Abstract
Background
Cytokinesis in many eukaryotes involves the function of an actomyosin-based contractile ring. In fission yeast, actomyosin ring maturation and stability require a conserved signaling pathway termed the SIN (septation initiation network). The SIN consists of a GTPase (Spg1p) and three protein kinases, all of which localize to the mitotic spindle pole bodies (SPBs). Two of the SIN kinases, Cdc7p and Sid1p, localize asymmetrically to the newly duplicated SPB in late anaphase. How this asymmetry is achieved is not understood, although it is known that their symmetric localization impairs cytokinesis.
Results
Here we characterize a new Forkhead-domain-associated protein, Csc1p, and identify SIN-Inhibitory PP2A-complex (SIP), which is crucial for the establishment of SIN asymmetry. Csc1p localizes to both SPBs early in mitosis, is lost from the SPB that accumulates Cdc7p, and instead accumulates at the SPB lacking Cdc7p. Csc1p is required for the dephosphorylation of the SIN scaffolding protein Cdc11p and is thereby required for the recruitment of Byr4p, a component of the GTPase-activating subunit for Spg1p, to the SPB.
Conclusions
Since Cdc7p does not bind to GDP-Spg1p, we propose that the SIP-mediated Cdc11p-dephosphorylation and the resulting recruitment of Byr4p are amongst the earliest steps in the establishment of SIN asymmetry.
INTRODUCTION
Cytokinesis is the terminal step in cell division during which a single mother cell is physically divided into two daughters. Cytokinesis in many eukaryotes, ranging from yeasts and fungi to metazoans, involves the function of an actomyosin-based contractile ring. The actomyosin ring is assembled upon entry into mitosis and constricts following completion of anaphase. Although detailed mechanisms regulating actomyosin ring positioning and assembly have emerged in recent years [1–3], the molecular controls that ensure that cytokinesis occurs only once per cell cycle and precisely after chromosome segregation are not understood.
Over the last two decades, the fission yeast Schizosaccharomyces pombe has emerged as an attractive model for the study of cytokinesis, since it divides using an actomyosin ring and due to the well developed experimental approaches applicable to this organism [2, 4, 5]. In fission yeast, a signaling cascade termed the Septation Initiation Network (SIN) plays a key role in the regulation of cytokinesis [6–8]. Fission yeast SIN-like signaling cascades have been identified in budding yeasts (termed the MEN-mitotic exit network), fungi, and metazoans [3, 6, 7, 9, 10]. SIN includes three protein kinases (Cdc7p, Sid1p, and Sid2p), a small GTPase (Spg1p), a pair of spindle pole body (SPB) resident proteins Cdc11p and Sid4p, and binding-partners of Sid1p and Sid2p, termed Cdc14p and Mob1p, respectively [9, 11–20]. Loss of function mutations in any of these genes leads to defects in actomyosin ring maturation, constriction and septation and to the generation of multinucleate cells [9, 12, 14–16, 18, 20]. By contrast, ectopic activation of the SIN through mutations in the two component GTPase activating protein for Spg1p (Cdc16p and Byr4p are the subunits) [21–24] or upon overexpression of Spg1p [18] leads to uncontrolled septation without intervening mitosis. These and other observations have led to the idea that the SIN coordinates chromosome segregation with cytokinesis [5, 15, 25–29].
Protein localization studies have shown that all SIN components localize to the SPB in a manner dependent on the proteins Cdc11p and Sid4p [11, 19, 30]. Of these, Spg1p, Sid2p, and Mob1p are detected at the SPB throughout the cell cycle [9, 16, 18, 20], whereas Sid1p, Cdc7p and Cdc14p localize to only one of the two SPBs (the newly generated SPB) during late mitosis and do not localize to the SPB in interphase [14, 15, 31]. Innovative cell biological experiments have shown that Cdc7p, Sid1p, and Cdc14p reside on the SPB containing GTP bound Spg1p [31, 32]. Conversely, Byr4p and Cdc16p localize to the SPBs throughout interphase and then specifically to the old SPB during late mitosis [25, 31–33]. Importantly, the ectopic localization of Cdc7p, Sid1p, and Cdc14p [15] [32] to either the interphase SPB or to both SPBs in mitosis correlates with repeated rounds of septation in the absence of intervening mitosis. These findings have led to the proposal that hyperactivation of the SIN (via the symmetric localization of Cdc7p, Sid1p, and Cdc14p) leads to defective cytokinesis characterized by uncontrolled septation [21, 22, 32]. However, the molecular mechanisms behind the generation of asymmetry in the localization of these proteins are not fully understood.
In this study we identify SIP (SIN-inhibitory phosphatase complex) containing at least 6 proteins, including a previously uncharacterized catalytic subunit of protein phosphatase 2A (Ppa3p) and the A-subunit of the PP2A holoenzyme Paa1p. SIP components localize to the SPBs early in mitosis and transiently localize asymmetrically to the old SPB and promote localization of Byr4p to the old SPB. This in turn leads to the establishment and propagation of Cdc7p and Sid1p at the new SPB.
RESULTS
Csc1p localizes to the SPB in early mitosis
Since all known SIN components localize to the SPBs during mitosis, we hypothesized that additional new regulators of the SIN can be identified by characterizing proteins that specifically localize to the mitotic SPBs [6, 7, 34, 35]. To this end we searched the GFP-ORFeome [36] and identified a previously uncharacterized Forkhead-associated domain protein (SPBC3H7.13), which we have named as Csc1p (Component of SIP Complex 1). Csc1p is a 301 amino-acid protein that contains an N-terminal FHA-domain (Figure 1A), and is most related to Saccharomyces cerevisiae Far10p (data not shown).
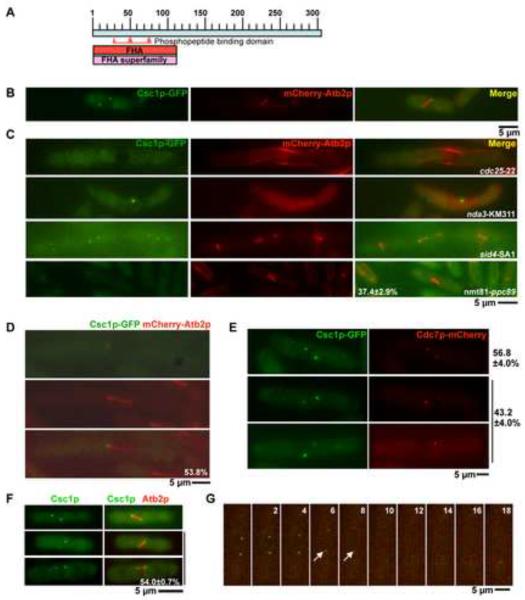
Figure 1. Csc1p, a new Forkhead-associated domain containing protein localizes to both SPBs and then transiently to one SPB during early mitosis.
