Abstract
Chromosomal instability (CIN) is associated with poor outcome in epithelial malignancies including breast carcinomas. Evidence suggests that prognostic signatures in estrogen receptor-positive (ER+) breast cancer define tumors with CIN and high proliferative potential. Intriguingly, CIN induction in lower eukaryotic cells and human cells is context-dependent, typically resulting in a proliferation disadvantage but conferring a fitness benefit under strong selection pressures. We hypothesised that CIN permits accelerated genomic evolution through the generation of diverse DNA copy number events that may be selected during disease development. In support of this hypothesis, we found evidence for selection of gene amplification of core regulators of proliferation in CIN-associated cancer genomes. Stable DNA copy number amplifications of the core regulators TPX2 and UBE2C were associated with expression of a gene module involved in proliferation. The module genes were enriched within prognostic signature gene sets for ER+ breast cancer, providing a logical connection between CIN and prognostic signature expression. Our results provide a framework to decipher the impact of intratumor heterogeneity on key cancer phenotypes, and they suggest that CIN provides a permissive landscape for selection of copy number alterations which drive cancer proliferation.
Introduction
Induction of aneuploidy affects cellular fitness in various organisms including yeast (1), mice (2) and human cells (3). Most aneuploid cells exhibit reduced proliferation rates, but studies in yeast have shown that aneuploidy can also be beneficial for the adaptation to stressful conditions not previously experienced by the cell population (4). Highly aneuploid cancers are characterised by a large number of structural and numerical DNA copy number changes altering expression of most genes located in these regions. Aneuploidy is frequently accompanied by chromosomal instability (CIN), defined as an increased rate of gains and losses of whole chromosomes or fractions of chromosomes (5). CIN generates intercellular chromosomal heterogeneity that may facilitate selection of genotypes tolerant to aneuploidy and strong selective pressures in the tumour, and accelerate the proliferation of aneuploid cancer cells (6,7). The specific chromosomal changes and expression patterns that are implicated in this adaptive response remain unclear.
Several breast cancer gene expression signatures forecasting clinical outcome and response to chemotherapy are strongly associated with proliferation (8–11). It is still being debated whether the prognostic value of these diverse gene lists reflects similar underlying biological processes and pathways (12). Intriguingly, there is increasing evidence that many breast cancer prognostic signatures are associated with CIN (13,14). CIN is associated with inferior prognosis across multiple cancer types, including ER-positive breast cancer (15). The CIN70 signature (16) and a 12-gene genomic instability signature (13), both derived from their associations with aneuploidy and chromosomal instability, have prognostic value in many cancer types and also correlate with proliferation markers. However, little is known about possible genotypes crucial for the CIN phenotype or the relationships between CIN and proliferation in vivo. Indeed it has been proposed that CIN is a consequence, rather than a cause of the selective pressure for proliferative drive in tumours.
In this study we sought to better characterise the relationships between CIN and proliferation through an analysis of the effects of copy number alterations associated with CIN and the downstream consequences of such alterations upon prognostic signature gene expression.
Materials and Methods
SNP and expression data processing
Microarray expression data and SNP array based copy number data of 264 ER-positive breast cancer patients with pathologist estimates of more than 60% tumour cell content or more than 60% tumour nuclei were selected from the Cancer Genome Atlas (TCGA). Agilent 244K Custom Gene Expression G4502A-07 two-colour microarrays were normalized (print-tip-group loess normalization, Bioconductor package marray (17)). Probes with missing values in more than 15% of the samples were excluded and duplicated probes were averaged. Gene mappings based on NCBI build 36.3 were downloaded from the TCGA data portal (18) For probes mapped to the same ENTREZ Gene ID, the probe with the highest Pearson correlation coefficient of gene expression with wGII score was chosen. For the remaining genes, missing gene expression data was imputed with nearest neighbour (k=10) averaging implemented in the R package impute (19). Genes with standard deviation lower than 0.25 were removed. The expression values of all remaining genes were standardised.
SNP 6.0 samples were normalized with the Affymetrix Genotyping Console using standard settings. Samples not passing the quality check criteria were excluded. Integer copy numbers were estimated by the GAP algorithm (20).
RNAi Screening data
Whole genome RNAi screening data (21) analysing cell numbers after gene silencing was available for five cell lines: PC9 (lung), RCC4 (kidney), HCT116 (colon), MCF-10A (breast) and HT1080 (fibrosarcoma). The PC9 and RCC4 screening data was normalised with respect to the plate median, and then smoothed using the well position in the following manner: normalised well value = (well value - plate median)/(plate median absolute deviation) and smoothed well value = (normalised well value - well position median)/(well position median absolute deviation). For HT1080, the data was logged and then normalised with respect to the plate median: normalised well value = log (well value - plate median)/(plate median absolute deviation). For MCF-10A the data was normalised as: normalised well value = (well value - plate median)/(batch median absolute deviation). The median over the replicates (2-3) was taken as the normalised Z-score and the 82 genes that impair cell viability following siRNA transfection (Z-Score < −2) in at least three of the five cell lines were defined as proliferation genes (see Supplemental Table S1).
Genome wide shRNA data including p-values for impaired cell survival for the 11 ER-positive cell lines BT-474, ZR-75-1, T47D, MCF7, MDA-MB-361, KPL-1, HCC1500, HCC1428, HCC1419, EFM-19, CAMA-1 were downloaded from the COLT data base (22). Genes inducing cell death (p-value < 0.05) in at least four cell lines were considered as proliferation genes in ER-positive breast cancer.
