acetylcholine receptor. Sci. Rep. 2011;1:69. doi: 10.1038/srep00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bellomio A., Morante K., González-Mañas J.M. Purification, cloning and characterization of fragaceatoxin C, a novel actinoporin from the sea anemone Actinia fragacea. Toxicon. 2009;54:869–880. doi: 10.1016/j.toxicon.2009.06.022. [DOI] [PubMed] [Google Scholar]
- 40.Caaveiro J.M., Echabe I., González-Mañas J.M. Differential interaction of equinatoxin II with model membranes in response to lipid composition. Biophys. J. 2001;80:1343–1353. doi: 10.1016/S0006-3495(01)76107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Epand R.M., Maekawa S., Epand R.F. Protein-induced formation of cholesterol-rich domains. Biochemistry. 2001;40:10514–10521. doi: 10.1021/bi010897s. [DOI] [PubMed] [Google Scholar]
- 42.Epand R.F., Thomas A., Epand R.M. Juxtamembrane protein segments that contribute to recruitment of cholesterol into domains. Biochemistry. 2006;45:6105–6114. doi: 10.1021/bi060245+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, F., Y. Xia, J. Meiler, and S. Ferguson-Miller. 2013. Characterization and modeling of the oligomeric state and ligand binding behavior of purified translocator protein 18 kDa from Rhodobacter sphaeroides. Biochemistry 52:5884–5899. [DOI] [PMC free article] [PubMed]
- 44.Langer M., Langosch D. Is lipid flippase activity of SNARE transmembrane domains required for membrane fusion? FEBS Lett. 2011;585:1021–1024. doi: 10.1016/j.febslet.2011.02.033. [DOI] [PubMed] [Google Scholar]
- 45.Rajasekharan A., Gummadi S.N. Inhibition of biogenic membrane flippase activity in reconstituted ER proteoliposomes in the presence of low cholesterol levels. Cell. Mol. Biol. Lett. 2012;17:136–152. doi: 10.2478/s11658-011-0042-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langer M., Sah R., Langosch D. Structural properties of model phosphatidylcholine flippases. Chem. Biol. 2013;20:63–72. doi: 10.1016/j.chembiol.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 47.Smeijers A.F., Pieterse K., Hilbers P.A.J. Coarse-grained transmembrane proteins: hydrophobic matching, aggregation, and their effect on fusion. J. Phys. Chem. B. 2006;110:13614–13623. doi: 10.1021/jp062012y. [DOI] [PubMed] [Google Scholar]
- 48.Kol M.A., de Kroon A.I.P.M., de Kruijff B. Transbilayer movement of phospholipids in biogenic membranes. Biochemistry. 2004;43:2673–2681. doi: 10.1021/bi036200f. [DOI] [PubMed] [Google Scholar]
- 49.Mineo C., Gill G.N., Anderson R.G. Regulated migration of epidermal growth factor receptor from caveolae. J. Biol. Chem. 1999;274:30636–30643. doi: 10.1074/jbc.274.43.30636. [DOI] [PubMed] [Google Scholar]
- 50.Pike L.J., Casey L. Cholesterol levels modulate EGF receptor-mediated signaling by altering receptor function and trafficking. Biochemistry. 2002;41:10315–10322. doi: 10.1021/bi025943i. [DOI] [PubMed] [Google Scholar]
- 51.Ringerike T., Blystad F.D., Stang E. Cholesterol is important in control of EGF receptor kinase activity but EGF receptors are not concentrated in caveolae. J. Cell Sci. 2002;115:1331–1340. doi: 10.1242/jcs.115.6.1331. [DOI] [PubMed] [Google Scholar]
- 52.Pike L.J. Lipid rafts: bringing order to chaos. J. Lipid Res. 2003;44:655–667. doi: 10.1194/jlr.R200021-JLR200. [DOI] [PubMed] [Google Scholar]
- 53.Nanjundan M., Sun J., Wiedmer T. Plasma membrane phospholipid scramblase 1 promotes EGF-dependent activation of c-Src through the epidermal growth factor receptor. J. Biol. Chem. 2003;278:37413–37418. doi: 10.1074/jbc.M306182200. [DOI] [PubMed] [Google Scholar]
- 54.Zhou Q., Ben-Efraim I., Sims P.J. Phospholipid scramblase 1 binds to the promoter region of the inositol 1,4,5-triphosphate receptor type 1 gene to enhance its expression. J. Biol. Chem. 2005;280:35062–35068. doi: 10.1074/jbc.M504821200. [DOI] [PubMed] [Google Scholar]
- 55.Webb Y., Hermida-Matsumoto L., Resh M.D. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J. Biol. Chem. 2000;275:261–270. doi: 10.1074/jbc.275.1.261. [DOI] [PubMed] [Google Scholar]
- 56.Moffett S., Brown D.A., Linder M.E. Lipid-dependent targeting of G proteins into rafts. J. Biol. Chem. 2000;275:2191–2198. doi: 10.1074/jbc.275.3.2191. [DOI] [PubMed] [Google Scholar]