A. Schematic representation of Csc1p with the FHA domain at the N-terminus. The phosphopeptide binding domain is highlighted between amino acids 30 and 80. B. Csc1p localizes to both SPBs in cells with short mitotic spindles. csc1-GFP mCherry-atb2 cells were grown at 24°C till mid-log phase, fixed with methanol and the localization of Csc1p imaged. C. Csc1p localization depends on entry into mitosis and the spindle pole body component Ppc89p, but does not require microtubules and SIN activity. cdc25-22 cells and sid4-SA1 cells expressing Csc1p-GFP and mCherry-Atb2p were grown at 24°C, shifted to the semi-permissive temperature of 33°C for 5 hours and imaged for the two fluorescent proteins. nda3-KM311 cells expressing Csc1p-GFP and mCherry-Atb2p were grown at 30°C, shifted to the non-permissive temperature of 19°C and imaged for the two fluorescent proteins. nmt81-ppc89 cells expressing Csc1p-GFP and mCherry-Atb2p were grown in EMM medium containing thiamine for 36 hours. The cells were then fixed with methanol imaged and for the two fluorescent proteins.(n=50×3) D. Csc1p shows a transient asymmetric localization to one SPB. cdc25-22 cells expressing Csc1p-GFP and mCherry-Atb2p were arrested at G2 by incubation at 33°C for 3 hours. The cells were then released at 24°C and grown for 1 hour. Cells were fixed with methanol and the localization of Csc1p and microtubules imaged. The percentage of cells with asymmetric distribution of Csc1p is shown at the bottom of the figure (n=26). E–G. Csc1p and Cdc7p segregate to opposite SPBs. E. cdc25-22 cells expressing Csc1p-GFP and Cdc7p-mCherry were grown at the permissive temperature and fixed with methanol. The fluorescence patterns of Csc1p and Cdc7p were scored. Cells that showed signals of both Csc1p and Cdc7p were imaged to analyze the localization relationship of the two proteins at SPB(s). The percentage of cells showing symmetric distribution (top cell) and asymmetric distribution (bottom two cells) of Cdc7p and Csc1p are indicated (n=50×3). F. Csc1p-GFP asymmetry is observed in wild-type cells. Wild-type cells expressing Csc1p-GFP and mCherry-Atb2p were grown at 24°C till mid-log phase, fixed with methanol and scored for the symmetric or asymmetric distribution of Csc1p in cells with short spindles (n=50×3). G. cdc25-22 cells expressing Csc1p-GFP and Cdc7p-mCherry were grown at the permissive temperature and imaged by time-lapse microscopy. Elapsed time (in minutes) after start of the imaging is indicated above the panels. The arrows indicate positions when asymmetry of Cdc7p and Csc1p are clearly apparent.
To further analyze the localization of Csc1p, we generated a Csc1p-GFP (green fluorescent protein) expressing strain. We deemed the Csc1p-GFP to be fully functional based on criteria described in a subsequent section. Csc1p-GFP was detected at both SPBs in early mitotic cells (Figure 1B), but was not clearly detected on either of the SPBs following initiation of anaphase B (data not shown). We utilized several conditional mutants to further analyze its localization. Csc1p was not detected on the SPBs in G2 arrested cdc25-22 cells [37] (Figure 1C), but was detected on the non-separated SPBs in prometaphase arrested nda3-KM311 [38] cells lacking a mitotic spindle (Figure 1C). Localization of Csc1p to the SPBs did not require signaling through the SIN, since Csc1p localization was unaffected in SIN scaffold mutants-cdc11-123, sid4-SA1, and the downstream kinase mutant sid2-250 (Figure 1C; data not shown for cdc11-123 and sid2-250). It has been shown previously that SIN components require the SPB resident protein Ppc89p for their localization to the SPB [39]. We therefore tested if Ppc89p was required for the SPB localization of Csc1p. To this end, Csc1p-GFP was expressed in a yeast strain in which the ppc89 gene was placed under control of the thiamine repressible nmt81 promoter. In this strain, upon growth in repressing conditions, Csc1p-GFP failed to localize to the SPBs in approximately 37% of the cells (Figure 1C). In other experiments, we found that the localization of a Csc1p interacting protein (Csc3p; described in a later section) was fully abolished in germinated spores deleted for ppc89, establishing that the localization of Csc1p (and associated factors) to the SPB depended on Ppc89p. Collectively, these experiments established that Csc1p localized to the SPB early in mitosis in a Ppc89p-dependent manner and exited the SPB upon anaphase progression.
Csc1p becomes asymmetrically localized transiently to the mother SPB not containing Cdc7p
In some cells Csc1p displayed a slight asymmetry in localization before disappearing from both SPBs. To address if Csc1p indeed exhibited a transient asymmetric localization at the SPBs we studied its localization in synchronous cdc25-22 mutant cells, which undergo a prolonged mitosis with a mitotic spindle nearly two and a half times longer than that in wild-type cells. From this analysis it was apparent that approximately 54% cells showed unequal distribution of Csc1p to one of the two SPBs (Figure 1D). Since several components of the SIN are known to localize to one of the two SPBs [14, 15, 17, 22], we tested if Csc1p levels were higher on the new SPB containing the SIN effector kinases Cdc7p and Sid1p [31], or on the old SPB that does not contain these effector kinases. To investigate this, we generated a strain expressing Csc1p-GFP and Cdc7p-mCherry. Csc1p was detected in two patterns: in ~ 57% of cells Csc1p signal was of equal intensity on both SPBs and in ~ 43 % of cells Csc1p signal was weaker or absent on one SPB and stronger on the other SPB. In cells containing Csc1p signals of equal intensity, Cdc7p also localized to both SPBs with equal intensity (Figure 1E). Interestingly, when Csc1p displayed unequal localization, Cdc7p signal intensity was reciprocal to that of Csc1p (i.e. if Csc1p levels were high on one SPB, Cdc7p levels were low on that SPB and vice versa).
Since many of our studies utilized the cdc25-22 mutant, albeit at permissive temperature, it was possible that the asymmetry was an artifact of the cdc25-22 mutation. To rule out this possibility we investigated the localization of Csc1p in otherwise wild-type cells. Consistent with our previous studies, Csc1p was asymmetrically localized in approximately 54% of cells with a short mitotic spindle (Figure 1F; 2 cells in the bottom; short spindle defined as those less than 5 μm in size), whereas in the rest of cells with short spindles Csc1p was symmetric in localization (Figure 1F; 1 cell on top). Based on these results, we conclude that Csc1p localizes to early mitotic SPBs and then is redistributed at the onset of anaphase such that Csc1p and Cdc7p segregate to opposite SPBs.
We performed time-lapse analysis of the distribution of Csc1p-GFP and Cdc7-mCherry to gain better insight into the dynamics of these proteins in relation to each other. Time-lapse imaging of > 10 cells expressing Csc1p-GFP and Cdc7p-mCherry further confirmed that Csc1p-GFP and Cdc7p-mCherry segregate to opposite SPBs (Figure 1G; note at time-points 6' and 8', indicated with an arrow, when asymmetry of Csc1p and Cdc7p are established and propagated). Interestingly, we found in these studies that Csc1p was fully lost from the SPBs only when the mitotic spindle was approximately 7–8 μm in length (Figure 1G; time points 8' and 10').
Our analysis of fixed cells revealed that Cdc7p and Csc1p asymmetry was apparent in cells with short spindles, although complete loss of Csc1p from the SPB occurred in the mid-anaphase B. The mitotic B-type cyclin Cdc13p degradation (and cyclin dependent kinase inactivation) is completed only in the middle of anaphase B [26, 40]. Therefore we considered the possibility that removal of Csc1p from the SPB might be initiated at onset of cyclin B proteolysis and completed upon full degradation of Cdc13p. We could not test the relationship between cyclin-dependent-kinase inactivation and Csc1p localization to the SPB since Csc1p-GFP fluorescence was dampened at 36°C, the restrictive temperature for cdc13-117 [41] mutants. However, the localization of a Csc1p-associated protein (Csc3p, described in a later section) to the SPB was lost when metaphase arrested cdc13-117 cells were shifted to the restrictive temperature to inactivate CDK-function (Figure S1A; metaphase arrest was achieved by overproduction of the mitotic checkpoint protein Mad2p in cdc13-117 cells cultured at the permissive temperature). Furthermore, Csc1p was detected at the SPB in a high proportion of cells with elongated mitotic spindle in cells expressing a Cdc13Δ81, a non-degradable version of Cdc13p (Figure S1B). These experiments strongly suggested that the removal of Csc1p and associated factors from the SPB was linked with cyclin B degradation.