Microarray expression analysis after UBE2C and TPX2 silencing in T47D cells
T47D cells were maintained in 5% CO2 at 37° C, in RPMI medium supplemented with 10% FBS, glutamine and penicillin/streptomycin. All siRNA (siRNA; Dharmacon, Thermo Scientific) were performed at 40 nM final concentrations by reverse transfection with Dharmafect2 reagent according to the manufacturers instructions. Transfections were performed using TPX2 (L-010571-00-0005 and M-010571-00-0005 and UBE2C (L-004693-00-0005 and M-004693-03-0005) siRNA pools. Non-targeting control siRNA and scrambled control 2 were used. At 72 hours post transfection, RNA was extracted and knockdowns validated by quantitative PCR to be at least 85% - 90%. RNA was extracted and samples were hybridized to HG_U133 Plus 2.0 arrays. Expression calls were generated by the MAS5.0 algorithm (R-package simpleaffy (23)). Gene expression values were subtracted from the gene expression values after silencing TPX2 and UBE2C respectively from the siRNA control experiment. This difference was multiplied by the sign of the Spearman correlation of the gene expression with TPX2 or UBE2C in the tumour samples. Negative values indicate that silencing of TPX2 or UBE2C results in down-regulation of genes positively correlated and in up-regulation of genes negatively correlated with TPX2 or UBE2C expression in tumours. Deviations from zero were tested with Wilcoxon’s signed rank test.
Weighted Genome Integrity Index (wGII)
The ploidy of a tumour sample was determined as the weighted median integer copy number, with weights equal to the lengths of the copy number segments. For each sample and each of the 22 autosomal chromosomes, the percentage of gained and lost genomic material was calculated relative to the ploidy of the sample. The use of percentages eliminates the bias induced by differing chromosome sizes (24). The wGII score of a sample is defined as the average of this percentage value over the 22 autosomal chromosomes.
Identification of CIN Associated Core Regulators and Their Co-Expressed Gene Modules
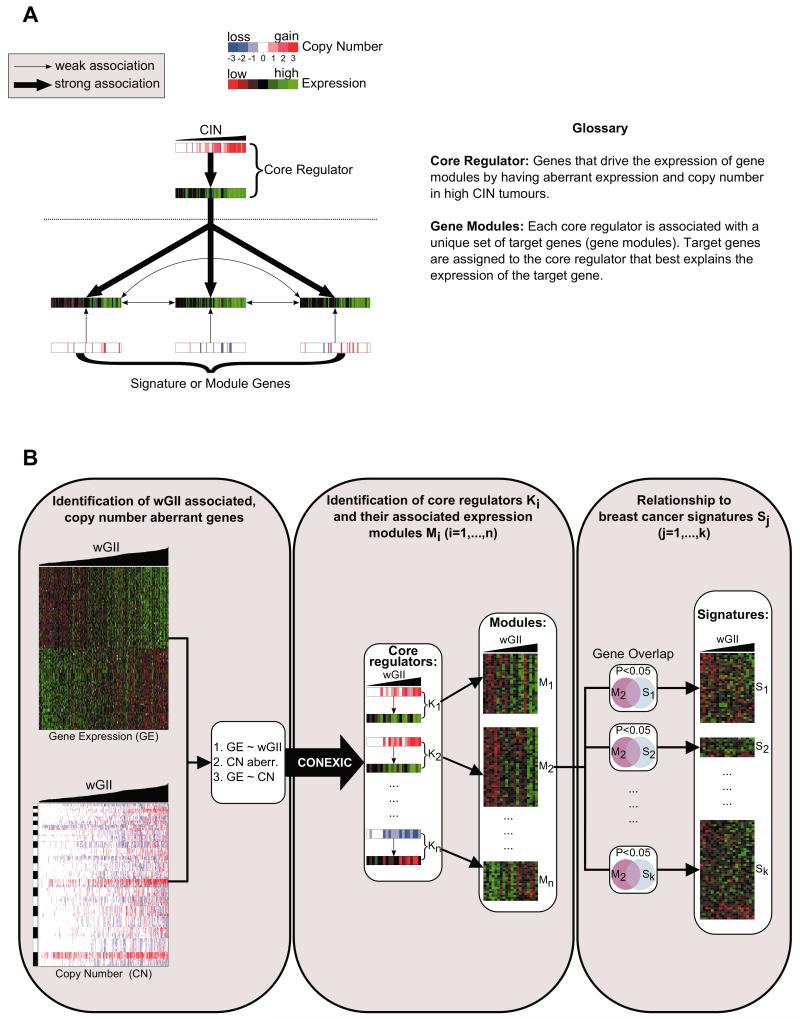
We defined a core regulator for CIN as a gene or transcript driving the expression of a co-regulated gene set (termed an expression module) by having aberrant expression and copy number in high CIN tumours.
Step1: Identification of genes whose expression is correlated with CIN
From all genes passing the thresholds for SNP and gene expression data processing, the 500 genes with the highest positive correlation (Kendall’s Tau τ) and the 500 genes with the lowest negative correlation between wGII score and gene expression were selected. All these genes had |τ| > 0.15 and a P-value < 0.05.
Step2: Identification of genes passing step 1 and whose expression is correlated with CIN
A modified GISTIC algorithm (25) for integer copy numbers was used to find genes significantly gained or lost (q-value threshold of 25%) in high CIN tumour samples (wGII > median(wGII)). For each gene passing the filtering step 1, we computed a two sample t-statistics for expression differences between samples where the gene was lost (copy number < ploidy) and samples where the gene was not lost. Analogously, a two sample t-test for expression differences between samples where the gene was gained (copy number > ploidy) and samples where it was not gained was performed. Only genes passing step 1 and having a two sided p-value<0.05 for gains or losses respectively were used for further analysis.
Step 3: CONEXIC Analysis to detect core regulators and their expression modules
Genes passing all three criteria were taken as candidates in the Single Modulator Step of the CONEXIC algorithm (26). This algorithm splits the gene expression values of each candidate regulator across the different samples into two groups of low and high expression. For a given candidate regulator, the resulting expression threshold separating these two groups is then used to split all ’target genes’ (all genes including other candidate regulators) into two classes. CONEXIC uses a Normal Gamma score to compute the performance of all pairwise splits of candidate regulators and ’target genes’ and uses permutation based sampling to assign a p-value (26). All genes with a p-vale<0.001 are then assigned to the candidate regulator with the highest score. Nonparametric bootstrapping (100 bootstrap samples) is used to filter out spurious associations and all candidate regulators with more than 20 module genes in 90% of the bootstrap runs are used for a final run of the Single Modulator step, resulting in a list of potential core regulators. The top 30 genes with highest Normal Gamma score were selected as core regulators.