Csc1p is a new negative regulator of the SIN
To check if csc1 functioned in cytokinesis, we generated a gene-deletion strain and investigated the phenotype. In wild type cells, septum deposition was seen only in binucleated cells that had completed mitosis. However, in cells deleted for csc1 a low proportion of uninucleate cells displayed division septa (Figure 2A; <1% of n>500). In addition, binucleate cells with more than one septum were also detected (Figure 2A). The presence of uninucleate cells with a septum and binucleate cells with more than one septum suggested that Csc1p might participate in inhibiting the SIN. Previous studies have shown that the inactivation of SIN inhibitors leads to septation even in interphase [21, 24, 27]. Consistent with Csc1p participating as an inhibitor of the SIN, 4–6% of S phase arrested csc1Δ cells contained a division septum (Figure 2B). The lower percentage of uninucleate cells with a division septum suggested that Csc1p participated in, but was not essential for, SIN inactivation.
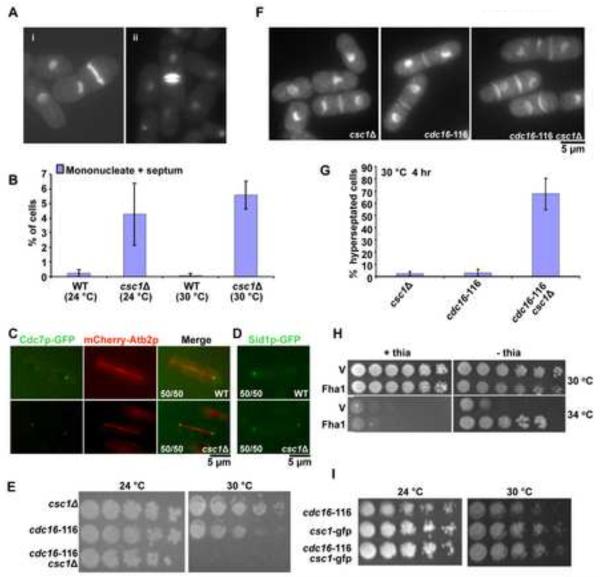
Figure 2. Csc1p promotes asymmetric localization of SIN-kinases Cdc7p and Sid1p.
A. csc1Δ generates uninucleate cells with a division septum and binucleate cells with more than one septum. csc1Δ cells were grown at 30°C, fixed, and stained with DAPI (for DNA) and aniline blue (for cell wall). B. Loss of Csc1p function can cause inappropriate septation in interphase. Wild type (WT) and csc1Δ cells were grown at 24°C or 30°C, treated with 12 mM HU for 6 hrs to arrest the cells at S-phase. Interphase cells with a division septum were quantitated (n=500×3). Cells with septum deposition were counted for WT as well as csc1Δ. C–D. Cdc7p and Sid1p localize symmetrically in csc1Δ cells. Wild type cells expressing Cdc7p OR Sid1p-GFP and mCherry Atb2p were fixed with methanol and the localization of the fluorescent proteins investigated. The fraction of cells displaying Cdc7p or Sid1p in an asymmetric configuration or the fraction of csc1Δ cells displaying a symmetric distribution of Cdc7p or Sid1p are indicated in the figure. E. Simultaneous compromise of Csc1p and Cdc16p function leads to growth defects. csc1Δ, cdc16-116 and csc1Δ cdc16-116 cultures were serially diluted 5 fold for 4 times and spotted on YES-agar medium and grown at 24°C and 30°C. The two parental strains were capable of colony formation at 24°C and 30°C. The csc1Δ cdc16-116 double mutant was capable of colony formation at 24°C, but not at 30°C. F. Simultaneous compromise of Csc1p and Cdc16p generates cells with multiple septa. csc1Δ, cdc16-116 and csc1Δ cdc16-116 mutants were grown at 24°C till early log phase, shifted to 30°C for 3 hours, fixed and stained with DAPI and aniline blue. G. Graphical representation of the percentage of hyperseptated cells in strains described F (n=500×3). H. Rescue of cdc16-116 growth defect with Csc1p overexpression (v-pREP41-GST and csc1-pREP41-csc1-GST). I. Csc1p-GFP cells do not display synthetic lethality with cdc16-116. csc1-GFP, cdc16-116 and csc1-GFP cdc16-116 cultures were serially diluted 5 fold and spotted on YES-agar plates and incubated at 24°C and 30°C.
SIN involves a linear cascade of three kinases - Cdc7p, Sid1p and Sid2p, in the order of their activation [6, 14–16]. Of these, Cdc7p and Sid1p localize only to the new SPB late in anaphase [17, 32]. Constitutive activation of the SIN, by the inactivation of inhibitors of the SIN, leads to localization of Cdc7p and Sid1p to both SPBs in mitotic cells and to the single SPB in interphase cells [17, 32]. Since deletion of csc1 led to phenotypes consistent with weak SIN activation we checked the localization of Cdc7p and Sid1p in csc1Δ cells (Figure 2C and D). While Cdc7p and Sid1p were only detected on one SPB in late anaphase in wild-type cells, Cdc7p and Sid1p were found on both SPBs in 100% of late anaphase csc1Δ cells (Figure 2C and D). These experiments establish that Csc1p is essential for the asymmetric localization of Cdc7p and Sid1p.
Previous work has shown that inactivation of SIN (using loss of Cdc7p from the SPB as an assay) occurs in coordination with actomyosin ring closure and division of the cytoplasm [28]. Since Cdc7p was detected on both SPBs in csc1Δ cells, but was not detected on interphase SPBs, we tested if loss of Cdc7p from the SPBs coincided with full constriction of the actomyosin ring. As in wild-type cells, Cdc7p signals were lost from both the SPBs before or upon completion of actomyosin ring constriction and septum assembly and did not persist after septation (Figure S2; approximately after time-point 60'). Curiously, we found that Cdc7p was not lost simultaneously from both SPBs, but was lost from one SPB first, followed by its loss from the other. Nevertheless, these experiments further substantiate the view that division of the cytoplasm promotes the removal of Cdc7p from the SPB.
To assess if Csc1p functioned in concert with the two-component GTPase activating protein (Cdc16p-Byr4p) [21, 33, 42] for Spg1p we generated double mutants defective in cdc16 and csc1. While each of the single mutants was capable of colony formation at the semi-permissive temperature of 30°C, the double mutant was unable to do so (Figure 2E) and died due to hyperactivation of the SIN (> 80% of cells displaying multiple septa; Figure 2F and G). The synthetic genetic interaction suggested that Csc1p function was crucial when the major inhibitors of SIN were even partially compromised.
Previous work has shown that over production of many negative regulators of the SIN, such as Byr4p, Zfs1p, and Dma1p, causes rescue of cdc16-116 mutants [23, 43, 44]. In light of this, we tested if over production of Csc1p, a new negative regulator of SIN, could suppress cdc16-116. A plasmid capable of expressing high levels of Csc1p and a control plasmid were introduced into cdc16-116 cells. We noticed that, consistent with the role of Csc1p in inhibition of SIN, overproduction of Csc1p allowed cdc16-116 to form colonies under conditions in which the control plasmid expressing cells were unable to do so (Figure 2H).
The discovery of a phenotype associated with csc1Δ allowed us to test if Csc1p-GFP was a functional version of this protein. Synthetic lethality was not detected in cdc16-116 csc1-GFP strains (Figure 2I). Moreover, since Cdc7p asymmetry was not compromised in Csc1p-GFP expressing cells (Figure 1G), we concluded that Csc1p-GFP was fully functional in our assay conditions.
Csc1p is present in a multi protein complex containing a new catalytic subunit of protein phosphatase 2A, Ppa3p
To gain biochemical insights into Csc1p functions, we carried out an immunoaffinity purification of Csc1p-Myc13 (Figure S3), in which four major Coomassie stained bands were detected. Proteins that associated with Myc13-beads were identified by tandem mass spectrometry. In this analysis, we identified 4 additional proteins, Csc2p (SPBC27B12.04c), Csc3p (SPBC1773.02), Csc4p (SPAC2C4.10c), and the scaffold A-subunit of PP2A, Paa1p [45]. Csc2p is most related to the budding yeast Far11p [46], Csc3p is most related to budding yeast Far8p [46], and Csc4p is an uncharacterized protein with no known orthologs. Csc2p contains the DUF3402 domain, while Csc3p is related to the conserved striatin-family of proteins [47, 48]. To confirm the physical interactions, lysates extracted from cells expressing Csc1p-Myc13 and one of the following proteins - Glutathine S-transferase (GST), Csc2p-GST, Csc3p-GST, or Paa1p-GST were incubated with glutathione-sepharose. Csc1p was not detected in pull-downs from cells expressing GST alone, but was detected in pull-downs from cells expressing Csc2p-GST, Csc3p-GST and Paa1p-GST (Figure 3A). We purified Csc3p-TAP and Paa1p-TAP, and used LC-MS/MS to identify proteins interacting with these proteins (Table S1). In both cases, Csc1p, Csc2p, and Csc4p were identified. In addition, Csc3p was detected in the Paa1p-TAP complex and Paa1p was detected in the Csc3p-TAP complex. Although a PP2A catalytic subunit was not detected in our original Csc1p purification, a novel catalytic subunit of PP2A (which we have termed Ppa3p; ORF SPAC22H10.04) was identified amongst proteins co-purifying with Csc3p-TAP and Paa1p-TAP. These experiments established that Csc1p physically associated with Csc2p, Csc3p, Csc4p, Paa1p, and Ppa3p.
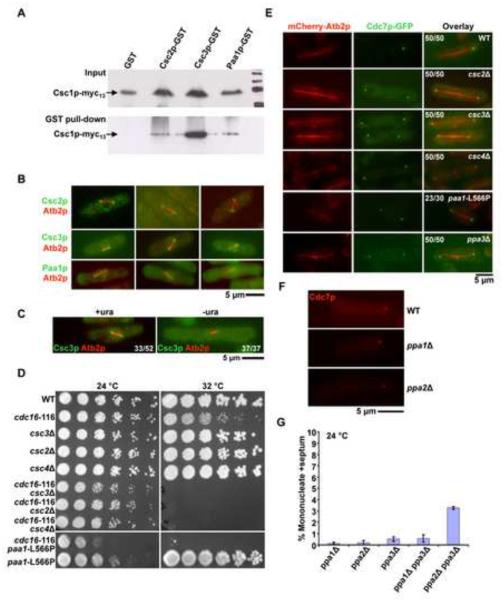
Figure 3. Identification of a multiprotein complex (SIP) required for generation of Cdc7p asymmetry.
A. Csc2p, Csc3p, and Paa1p bind Csc1p-Myc13. GST, Csc3-GST, Csc2-GST and Paa1-GST were isolated from yeast cells expressing Csc1p and one of these proteins using glutathione linked agarose beads. The isolated proteins were immunoblotted with an antibody against the Myc-epitope. The top panel shows the total lysates probed with the anti-Myc antibody, whereas the bottom panel shows an immunoblot of the proteins isolated from glutathione-agarose beads and probed with the anti-Myc antibody. B. Csc2p, Csc3p, and Paa1p localize to early mitotic SPBs. Cells individually mCherry-Atb2p and one of the following fusion proteins, Csc2p-GFP, Csc3p-GFP and Paa1p-GFP were grown at 24°C and the localization patterns of the fluorescent proteins investigated. Shown are 3 examples in each case, of cells with short mitotic spindles. C. The SPB component Ppc89p is required for Csc3p localization to SPB. Spores from a ppc89::ura4+/ppc89+ strain expressing Csc3p-GFP were germinated in minimal media in the presence or absence of uracil. The cells were then observed for the localization of Csc3p during early mitosis. The proportion of spores germinated in medium containing uracil and exhibiting and exhibiting a short spindle and Csc3p on SPBs is indicated in the figure. In addition, the proportion of spores germinated in the absence of uracil and exhibiting a short spindle and Csc3p on the SPB is indicated in the figure. D. csc2Δ, csc3Δ, csc4Δ, and paa1-L566P display synthetic lethal interactions with cdc16-116. Serial dilutions of csc2Δ cdc16-116, csc3Δ cdc16-116, csc4Δ cdc16-116 and paa1-L566P cdc16-116 and the corresponding single mutants were spotted on YES-agar medium and incubated at 24°C or at 30°C. E. Csc2p, Csc3p, Csc4p, Paa1p, and Ppa3p are required for the generation / maintenance of Cdc7p asymmetry. csc2Δ, csc3Δ, csc4Δ, and ppa3Δ cells expressing Cdc7p-GFP and mCherry-Atb2p were grown at 24°C, fixed with methanol and imaged for Cdc7p localization during late anaphase. paa1-L566P cells expressing Cdc7p-GFP and mCherry-Atb2p were grown at 24°C and then shifted to 36°C and the localization of Cdc7p during late anaphase investigated. The proportion of cells exhibiting symmetric localization of Cdc7p in the various mutants and the proportion of wild-type cells showing asymmetric distribution of Cdc7p are indicated. F. Cdc7p asymmetry is not compromised in the absence of Ppa1p and Ppa2p. Wild-type, ppa1Δ, and ppa2Δ cells expressing mCherry-Cdc7p were grown at 24°C and the localization of Cdc7p during late anaphase investigated. G. Combined loss of Ppa2p and Ppa3p leads to a modest increase in uninucleate cells with division septum. ppa1Δ, ppa2Δ, ppa3Δ, ppa1Δ ppa3Δ, and ppa2Δ ppa3Δ cells were cultured in the presence of 12 mM hydroxyurea and the proportion of uninucleate cells with a division septum estimated (n=500×3).
We then investigated the cellular localization of these Csc1p-associated proteins using strains expressing Csc2p, Csc3p, Csc4p, Paa1p, and Ppa3p as GFP fusions under the control of endogenous regulatory sequences. Signals of Ppa3p and Csc4p were undetectable (data not shown). However, we were able to visualize the other four proteins. Csc2p and Csc3p, like Csc1p, were detected in early mitotic cells on both SPBs (Figure 3B). In addition, Csc2p and Csc3p were also detected around the nuclear envelope (Figure 3B). In approximately 25–30 % of cells with short spindles Csc2p and Csc3p segregated asymmetrically to the SPB containing less Cdc7p (Figure S4) and in approximately 40–50% of cells Csc2p and Csc3p signals were of equal intensity on both SPBs (Figure S4). We were unable to ascertain the fate of Csc2p and Csc3p in the rest of the cells due to the strong nuclear envelope fluorescence of Csc2p and Csc3p (data not shown). Paa1p was weakly detected on mitotic SPBs, and was also detected at the actomyosin ring late in mitosis (Figure 3B). The localization experiments therefore established that, Csc2p, Csc3p, and Paa1p colocalized significantly during early mitosis and that Csc2p and Csc3p exhibited asymmetric segregation to the SPB containing less Cdc7p.
We have shown that Csc1p required Ppc89p to localize to the SPB (Figure 1C). We then tested if other Csc1p-interactors also depended on Ppc89p to localize to the SPB. To this end, we investigated the localization of Csc3p in ppc89 mutants. Csc3p localized to the SPB in 33/52 mitotic cells (Figure 3C) from the diploid ppc89Δ/ppc89+, upon spore germination in medium containing uracil (which allows germination of ppc89+ and ppc89Δ spores). However, Csc3p failed to localize to the SPB in 37/37 cases (Figure 3C) when spores were germinated in medium lacking uracil (which selectively allows germination of ppc89Δ spores). Thus Csc3p, like Csc1p, requires functional Ppc89p to localize to the SPB.