Association of Key Regulator Copy Number with Signature Expression
The correlation (Spearman) between copy number of all core regulators and expression of all signature genes was compared to the corresponding copy number –gene expression correlations of all signature genes and the percentage of significant (p<0.05) correlations was computed. The connectedness of a gene is the mean over all these correlation coefficients. For Oncotype Dx®, all loading control genes were removed prior to the analysis.
Enrichment of GO Terms
Several sets of genes were tested for an enrichment of GO (gene ontology) terms using version v3.0 (27,28). A 2×2 contingency table was created by overlapping the ENTREZ gene identifiers of the gene lists with GO term associated gene sets. GO term enrichment was tested by one sided Fisher’s exact tests.
Somatic Mutations
For the analysis of somatic gene mutations with next-generation DNA sequencing data, unvalidated (level 2) Mutation Annotation Format (MAF) files were downloaded from the TCGA data portal (18). The MAF files contain information about mutation type, gene names, validation status and the type of sequencing platform. Matching copy number and somatic mutation data was available for 254 ER-positive breast tumours. To detect genes, which show an association of the probability of a somatic mutation with increasing or decreasing degrees of wGII scores, a univariate logistic regression model was applied for each gene separately and genes were ranked by P-values.
Data Access
Microarray expression data and SNP array data were selected from the Cancer Genome Atlas (TCGA,(18)). Genome wide shRNA data for ER-positive cell lines were downloaded from the COLT data base (22).. The RNAi Screening data for HCT116, PC9, RCC4, HT1080, MCF-10A are available from the High throughput screening database (21).
Results
Prognostic signature expression correlates with proliferation
The association of prognostic gene expression signatures with outcome may be explained in part by their correlation with measures of cellular proliferation (10,29,30). We investigated this relationship for the five prognostic gene signatures CIN70, Oncotype Dx®, MammaPrint®, PAM50, and Gene expression Grade Index (GGI) (16,29–32) in 264 ER-positive breast tumour samples from the Cancer Genome Atlas (TCGA) (33). Oncotype Dx® (16 genes + 5 genes for baseline normalization), MammaPrint® (70 genes), PAM50 (50 genes) and GGI (97 genes) are all breast cancer specific prognostic gene expression signatures. The CIN70 signature was derived from a measure of total functional aneuploidy, indicative of chromosomal instability, and is associated with prognosis in multiple tumour types, including breast cancer (16).
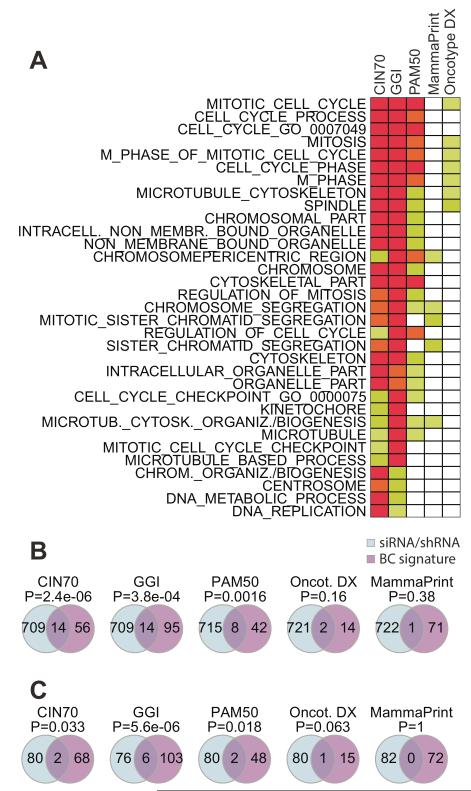
Based on a gene ontology (GO) analysis we find many genes in these five signatures to be involved in cell cycle and mitotic processes (Fig. 1A). Consistent with previous studies, the expression of most signature genes of all five of the prognostic signatures is highly correlated with the mRNA expression of the MKI67 proliferation marker (Supplemental Fig. S1A).
Fig. 1. Association of breast cancer prognostic signatures with proliferation.
(A) Enrichment of GO terms in the five breast cancer prognostic signatures. The rows are ordered by increasing sums of p-value across the breast cancer signatures. (B) Overlap of breast cancer signature genes with genes essential (p<0.05) in at least 4/11 whole genome shRNA screens from the COLT data base (22,34). (C) Overlap of breast cancer signature genes with proliferation genes whose siRNAs reduced cell viability by a Z-Score of at least −2 in 3/5 cell lines with diverse cell type or tissue of origin.
MKI67 is a member of the three signatures GGI, PAM50 and Oncotype DX®. Prognostic signature gene expression is also significantly correlated with expression of a gene set that correlates with the key proliferation gene PCNA (proliferating cell nuclear antigen) – the meta-PCNA signature (10), see Supplemental Fig. S1B.
Next, we checked for a functional role of the individual genes within the breast cancer prognostic signatures with cellular proliferation through an independent analysis of two whole genome RNA interference screening datasets (see Materials and Methods). Genes inducing cell death (p-value < 0.05) in at least four out of 11 ER-positive breast cancer cell lines (22,34) were defined as proliferation genes in breast cancer (721 genes). A second set of 82 proliferation genes was derived from inhouse RNA interference screens performed in a panel of 5 human cell lines: HCT116 (colon carcinoma), PC9 (lung cancer), RCC4 (renal cell carcinoma), HT1080 (fibrosarcoma), MCF-10A(mammary epithelial cells). For both proliferation gene sets, we found a significant (p<0.05) overlap with the CIN70, GGI and PAM50 signatures (Fig 1B,C). The relatively small overlap with Oncotype DX® signature genes might be attributable to the small number of genes in Oncotype DX®. The exception is MammaPrint®, where no significant overlap with any proliferation gene set was observed. This might be related to the fact that the 70 signature genes in MammaPrint® were obtained from genes differentially expressed in metastatic versus non metastatic tumours.