Next, we tested if Csc2p, Csc3p, Csc4p, Paa1p, and Ppa3p functioned as inhibitors of the SIN. Towards this goal, we generated strains deleted for the csc2, csc3, csc4, and ppa3 genes. All four genes were found to be dispensable for cell growth. Since Paa1p is essential for cell viability [45] we generated a temperature-sensitive mutant paa1-L566P, which was capable of colony formation at 24°C and weakly at 32°C, but died at 36°C. As in the case of csc1Δ cdc16-116 double mutants, double mutants of the genotypes cdc16-116 csc2Δ, cdc16-116 csc3Δ, cdc16-116 csc4Δ, and cdc16-116 paa1-L566P were all unable to grow and form colonies under conditions in which the parental single mutants were capable of colony formation (Figure 3D). Furthermore, as in csc1Δ cells, Cdc7p localized to both SPBs in 100% of late mitotic csc2Δ, csc3Δ, csc4Δ, paa1-L566P, and ppa3Δ (Figure 3E). Double mutants (in all possible pairwise combinations) defective in two of the six members of the SIP were generated. These double mutants did not display significant additive effects (Table S2). In addition, loss of function of any of these genes led to a failure of localization of each of the members of the multiprotein complex to the SPB (Table S3). Collectively, the biochemical, genetic, and cytological analyses establish that Csc1p functioned in a PP2A-associated multiprotein complex containing at least 5 other proteins-Csc2p, Csc3p, Csc4p, Paa1p, and Ppa3p-which functions as a weak inhibitor of the SIN and promotes SIN asymmetry. We refer to this complex as the SIN-inhibitory-PP2A-containing complex (SIP).
Having identified Ppa3p, a new PP2A catalytic subunit, as a member of the SIP we tested if the known catalytic subunits Ppa1p and Ppa2p also played a role in the generation of Cdc7p asymmetry. As in wild-type cells, Cdc7p localized to only one SPB in ppa1Δ and ppa2Δ mutants (Figure 3F), suggesting that Ppa1p and Ppa2p do not directly participate in the generation of Cdc7p asymmetry. However, it is possible that Ppa2p might play a minor role in SIN inactivation, since ppa2Δ ppa3Δ double mutants, but not ppa1Δ ppa3Δ double mutants, showed a modest increase in the number of septated interphase cells (Figure 3G).
SIP promotes dephosphorylation of Cdc11p
In the course of experiments aimed at studying potential genetic interactions between SIP mutants and SIN mutants, we found that the inability of cdc11-136 [49] to form colonies at 36°C was reversed by the introduction of csc1Δ mutation (Figure 4A). Unlike in the cdc11-136 single mutants, septa of normal appearance were detected in cdc11-136 csc1Δ (Figure 4B). Furthermore, the percentage of cells with either multiple nuclei or two clustered nuclei (denoted SIN inactive in Figure 4C) were dramatically reduced in cdc11-136 csc1Δ (Figure 4C). A similar suppression of the temperature sensitive lethality of cdc11-136 was also observed in cdc11-136 csc4Δ double mutants (Figure 4A–C). Furthermore, csc2 and csc3 mutants also suppressed cdc11-136 mutants, although the efficiency was reduced compared to the suppression by csc1 and csc4 (data not shown). These observations suggested that SIP might function in the generation of Cdc7p asymmetry by modulating Cdc11p function.
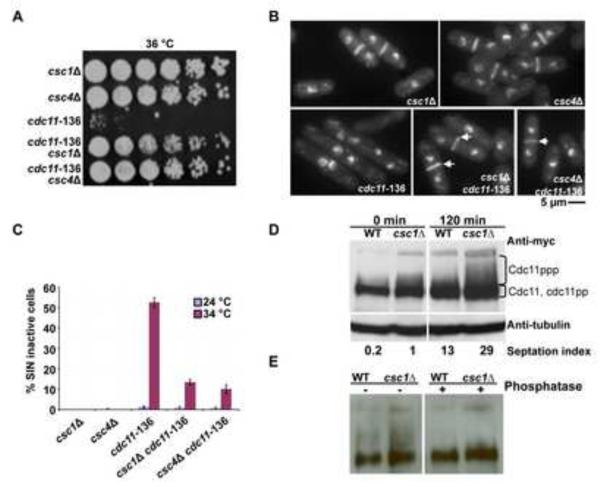
Figure 4. SIP complex interacts physically with the SIN scaffold protein Cdc11p and promotes dephosphorylation of Cdc11p.
A. Restoration of colony formation ability of cdc11-136 upon deletion of csc1 or csc4 genes. Overnight cultures of csc1Δ, csc4Δ, cdc11-136, csc1Δ cdc11-136, and csc4Δ cdc11-136 grown at 24°C were serially diluted 5 fold for 5 times and spotted on YES-agar plates and incubated at 36°C. B. cdc11-136 csc1Δ and cdc11-136 csc4Δ cells assemble normal-appearing septa. csc1Δ, csc4Δ, cdc11-136, csc1Δ cdc11-136, and csc4Δ cdc11-136 cells were grown at 24°C and shifted to 36°C. The cells were fixed and stained with aniline blue (cell wall) and DAPI (DNA). C. The percentage of multinucleate cells in all strains described in B. were estimated and plotted to gain a measure of the extent of suppression of cdc11-136 by loss of function of csc1 or csc4 (n=500×3). D. Cdc11p extracted from csc1Δ cells migrates slower than Cdc11p from wild type cells. cdc11-myc13 and cdc11-myc13 csc1Δ were arrested at S-phase using 12 mM hydroxyurea for four hours. The cells were released by washing three times with YES and samples were collected at indicated times. Cell extracts were resolved on 8% SDS-PAGE and immunoblot analysis was carried out using an anti-Myc antibody. E. SIP promotes dephosphorylation of Cdc11p. nda3-KM311 cdc11-myc13 and nda3-KM311 cdc11-myc13 csc1Δ cells were arrested at prometaphase by shift to 18°C for 4 hours and released to 32°C. The phosphorylation state of Cdc11p was then analysed after 50 min of release by immunoblot analysis using anti-Myc antibody. Cell lysates were prepared and immunoblotted with antibodies against the Myc-epitope (left panel). To establish that the slower migration observed in the csc1Δ cells was a result of increased phosphorylation of Cdc11p, lysates were treated with Lambda phosphatase (right panel).
Previous work has shown that Cdc11p becomes phosphorylated during progression through mitosis and PP2A is required for Cdc11p dephosphorylation [50]. We therefore considered the possibility that SIP might promote dephosphorylation of Cdc11p, thereby negatively regulating SIN. To address this possibility, we generated hydroxyurea synchronized cultures of wild-type and csc1Δ. As shown by other investigators, a slower migrating smear of Cdc11p appeared with progression through mitosis in wild-type cells (Figure 4D). Importantly, the intensity of the slower migrating smear of Cdc11p was increased in csc1Δ cells (Figure 4D). We also confirmed decreased mobility and increased smear of Cdc11p in csc1Δ cells synchronized using a metaphase block-and-release using the nda3-KM311 mutation. Since the smearing was lost upon treatment with phosphatase treatment (Figure 4E), we concluded that the smear pattern of was a result of an increased phosphorylation of Cdc11p and that Csc1p (and the SIP) participated in the dephosphorylation of Cdc11p.
Byr4p localization to the SPB is compromised in SIP-complex mutants
Byr4p, an essential component of the Spg1p GAP complex, has been shown to bind to dephosphorylated Cdc11p [50]. Since the SIP concentrates on the SPB lacking Cdc7p, we hypothesized that a potential function of SIP was to dephosphorylate Cdc11p, thereby promoting Byr4p re-localization to the SPB containing the increased amount of SIP. If this were the case, Byr4p should not be detected on either SPB during anaphase in SIP mutants. To address this issue, wild-type and csc1Δ-3xHA were fixed and stained with antibodies against HA and Byr4p. In late mitotic wild-type cells Cdc7p and Byr4p were detected on opposite SPBs (Figure 5A). By contrast, in csc1Δ cells Cdc7p was detected at both anaphase SPBs and importantly, Byr4p was not detected on either SPB in mitotic cells (Figure 5A). Byr4p was also undetectable at the SPB in csc3Δ, paa1-L566P, and ppa3Δ cells (Figure 5B). In wild-type cells Byr4p localizes to SPBs during interphase [50]. Curiously, Byr4p was readily detected on the SPB in uninucleate (interphase) wild-type, csc1Δ, csc3Δ, paa1-L566P, and ppa3Δ cells (see interphase cells in Figure 5 A and B), suggesting that the regulation of Byr4p localization by the SIP is specific to mitosis.
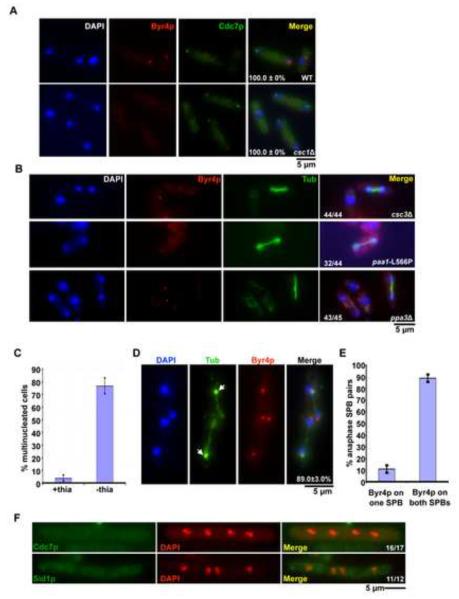
Figure 5. Modulation of SIP affects the localization of SIN and its regulators.
A, B. Byr4p localization to the mitotic SPB depends on SIP. A. Wild-type and csc1Δ cells expressing Cdc7p-3xHA were stained using antibodies against the HA-epitope and against Byr4p. After staining, cells were mounted in DAPI and images acquired (n=50×3). B. csc3Δ, ppa3Δ cells grown at 24°C and paa1-L566P cells grown at 36°C were stained with antibodies against tubulin and Byr4p, mounted in DAPI medium and images acquired. C. Overexpression of Csc1p leads to a failure in cytokinesis. Csc1p was overexpressed by growing nmt1-csc1-GFP cells in minimal medium lacking thiamine for 16 hours at 24°C, fixed and stained with aniline blue (cell wall) and DAPI (DNA). The percentage of multinucleated cells was then determined (n=500×3). D. Overproduction of Csc1p leads to recruitment of Byr4p to both SPBs in mitotic cells. nmt1-csc1-GFP cells were grown and fixed as in (C) above. The cells were then stained using antibodies against Byr4p and tubulin TAT-1 antibody. Arrows indicate an increased accumulation of GFP-Csc1p (n=50×3). E. Quantitation of the percentage of anaphase SPB pairs with Byr4p on one or both the SPBs (n=50×3). F. Cdc7p and Sid1p are displaced from mitotic SPBs upon Csc1p overexpression. nmt1-csc1-GFP expressing Cdc7p-mCherry were grown in EMM medium lacking thiamine for 16 hours at 24°C, fixed with methanol and imaged. To visualize Sid1p, nmt1-csc1-GFP cells expressing Sid1p-Myc13 were stained with antibodies against the Myc-epitope.
If SIP plays a role in recruiting Byr4p to the SPB, it would be expected that overproduction of Csc1p and/or other members of the SIP might cause inappropriate recruitment of Byr4p to the SPB leading to failure of cytokinesis. To this end, Csc1p-GFP was overexpressed in wild-type cells. Under inducing conditions nearly 75% of the cells became multinucleated (Figure 5C). Furthermore, unlike in wild-type cells, Byr4p was recruited to each of the two SPBs connecting elongated anaphase B spindles in more than eighty five percent of the cells with 2 mitotic spindles (Figure 5D and 5E). Conversely, in Csc1p overexpressing cells, Cdc7p and Sid1p, failed to localize to any of the SPBs in mitotic cells with 4 nuclei (Figure 5F). Sid2p localization was unaltered upon Csc1p over production (data not shown). We conclude that excess Csc1p promoted inappropriate retention of Byr4p at SPBs into anaphase and failure of retention of Cdc7p and Sid1p at the SPBs.
DISCUSSION
In this study we have identified a SIN-inhibitory phosphatase complex that we have termed SIP. SIP contains at least 6 proteins in unknown relative stoichiometry, including a new catalytic subunit of PP2A (Ppa3p), the A-subunit of PP2A (Paa1p) [45], 3 proteins related to the budding yeast proteins Far8p (Csc3p), Far10p (Csc1p), and Far11p (Csc2p) [46], and a novel protein Csc4p. Four out of the six components of the SIP localize to the SPB, while we were unable to determine the localization patterns of the other two through introduction of a C-terminal GFP tag. Interestingly, Csc1p and Csc3p depended on Ppc89p rather than the known SIN-scaffolds Cdc11p and Sid4p to localize to the SPB. As expected for a protein complex, the SPB localization of each SIP component depends on all other SIP components and double SIP mutants displayed phenotypes similar to single mutants, suggesting that the SIP functions as a single unit to regulate the SIN.
SIP localizes to both SPBs in early mitosis and then transiently asymmetrically localizes to the old SPB that would contain Byr4p (i.e. the SPB that loses Cdc7p and Sid1p), before disappearing from both SPBs prior to full elongation of the mitotic spindle. These observations suggest a role for SIP in the regulation of SIN asymmetry or function. We have found that loss of SIP function led to a weak SIN-activated phenotype, in that binucleate cells with 2 or more septa or uninucleate cells with septa were detected. Based on these observations we conclude that SIP functions as a negative regulator of the SIN consistent with previous genetic studies demonstrating suppression of SIN mutants by other PP2A mutants [51–53].
Loss of SIP function leads to symmetric localization of Cdc7p and Sid1p to both SPBs in 100% of mitotic cells. Despite the presence of Cdc7p and Sid1p on both SPBs in mitotic cells defective in SIP function, mutants defective in SIP are viable, unlike cdc16-116 and byr4Δ mutants [21, 24], in which Cdc7p has also been shown to localize to both SPBs. The fact that Cdc7p leaves both SPBs upon or before completion of septation in SIP mutants and the presence of a mechanism to prevent localization of Cdc7p to the SPBs in interphase SIP mutant cells, might account for the viability of SIP-mutants. It is likely that localization of Cdc7p and Sid1p to both SPBs may only activate SIN moderately over the wild-type levels (as gauged by a modest but reproducible increase in septated cells). By contrast, continued maintenance of Cdc7p and Sid1p on the SPB following completion of septation and in subsequent interphase might lead to hyperactivation of the SIN and uncontrolled septation.
Byr4p fails to localize to the SPB during mitosis in SIP mutants. However, Byr4p localization to the SPB during interphase was not compromised in SIP mutants. These observations indicate that distinct mechanisms regulate the SPB-localization of Byr4p during these different cell cycle stages. We have shown that over production of Csc1p leads to cytokinesis defects akin to that observed in SIN-mutants, in that Cdc7p and Sid1p fail to localize to mitotic SPBs. It is possible that Csc1p might function as a key member of the SIP, whose presence at the SPB might lead to recruitment of other members of the SIP to the SPB.
How does SIP regulate Cdc7p (and Sid1p-Cdc14p) asymmetry? Our studies, in conjunction with studies of Krapp and colleagues [50] provide a molecular framework for the first steps in the establishment and propagation of asymmetry in Cdc7p during mitosis and cytokinesis, and a mechanistic explanation for the ability of some PP2A mutants to suppress SIN mutants [51–53]. It is likely that the establishment and maintenance of an asymmetric state of Cdc7p (and Sid1p localization) [15, 32] might operate in multiple steps, with the innate asymmetry of the age of the spindle pole body [31] potentially playing an important role. We believe that entry into mitosis and cyclin-dependent kinase activation leads to the symmetric localization of Cdc7p [32] and the SIP. It is possible that the SPB scaffold Cdc11p might be more accessible for dephosphorylation by the SIP at the old SPB. Since Byr4p binds preferentially to dephosphorylated Cdc11p [50], it is likely that the decreased phosphorylation of Cdc11p might lead to further recruitment of Byr4p from the newer SPB to the older SPB. The fact that Cdc11p phosphorylation is increased in par1 [50] and csc1 mutants suggests that SIP (via PP2A catalytic activity) dephosphorylates Cdc11p, leading to loss of Cdc7p from the old SPB and the recruitment of Byr4p to it. Our finding that Byr4p fails to localize to either of the SPBs in csc1∆, csc3Δ, paa1-L566P, and ppa3Δ cells and that Byr4p localizes symmetrically in late mitotic cells overexpressing Csc1p, is consistent with this view. As Cdc7p departs from the old SPB, further phosphorylation of Cdc11p on the new SPB might further reinforce Cdc7p at the new SPB leading to a stable asymmetric state. A key question for the future is to understand how the age of the SPB promotes and/or inhibits phosphorylation events that are key to symmetry breaking.