These results confirm the functional role in cellular viability and proliferation for many of the genes incorporated in prognostic signatures, independent of cell type or endocrine responsive status (Fig. 1B,C).
Increased chromosomal complexity is associated with increased expression of proliferative gene expression markers
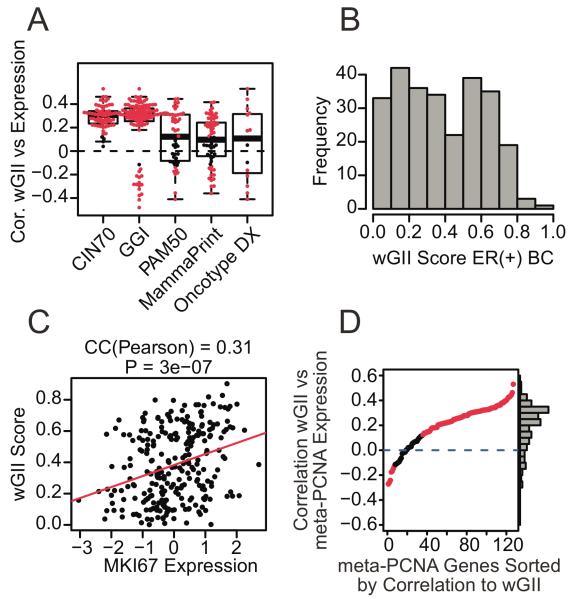
Prognostic signatures have been suggested to reflect CIN and proliferation (13,14). We explored this relationship in the cohort of 264 ER-positive breast tumour samples. To assess the CIN status of each tumour, we used a SNP array based surrogate score – the weighted genome integrity index score (wGII) (24,35). In comparison to the original GII score (35), the wGII score avoids the bias caused by gains and losses of large chromosomes relative to small chromosomes. The wGII score takes values between zero and one and is positively correlated with all genes in the CIN70 signature in the 264 tumour samples (Fig. 2A). This consistency supports the use of wGII to classify CIN status in breast tumours (36). In addition, the majority of genes in the four breast cancer specific gene signatures (Oncotype Dx®, MammaPrint®, PAM50 and GGI) are also significantly correlated with wGII (Fig. 2A). Together with the broad spectrum of wGII scores observed in these 264 tumours (Fig. 2B), these associations suggest that CIN status might define patient sub-groups with differential outcome (14,16,37,38).
Fig. 2. Association of wGII score with proliferation.
(A) Correlation of wGII score with prognostic signature gene expression. Each dot shows the correlation coefficient of wGII score with one gene of the particular signature (red: p<0.05 and black: p≥0.05). (B): Distribution of wGII scores in the 264 ER-positive breast cancer samples. (C) The wGII score versus MKI67 expression in each breast cancer sample. (D) Correlation of wGII score with expression of meta-PCNA genes. Each point corresponds to one gene in the meta-PCNA gene set.
We also examined the relationship between CIN and markers of proliferation in this cohort. We found a highly significant correlation between wGII score and both MKI67 expression (Fig. 2C, P=3×10-7), and the majority of genes in the meta-PCNA gene set (Fig. 2D, 78% with P < 0.05).
Identification of core regulators encoded within regions of somatic copy number alteration
Increased CIN requires reconfigurations in cancer cell gene expression programs. Based on the association of CIN and proliferation, we hypothesized that part of this CIN expression program may be regulated by a small number of ‘core regulators’. Specifically, we hypothesised that expression signatures functionally implicated with proliferation (see Glossary in Fig. 3) might be ‘hard wired’ in the CIN genome through the selection of recurrent copy number changes encoding such core regulators.
Fig 3. Identification of core regulators and modules.
(A) Core regulators of CIN are genes with altered copy number and expression in high CIN tumours which regulate a gene expression module. (B) First step: Candidate regulators are identified by filtering copy number aberrant genes whose expression (GE) is associated with both their copy number (CN) and with wGII score. Second step: Each core regulator is linked to expression modules which are regulated by the core regulator. The CONEXIC algorithm (26) is applied to rank these core regulator-module pairs. Third step: Core regulators and their associated modules are then compared with known breast cancer prognostic signatures. Copy number data is displayed in blue (loss), white (no change) and red (gain) and gene expression data is displayed in red (low), black (average) and green (high).
To identify possible core-regulators (Fig. 3) we first selected a set of candidate regulator genes whose expression and copy number is associated with increasing wGII score and whose somatic copy number gain or loss results in altered expression (see Materials and Methods). We found 180 candidate regulators located in regions of frequent genomic loss and 267 candidate regulators located in regions of frequent genomic gain in wGII high tumours (Supplemental Table S3).
In a second step, we searched for sets of co-expressed genes (collectively termed a ‘module’) belonging to each individual candidate regulator. During this search, the set of candidate regulators was reduced to the set of core regulators by eliminating candidates for which a sufficiently strong module could not be found. To find these regulator-module pairs we used the first step of the CONEXIC algorithm (26), which was originally developed to identify general drivers of cancer, not those specifically associated with CIN. In this approach, the pairing of a candidate regulator with a given individual module gene occurs only if the expression of the regulator best explains the expression of its module genes and if the expression of no module gene is better explained by any other candidate regulator. Each regulator and paired module of genes is assigned a normal gamma score (26), which measures the strength of the cohesion between the core regulator and its module. We selected the 30 regulator-module pairs with the highest normal gamma score (for both gained and lost chromosome regions) and defined these 30 regulators as core-regulators (see Table 1 and Supplemental Fig. S2).