Proteins related to 5 of 6 SIP members are conserved across evolution and are found from yeasts to human. A SIN-like network (termed the mitotic exit network: MEN) is also known to exist in budding yeast [3, 54]. Since members of the MEN localize asymmetrically to one of the two mitotic SPBs, future studies should test if SIP-like proteins participate in the generation or propagation of MEN asymmetry in budding yeast. Interestingly, in animal cells a protein complex termed the STRIPAK [55](related to the SIP) has been shown to negatively regulate the Hippo-Lats pathway [10](members of which are related to those of the SIN), although the mechanism of this negative regulation is not fully understood. Thus the mechanism of SIP function we describe in this study might be applicable to that of related protein complexes in other organisms.
MATERIALS AND METHODS
Materials and methods are described in detail in the supplemental material.
Supplementary Material
ACKNOWLEDGEMENTS
Many thanks are due to Drs. D. McCollum and M. Sato for yeast strains. We wish to thank members of Balasubramanian laboratory for helpful suggestions and Drs. S. Oliferenko, M. Mishra, R. Srinivasan, and Ms. D. Subramaniam for critical reading of the manuscript. This work was funded by funds from the Singapore Millennium Foundation (MKB) and the Howard Hughes Medical Institute, of which KLG is an investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Eggert US, Mitchison TJ, Field CM. Animal cytokinesis: from parts list to mechanisms. Annu Rev Biochem. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- 2.Pollard TD, Wu JQ. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balasubramanian MK, Bi E, Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr Biol. 2004;14:R806–818. doi: 10.1016/j.cub.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 4.Gould KL, Simanis V. The control of septum formation in fission yeast. Genes Dev. 1997;11:2939–2951. doi: 10.1101/gad.11.22.2939. [DOI] [PubMed] [Google Scholar]
- 5.Goyal A, Takaine M, Simanis V, Nakano K. Dividing the spoils of growth and the cell cycle: The fission yeast as a model for the study of cytokinesis. Cytoskeleton (Hoboken) 2011;68:69–88. doi: 10.1002/cm.20500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCollum D, Gould KL. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 2001;11:89–95. doi: 10.1016/s0962-8924(00)01901-2. [DOI] [PubMed] [Google Scholar]
- 7.Krapp A, Simanis V. An overview of the fission yeast septation initiation network (SIN) Biochem Soc Trans. 2008;36:411–415. doi: 10.1042/BST0360411. [DOI] [PubMed] [Google Scholar]
- 8.Guertin DA, Trautmann S, McCollum D. Cytokinesis in eukaryotes. Microbiol Mol Biol Rev. 2002;66:155–178. doi: 10.1128/MMBR.66.2.155-178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou MC, Salek J, McCollum D. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr Biol. 2000;10:619–622. doi: 10.1016/s0960-9822(00)00492-9. [DOI] [PubMed] [Google Scholar]
- 10.Ribeiro PS, Josue F, Wepf A, Wehr MC, Rinner O, Kelly G, Tapon N, Gstaiger M. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol Cell. 2010;39:521–534. doi: 10.1016/j.molcel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Tomlin GC, Morrell JL, Gould KL. The spindle pole body protein Cdc11p links Sid4p to the fission yeast septation initiation network. Mol Biol Cell. 2002;13:1203–1214. doi: 10.1091/mbc.01-09-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang L, Gould KL. Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc Natl Acad Sci U S A. 2000;97:5249–5254. doi: 10.1073/pnas.97.10.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehta S, Gould KL. Identification of functional domains within the septation initiation network kinase, Cdc7. J Biol Chem. 2006;281:9935–9941. doi: 10.1074/jbc.M600160200. [DOI] [PubMed] [Google Scholar]
- 14.Fankhauser C, Simanis V. The cdc7 protein kinase is a dosage dependent regulator of septum formation in fission yeast. Embo J. 1994;13:3011–3019. doi: 10.1002/j.1460-2075.1994.tb06600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guertin DA, Chang L, Irshad F, Gould KL, McCollum D. The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast. Embo J. 2000;19:1803–1815. doi: 10.1093/emboj/19.8.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sparks CA, Morphew M, McCollum D. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J Cell Biol. 1999;146:777–790. doi: 10.1083/jcb.146.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guertin DA, McCollum D. Interaction between the noncatalytic region of Sid1p kinase and Cdc14p is required for full catalytic activity and localization of Sid1p. J Biol Chem. 2001;276:28185–28189. doi: 10.1074/jbc.M103802200. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt S, Sohrmann M, Hofmann K, Woollard A, Simanis V. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 1997;11:1519–1534. doi: 10.1101/gad.11.12.1519. [DOI] [PubMed] [Google Scholar]
- 19.Krapp A, Schmidt S, Cano E, Simanis V. S. pombe cdc11p, together with sid4p, provides an anchor for septation initiation network proteins on the spindle pole body. Curr Biol. 2001;11:1559–1568. doi: 10.1016/s0960-9822(01)00478-x. [DOI] [PubMed] [Google Scholar]
- 20.Salimova E, Sohrmann M, Fournier N, Simanis V. The S. pombe orthologue of the S. cerevisiae mob1 gene is essential and functions in signalling the onset of septum formation. J Cell Sci. 2000;113(Pt 10):1695–1704. doi: 10.1242/jcs.113.10.1695. [DOI] [PubMed] [Google Scholar]
- 21.Minet M, Nurse P, Thuriaux P, Mitchison JM. Uncontrolled septation in a cell division cycle mutant of the fission yeast Schizosaccharomyces pombe. J Bacteriol. 1979;137:440–446. doi: 10.1128/jb.137.1.440-446.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fankhauser C, Marks J, Reymond A, Simanis V. The S. pombe cdc16 gene is required both for maintenance of p34cdc2 kinase activity and regulation of septum formation: a link between mitosis and cytokinesis? EMBO J. 1993;12:2697–2704. doi: 10.1002/j.1460-2075.1993.tb05931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furge KA, Wong K, Armstrong J, Balasubramanian M, Albright CF. Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr Biol. 1998;8:947–954. doi: 10.1016/s0960-9822(98)70394-x. [DOI] [PubMed] [Google Scholar]
- 24.Song K, Mach KE, Chen CY, Reynolds T, Albright CF. A novel suppressor of ras1 in fission yeast, byr4, is a dosage-dependent inhibitor of cytokinesis. J Cell Biol. 1996;133:1307–1319. doi: 10.1083/jcb.133.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C, Furge KA, Cheng QC, Albright CF. Byr4 localizes to spindle-pole bodies in a cell cycle-regulated manner to control Cdc7 localization and septation in fission yeast. J Biol Chem. 2000;275:14381–14387. doi: 10.1074/jbc.275.19.14381. [DOI] [PubMed] [Google Scholar]
- 26.Chang L, Morrell JL, Feoktistova A, Gould KL. Study of cyclin proteolysis in anaphase-promoting complex (APC) mutant cells reveals the requirement for APC function in the final steps of the fission yeast septation initiation network. Mol Cell Biol. 2001;21:6681–6694. doi: 10.1128/MCB.21.19.6681-6694.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krapp A, Collin P, Cano Del Rosario E, Simanis V. Homoeostasis between the GTPase Spg1p and its GAP in the regulation of cytokinesis in S. pombe. J Cell Sci. 2008;121:601–608. doi: 10.1242/jcs.022772. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Cortes JC, McCollum D. Proper timing of cytokinesis is regulated by Schizosaccharomyces pombe Etd1. J Cell Biol. 2009;186:739–753. doi: 10.1083/jcb.200902116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guertin DA, Venkatram S, Gould KL, McCollum D. Dma1 prevents mitotic exit and cytokinesis by inhibiting the septation initiation network (SIN) Dev Cell. 2002;3:779–790. doi: 10.1016/s1534-5807(02)00367-2. [DOI] [PubMed] [Google Scholar]
- 30.Morrell JL, Tomlin GC, Rajagopalan S, Venkatram S, Feoktistova AS, Tasto JJ, Mehta S, Jennings JL, Link A, Balasubramanian MK, et al. Sid4p-Cdc11p assembles the septation initiation network and its regulators at the S. pombe SPB. Curr Biol. 2004;14:579–584. doi: 10.1016/j.cub.2004.03.036. [DOI] [PubMed] [Google Scholar]
- 31.Grallert A, Krapp A, Bagley S, Simanis V, Hagan IM. Recruitment of NIMA kinase shows that maturation of the S. pombe spindle-pole body occurs over consecutive cell cycles and reveals a role for NIMA in modulating SIN activity. Genes Dev. 2004;18:1007–1021. doi: 10.1101/gad.296204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sohrmann M, Schmidt S, Hagan I, Simanis V. Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev. 1998;12:84–94. doi: 10.1101/gad.12.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerutti L, Simanis V. Asymmetry of the spindle pole bodies and spg1p GAP segregation during mitosis in fission yeast. J Cell Sci. 1999;112(Pt 14):2313–2321. doi: 10.1242/jcs.112.14.2313. [DOI] [PubMed] [Google Scholar]
- 34.Simanis V. Events at the end of mitosis in the budding and fission yeasts. J Cell Sci. 2003;116:4263–4275. doi: 10.1242/jcs.00807. [DOI] [PubMed] [Google Scholar]
- 35.Krapp A, Gulli MP, Simanis V. SIN and the art of splitting the fission yeast cell. Curr Biol. 2004;14:R722–730. doi: 10.1016/j.cub.2004.08.049. [DOI] [PubMed] [Google Scholar]
- 36.Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 37.Russell P, Nurse P. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 1986;45:145–153. doi: 10.1016/0092-8674(86)90546-5. [DOI] [PubMed] [Google Scholar]
- 38.Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- 39.Rosenberg JA, Tomlin GC, McDonald WH, Snydsman BE, Muller EG, Yates JR, 3rd, Gould KL. Ppc89 links multiple proteins, including the septation initiation network, to the core of the fission yeast spindle-pole body. Mol Biol Cell. 2006;17:3793–3805. doi: 10.1091/mbc.E06-01-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alfa CE, Booher R, Beach D, Hyams JS. Fission yeast cyclin: subcellular localisation and cell cycle regulation. J Cell Sci Suppl. 1989;12:9–19. doi: 10.1242/jcs.1989.supplement_12.2. [DOI] [PubMed] [Google Scholar]
- 41.Booher R, Beach D. Involvement of cdc13+ in mitotic control in Schizosaccharomyces pombe: possible interaction of the gene product with microtubules. EMBO J. 1988;7:2321–2327. doi: 10.1002/j.1460-2075.1988.tb03075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furge KA, Cheng QC, Jwa M, Shin S, Song K, Albright CF. Regions of Byr4, a regulator of septation in fission yeast, that bind Spg1 or Cdc16 and form a two-component GTPase-activating protein with Cdc16. J Biol Chem. 1999;274:11339–11343. doi: 10.1074/jbc.274.16.11339. [DOI] [PubMed] [Google Scholar]
- 43.Beltraminelli N, Murone M, Simanis V. The S. pombe zfs1 gene is required to prevent septation if mitotic progression is inhibited. J Cell Sci. 1999;112(Pt 18):3103–3114. doi: 10.1242/jcs.112.18.3103. [DOI] [PubMed] [Google Scholar]
- 44.Murone M, Simanis V. The fission yeast dma1 gene is a component of the spindle assembly checkpoint, required to prevent septum formation and premature exit from mitosis if spindle function is compromised. Embo J. 1996;15:6605–6616. [PMC free article] [PubMed] [Google Scholar]
- 45.Kinoshita K, Nemoto T, Nabeshima K, Kondoh H, Niwa H, Yanagida M. The regulatory subunits of fission yeast protein phosphatase 2A (PP2A) affect cell morphogenesis, cell wall synthesis and cytokinesis. Genes cells. 1996;1:29–45. doi: 10.1046/j.1365-2443.1996.02002.x. [DOI] [PubMed] [Google Scholar]
- 46.Kemp HA, Sprague GF., Jr. Far3 and five interacting proteins prevent premature recovery from pheromone arrest in the budding yeast Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:1750–1763. doi: 10.1128/MCB.23.5.1750-1763.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castets F, Bartoli M, Barnier JV, Baillat G, Salin P, Moqrich A, Bourgeois JP, Denizot F, Rougon G, Calothy G, et al. A novel calmodulin-binding protein, belonging to the WD-repeat family, is localized in dendrites of a subset of CNS neurons. J Cell Biol. 1996;134:1051–1062. doi: 10.1083/jcb.134.4.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moreno CS, Park S, Nelson K, Ashby D, Hubalek F, Lane WS, Pallas DC. WD40 repeat proteins striatin and S/G(2) nuclear autoantigen are members of a novel family of calmodulin-binding proteins that associate with protein phosphatase 2A. J Biol Chem. 2000;275:5257–5263. doi: 10.1074/jbc.275.8.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt S, Hofmann K, Simanis V. Sce3, a suppressor of the Schizosaccharomyces pombe septation mutant cdc11, encodes a putative RNA-binding protein. Nucleic Acids Res. 1997;25:3433–3439. doi: 10.1093/nar/25.17.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krapp A, Cano E, Simanis V. Mitotic hyperphosphorylation of the fission yeast SIN scaffold protein cdc11p is regulated by the protein kinase cdc7p. Curr Biol. 2003;13:168–172. doi: 10.1016/s0960-9822(02)01417-3. [DOI] [PubMed] [Google Scholar]
- 51.Jiang W, Hallberg RL. Correct regulation of the septation initiation network in Schizosaccharomyces pombe requires the activities of par1 and par2. Genetics. 2001;158:1413–1429. doi: 10.1093/genetics/158.4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lahoz A, Alcaide-Gavilan M, Daga RR, Jimenez J. Antagonistic roles of PP2A-Pab1 and Etd1 in the control of cytokinesis in fission yeast. Genetics. 186:1261–1270. doi: 10.1534/genetics.110.121368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Le Goff X, Buvelot S, Salimova E, Guerry F, Schmidt S, Cueille N, Cano E, Simanis V. The protein phosphatase 2A B'-regulatory subunit par1p is implicated in regulation of the S. pombe septation initiation network. FEBS Lett. 2001;508:136–142. doi: 10.1016/s0014-5793(01)03047-2. [DOI] [PubMed] [Google Scholar]
- 54.Seshan A, Amon A. Linked for life: temporal and spatial coordination of late mitotic events. Curr Opin Cell Biol. 2004;16:41–48. doi: 10.1016/j.ceb.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 55.Goudreault M, D'Ambrosio LM, Kean MJ, Mullin MJ, Larsen BG, Sanchez A, Chaudhry S, Chen GI, Sicheri F, Nesvizhskii AI, et al. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol Cell Proteomics. 2009;8:157–171. doi: 10.1074/mcp.M800266-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.