Table 1.
Top 30 core regulators (CONEXIC) ordered by decreasing CONEXIC scores. Amplified core regulators (AMP) showed recurrent copy number gains and deleted core regulators (DEL) showed recurrent copy number loss in high wGII tumours.
| Rank(*) | Core Regulator | Type | N(**) | Chrom. |
|---|---|---|---|---|
| 1 | PAQR4 | AMP | 63 | 16p |
| 2 | TPX2 | AMP | 111 | 20q |
| 3 | UBE2C | AMP | 108 | 20q |
| 4 | RPL12P14 | DEL | 70 | 1p |
| 5 | RPS3AP49 | DEL | 46 | 18q |
| 6 | EXO1 | AMP | 122 | 1q |
| 7 | PTTG3P | AMP | 44 | 8q |
| 8 | SHARPIN | AMP | 58 | 8q |
| 9 | RPL17 | DEL | 106 | 18q |
| 10 | TSEN54 | AMP | 64 | 17q |
| 11 | LSM14B | AMP | 117 | 20q |
| 12 | LOC253482 | DEL | 66 | 9p |
| 13 | PHB | AMP | 65 | 17q |
| 14 | DCAF13 | AMP | 126 | 8q |
| 15 | MYBL2 | AMP | 76 | 20q |
| 16 | EIF2C2 | AMP | 111 | 8q |
| 17 | C8orf76 | AMP | 92 | 8q |
| 18 | TACO1 | AMP | 68 | 17q |
| 19 | PSMD12 | AMP | 36 | 17q |
| 20 | UBE2T | AMP | 65 | 1q |
| 21 | MRPL12 | AMP | 78 | 17q |
| 22 | AURKA | AMP | 62 | 20q |
| 23 | PKMYT1 | AMP | 52 | 16p |
| 24 | ATP6V1C1 | AMP | 85 | 8q |
| 25 | RPS13P2 | DEL | 33 | 1p |
| 26 | SLC52A2 | AMP | 86 | 8q |
| 27 | THSD1 | DEL | 447 | 13q |
| 28 | TMEM189 | AMP | 100 | 20q |
| 29 | TPT1 | DEL | 57 | 13q |
| 30 | KIF14 | AMP | 112 | 1q |
Ranked by CONEXIC Score
Number Genes in Module
Amplified Core Regulators and their Gene Modules are Enriched for Proliferation GO Terms
A GO analysis revealed a significant association of the 30 core regulators with mitotic and cell cycle related processes (Supplemental Table S4). We also tested the individual modules of each core regulator separately for an enrichment of biological processes in GO and ranked them according to their p-values (see Supplemental Table S5 for the top 50 GO terms). The modules of the amplified core regulators PAQR4, TPX2, UBE2C, PTTG3P, PKMYT1 were among the top 50 associations and were enriched for genes involved in cell cycle and mitotic processes.
Only 7 of the 30 core regulators are located in regions of recurrent genomic loss (Table 1). With exception of THSD1 and TPT1 (39) (also known as TCTP), the core regulators in regions of recurrent genomic loss encode components of the ribosome. Four of the five ribosomal core regulators are pseudogenes, which might indicate a regulatory function of these transcripts, and only RPL17 encodes a protein product. Accordingly, the modules belonging to the lost core regulator pseudogenes RPS13P2, LOC253482 and RPL12P14 exhibit highly ranked associations with structural components of the ribosome (Supplemental Table S5). The lost core regulator TPT1 was recently shown to interact with p53 and to be important for DNA damage sensing and repair (40).
Expression of a large fraction of genes in the meta-PCNA proliferation signature is significantly correlated with expression of the core regulators (Supplemental Fig. S3). In addition, 8/30 core regulator modules overlapped significantly (p<0.05) with the meta-PCNA signature (Table 2). Three amplified core regulators TPX2, UBE2C and AURKA are themselves members of the meta-PCNA signature (Fishers exact test p=0.00097). The TPX2 module contained 20 genes from the meta-PCNA signature (P=1.4×10-23) and TPX2 was also classified as a proliferation gene in our meta-analysis of genome RNA interference screens (see Supplemental Table S1).
Table 2.
Overlap of the meta-PCNA signature with core regulator modules. The table shows all core regulator genes with at least 3 genes in the overlap. P-values were derived by Fishers exact tests.
| Core Regulator | N (overlap) | N (meta-PCNA) | N (gene module) | P-Value |
|---|---|---|---|---|
| TPX2 | 20 | 131 | 111 | 1.4e-23 |
| UBE2C | 14 | 131 | 108 | 1e-14 |
| PAQR4 | 11 | 131 | 63 | 2.8e-13 |
| EXO1 | 9 | 131 | 122 | 1.1e-07 |
| MYBL2 | 7 | 131 | 76 | 6.5e-07 |
| UBE2T | 5 | 131 | 65 | 6.6e-05 |
| PTTG3P | 3 | 131 | 44 | 0.003 |
| PKMYT1 | 3 | 131 | 52 | 0.0048 |
These strong associations suggest that copy number amplification of certain core regulators including TPX2 and UBE2C might regulate proliferation in high CIN tumours by regulating CIN specific gene expression modules functionally implicated in proliferation. Of note, we found a strong and significant association of p53 somatic mutations with wGII (Supplemental Fig. S4), reconfirming published associations of p53 with CIN (7,41).
Copy number of UBE2C and TPX2 is highly associated with prognostic signature gene expression
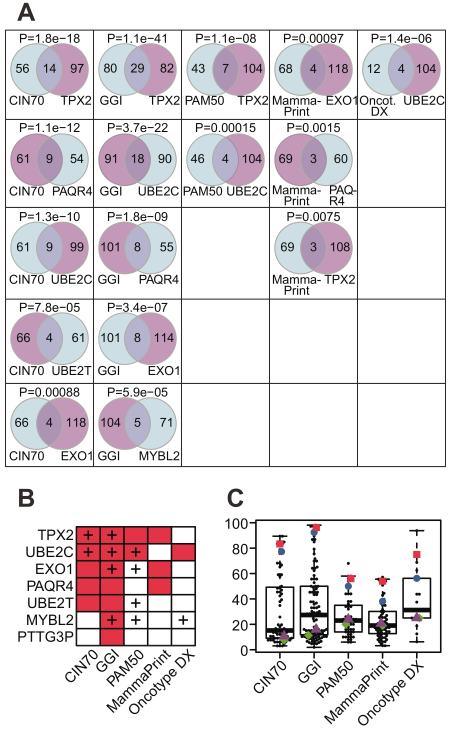
The modules of seven core regulators (TPX2, UBE2C, EXO1, PAQR4, PTTG3P, MYBL2, UBE2T) significantly (Fisher’s Exact test p < 0.05) overlap with the CIN70 signature (Fig 4A,B). More surprisingly, the expression modules of the seven amplified core regulators TPX2, UBE2C, EXO1, PAQR4, UBE2T, MYBL2, PTTG3P have a significant overlap (Fisher’s Exact test p < 0.05) with at least one of the four breast cancer specific prognostic gene signatures GGI, PAM50, MammaPrint® or Oncotype Dx® (Fig. 4A,B). The TPX2 and UBE2C expression modules were overrepresented in three breast cancer specific prognostic signatures (Fig. 4B) and in CIN70 with p-values ranging from 0.0075 to 1.1×10−41 for TPX2 and from 1.5×10−4 to 3.7×10−24 for UBE2C. The modules of EXO1 and PAQR4 were overrepresented in GGI and MammaPrint®. In addition, eight amplified core regulators (TPX2, AURKA, UBE2C, EXO1, MYBL2, TPT1, KIF14, UBE2T) are members of one or more prognostic signatures, and 4 out of 5 of the signatures contain one or more amplified core regulators (indicated as crosses in Fig. 4B, see also Supplemental Table S6). We did not find any such associations for core regulators or their modules located in regions of genomic loss.
Fig. 4. Association of core regulators with breast cancer prognostic signatures.
(A): The overlap of breast cancer signatures (columns) with core regulator modules (rows). The rows in each column are ordered by decreasing Fisher’s Exact Test p-values and only the top five significant core regulator associated modules are displayed per breast cancer signature. (B) Core regulators (rows) with modules enriched (p<0.05) for at least one breast cancer prognostic signature (red: significant enrichment; “+” sign: The core regulator is a member of the signature). (C) Percentage of significant correlations between copy number of signature genes with gene expression of each other signature gene (circles). The coloured symbols indicate the percentage of significant correlations between the copy number of the core regulators UBE2C (red squares), TPX2 (blue circles), PAQR4 (magenta triangles) and EXO1 (green diamonds) with the expression of all genes in the respective gene signature.
We next asked how the copy number of any amplified core regulator is related to the expression of the prognostic cancer signature genes (Fig. 3A). We restricted this analysis to TPX2, UBE2C, EXO1 and PAQR4, the only four core regulators whose modules were significantly overrepresented in at least two breast cancer specific prognostic signatures and in CIN70 (Fig. 4B). For a given signature, we computed the correlation coefficient between the copy number of each signature gene with the expression of all other signature genes. Similarly, the copy number of the four core regulators TPX2, UBE2C, EXO1 and PAQR4 was correlated with the expression of each signature gene. By comparing the percentage of significant correlations (Fig. 4C), we found the core regulators UBE2C and TPX2 to be highly ranked in the six signatures CIN70, GGI, PAM50, PAM50, MammaPrint® and Oncotype Dx® with UBE2C being found among the top 5 genes in all of them (red squares in Fig. 4C). In particular, UBE2C copy number is significantly correlated with the expression of more than 80% of the genes in the GGI signature and ranks second in Oncotype DX®, although it is not a member of that gene expression signature. The copy number of the core regulators PAQR4 and EXO1 had only moderate correlations with signature gene expression.
As an alternative summary measure of association between copy number and expression we computed the average connectedness (27) of a gene, which is given by its average absolute correlation coefficient between its copy number and expression taken over all genes in all signatures. We found UBE2C and TPX2 copy numbers to be strongly connected with the expression of the signature genes (Supplemental Fig. S5). In summary, copy number amplifications of the core regulators UBE2C and TPX2 are strongly associated with prognostic breast cancer signature expression in ER-positive CIN tumours.
Effects of silencing UBE2C or TPX2 by RNAi in T47D cells
We tested the effects of silencing UBE2C and TPX2 upon gene expression in the ER-positive T47D cell line (Fig. 5). T47D cells have increased UBE2C and TPX2 expression and copy number. For each gene in the respective module or signature, we computed the expression difference of the RNA-interference non-targeting control experiment with the expression value in cells silenced for the core regulator TPX2 or UBE2C. This difference was then multiplied by the sign of the correlation coefficient between the core regulator and the respective gene in the tumour data. Thus, a negative sign indicates expression changes in T47D concordant with the changes predicted from the tumour data, whereas positive values correspond to discordant changes.
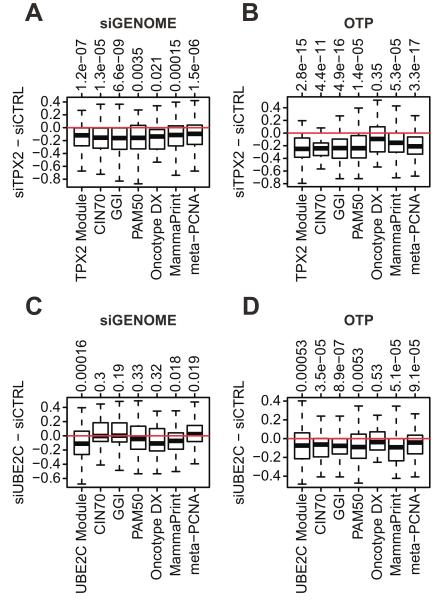
Fig. 5. Silencing TPX2 or UBE2C in the T47D cell line.
Silencing of TPX2 (A,B) and UBE2C (C,D) using two different siRNA agents (siGenome and ON-Targetplus) and effect on the expression of the respective core regulator modules and on gene expression signatures. The boxplots display signed differences in expression between siRNA and control samples for the genes in the respective core regulator module or gene signature. Negative values of these signed differences correspond to expression changes concordant with the corresponding associations in the tumour data and positive values to discordant results (see text for details). The overall p-value indicates the overall significance of the signed expression differences.
The majority of the gene expression changes of core regulator modules and gene signatures is concordant with the expected changes in the tumour data (Fig. 5). The only changes discordant appear for CIN70, GGI and the meta- PCNA proliferation signature in response to UBE2C silencing with siGENOME (Fig. 5C). The p-values for the signed expression changes of Oncotype DX® are relatively large, a finding that is not unexpected given that there are only 16 genes in this signature. Taken together the expression changes for the signatures and modules follow the same pattern (Fig. 5) as observed for the tumour data. These data support the effect of TPX2 and UBE2C core regulator expression on their modules and the relation with prognostic and proliferation signatures.
Discussion
In this study, we worked from the hypothesis that CIN tumour cells have evolved a specific gene expression programme conferring a selective advantage (41,42). We focused on the detection of specific copy number aberrations which are both strongly associated with CIN (as assessed by wGII) and with the altered expression activity of characteristic expression modules (Fig. 3B). For ER-positive breast tumours we provide evidence that part of the CIN expression program is indeed hard wired in the CIN genome by specific copy number aberrations of core regulators such as TPX2 and UBE2C. In particular, the amplified core regulators TPX2 and UBE2C and their respective modules are associated with both proliferation markers and cell cycle or mitosis related processes.
Whilst the association of TPX2 and UBE2C copy number gain and CIN has been previously reported (43,44), we show that the copy number of these core regulators is also strongly linked to the expression of prognostic signature gene sets for ER-positive breast cancer, which themselves are associated with proliferation and CIN. These results are supported by the changes in the expression patterns of the breast cancer signatures following siRNA silencing of UBE2C and TPX2.
Whilst most core regulators were amplified, we also found seven core regulators in regions of genomic loss. The lost core regulator TPT1 encodes a multifunctional protein involved in the regulation of cell death and proliferation, as well as DNA damage sensing and repair and is part of a reciprocal feedback loop with p53 enabling tolerance of ongoing DNA damage and repair (39). Five of the seven lost core regulators are related to the ribosome, which might reflect evolutionary adaptation to the increased transcriptional and translational load of CIN cells, due to increased ploidy, or other extra-ribosomal functions such as DNA repair, apoptosis and cellular homeostasis (45). The fact that four of the five ribosomal core regulators are pseudogenes might also hint to a potential regulatory function, possibly similar to the role of the PTENP1 pseudogene as a growth suppressor (46).
A direct measurement of both CIN and proliferation, in conjunction with copy number and gene expression data acquisition, is currently not feasible. Increased proliferation was only indirectly assessed by correlation with proliferation markers. We can not exclude potential technical bias caused by comparing data from different expression microarray platforms. The wGII surrogate score is also not a direct measure for CIN status, but its predictive capability is well tested in various data sets (35,36). By design, we might have missed regulators of gene expression in CIN cells that are not located in regions of aberrant copy number. A similar analysis using alternative aberrations as filters, such as methylation patterns or frequent somatic mutations, is a direction for future research.
The relationship between aneuploidy, CIN and proliferation remains a complex research question (42,47). Our results cannot explain the development of CIN and tumour heterogeneity. However, one emerging picture is that a certain level of CIN provides a way to sample many aneuploid karyotypes and, by means of Darwinian selection, to increase the chance of acquiring genomic configurations associated with higher fitness. On the basis of results presented here, we suggest that the identification of core regulators driving gene expression in CIN tumours contributes to the understanding of these fitness states, which might facilitate the identification of therapeutic strategies to attenuate CIN.
Supplementary Material
Acknowledgments
CS is a senior Medical Research Council clinical research fellow. CS, PP, JD, RB and DE were funded by Cancer Research UK. CS receives funding from the EU Framework Program 7, The Prostate Cancer Foundation, and the Breast Cancer Research Foundation, and supported by researchers at the National Institute for Health Research University College London Hospitals Biomedical Research Centre. The results published here are in part based upon data generated by The Cancer Genome Atlas pilot project established by the NCI and NHGRI. Information about The Cancer Genome Atlas and the investigators and institutions that constitute The Cancer Genome Atlas Research Network can be found at http://cancergenome.nih.gov/. The data were retrieved through dbGaP authorization (accession numbers phs000178.v4.p4 and phs000178.v5.p5).
Footnotes
The authors have no conflicts of interest to disclose.
References
- 1.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–24. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 2.Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, et al. Aneuploidy Affects Proliferation and Spontaneous Immortalization in Mammalian Cells. Science. 2008;322:703–9. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol. 2012;8 doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–5. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR. Defining “chromosomal instability”. Trends Genet. 2008;24:64–9. doi: 10.1016/j.tig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 7.Coschi CH, Dick FA. Chromosome instability and deregulated proliferation: an unavoidable duo. Cell Mol Life Sci. 2012;69:2009–24. doi: 10.1007/s00018-011-0910-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desmedt C, Haibe-Kains B, Wirapati P, Buyse M, Larsimont D, Bontempi G, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14:5158–65. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- 9.Haibe-Kains B, Desmedt C, Sotiriou C, Bontempi G. A comparative study of survival models for breast cancer prognostication based on microarray data: does a single gene beat them all? Bioinformatics. 2008;24:2200–8. doi: 10.1093/bioinformatics/btn374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venet D, Dumont JE, Detours V. Most random gene expression signatures are significantly associated with breast cancer outcome. Plos Comput Biol. 2011;7:e1002240. doi: 10.1371/journal.pcbi.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wirapati P, Sotiriou C, Kunkel S, Farmer P, Pradervand S, Haibe-Kains B, et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10:R65. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drier Y, Domany E. Do two machine-learning based prognostic signatures for breast cancer capture the same biological processes? Plos One. 2011;6:e17795. doi: 10.1371/journal.pone.0017795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Habermann JK, Doering J, Hautaniemi S, Roblick UJ, Bündgen NK, Nicorici D, et al. The gene expression signature of genomic instability in breast cancer is an independent predictor of clinical outcome. Int J Cancer. 2009;124:1552–64. doi: 10.1002/ijc.24017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roylance R, Endesfelder D, Gorman P, Burrell RA, Sander J, Tomlinson I, et al. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol Biomarkers Prev. 2011;20:2183–94. doi: 10.1158/1055-9965.EPI-11-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 2012;13:528–38. doi: 10.1038/embor.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–8. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 17.Yang YH, Paquet A, Dudoit S. marray: Exploratory analysis for two-color spotted microarray data. 2009 http://www.maths.usyd.edu.au/u/jeany/ [Google Scholar]
- 18.TCGA The Cancer Genome Atlas. 2011 [Internet] Available Httpwwwcancergenomenihgov.
- 19.Hastie T, Tibshirani R, Narasimhan B, Chu G. impute: Imputation for microarray data. 2012 http://www.bioconductor.org/packages/release/bioc/html/impute.html. [Google Scholar]
- 20.Popova T, Manié E, Stoppa-Lyonnet D, Rigaill G, Barillot E, Stern MH. Genome Alteration Print (GAP): a tool to visualize and mine complex cancer genomic profiles obtained by SNP arrays. Genome Biol. 2009;10:R128. doi: 10.1186/gb-2009-10-11-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.High throughput screening database. [Internet] Available from: http://hts.cancerresearchuk.org/db/public/
- 22.Koh JLY, Brown KR, Sayad A, Kasimer D, Ketela T, Moffat J. COLT-Cancer: functional genetic screening resource for essential genes in human cancer cell lines. Nucleic Acids Res. 2012;40:D957–D963. doi: 10.1093/nar/gkr959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller CJ. simpleAffy-Very simple high level analysis of Affymetrix data [Internet] Available from: http://www.bioconductor.org/packages/2.12/bioc/html/simpleaffy.html.
- 24.Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller M-C, Shaikh N, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–6. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S. 2007;104:20007–12. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akavia UD, Litvin O, Kim J, Sanchez-Garcia F, Kotliar D, Causton HC, et al. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–17. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.GSEA. Internet Broadinstitute; 2012. Available from: http://www.broadinstitute.org/gsea/ [Google Scholar]
- 29.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–72. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 30.Van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AAM, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 31.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 32.Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcotte R, Brown KR, Suarez F, Sayad A, Karamboulas K, Krzyzanowski PM, et al. Essential Gene Profiles in Breast, Pancreatic, and Ovarian Cancer Cells. Cancer Discov. 2012;2:172–89. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chin SF, Teschendorff AE, Marioni JC, Wang Y, Barbosa-Morais NL, Thorne NP, et al. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8:R215. doi: 10.1186/gb-2007-8-10-r215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee AJX, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011;71:1858–70. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walther A, Houlston R, Tomlinson I. Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut. 2008;57:941–50. doi: 10.1136/gut.2007.135004. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe T, Kobunai T, Yamamoto Y, Matsuda K, Ishihara S, Nozawa K, et al. Chromosomal Instability (CIN) Phenotype, CIN High or CIN Low, Predicts Survival for Colorectal Cancer. J Clin Oncol. 2012;30:2256–64. doi: 10.1200/JCO.2011.38.6490. [DOI] [PubMed] [Google Scholar]
- 39.Amson R, Pece S, Lespagnol A, Vyas R, Mazzarol G, Tosoni D, et al. Reciprocal repression between P53 and TCTP. Nat Med. 2011;18:91–9. doi: 10.1038/nm.2546. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, de Toledo SM, Pandey BN, Guo G, Pain D, Li H, et al. Role of the translationally controlled tumor protein in DNA damage sensing and repair. Proc Natl Acad Sci. 2012;109:E926–E933. doi: 10.1073/pnas.1106300109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability — an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–8. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 42.Bakhoum SF, Compton DA. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J Clin Invest. 2012;122:1138–43. doi: 10.1172/JCI59954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hao Z, Zhang H, Cowell J. Ubiquitin-conjugating enzyme UBE2C: molecular biology, role in tumorigenesis, and potential as a biomarker. Tumor Biol. 2011;33:723–30. doi: 10.1007/s13277-011-0291-1. [DOI] [PubMed] [Google Scholar]
- 44.Perez de Castro I, Malumbres M. Mitotic Stress and Chromosomal Instability in Cancer: The Case for TPX2. Genes Cancer. 2013;3:721–30. doi: 10.1177/1947601912473306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shenoy N, Kessel R, Bhagat T, Bhattacharya S, Yu Y, Mcmahon C, et al. Alterations in the ribosomal machinery in cancer and hematologic disorders. J Hematol OncolJ Hematol Oncol. 2012;5:32. doi: 10.1186/1756-8722-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roschke AV, Rozenblum E. Multi-Layered Cancer Chromosomal Instability Phenotype. Front Oncol [Internet] 2013:3. doi: 10.3389/fonc.2013.00302. [cited 2014 Feb 17] Available from: http://www.frontiersin.org/Journal/10.3389/fonc.2013.00302/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.