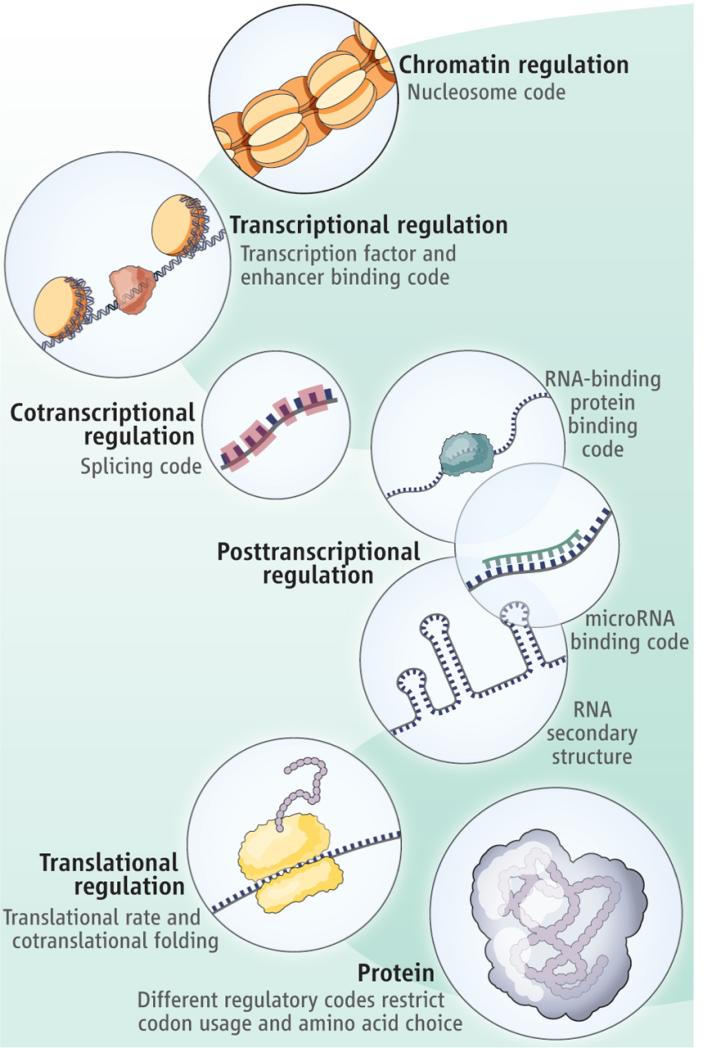
Despite redundancy in the genetic code (1), the choice of codons used is highly biased in some proteins, suggesting that additional constraints operate in certain protein-coding regions of the genome. This suggests that the preference for particular codons, and therefore amino acids in specific regions of the protein, is often determined by factors unrelated to protein structure or function (2, 3). On page 1367 in this issue, Stergachis et al. (4) reveal that transcription factors bind within protein-coding regions (in addition to nearby noncoding regions) in a large number of human genes. Thus, a transcription factor “binding code” may influence codon choice and, consequently, protein evolution. This “binding” code joins other “regulatory” codes that govern chromatin organization (3), enhancers (5, 6), mRNA structure (7), mRNA splicing (3), microRNA target sites (6, 8), translational efficiency (9), and cotranslational folding (10), all of which have been proposed to constrain codon choice, and thus protein evolution (see the figure).
Figure. Constraining codes. Regulatory elements within protein-coding regions (such as transcription factor binding) can influence codon choice and amino acid preference that are independent of protein structure or function.
Redundancy in the genetic code might facilitate the existence of multiple overlapping regulatory codes within protein-coding regions of the genome.
How widespread is the phenomenon of “regulatory” codes that overlap the genetic code, and how do they constrain the evolution of protein sequences? Stergachis et al. address these questions for the transcription factor–binding regulatory code. They use deoxyribonuclease I (DNase I) footprinting to map transcription factor occupancy (a protein bound to DNA can protect that region from enzymatic cleavage) at nucleotide resolution across the human genome in 81 diverse cell types. The authors determined that ~14% of the codons within 86.9% of human genes are occupied by transcription factors. Such regions, called “duons,” therefore encode two types of information: one that is interpreted by the genetic code to make proteins and the other, by the transcription factor–binding regulatory code to influence gene expression. This requirement for transcription factors to bind within protein-coding regions of the genome has led to a considerable bias in codon usage and choice of amino acids, in a manner that is constrained by the binding motif of each transcription factor.
To investigate whether single-nucleotide variants within duons affect transcription factor binding, Stergachis et al. mapped the known variants that are associated with a disease or a trait onto duons. Of those, 17.4% quantitatively skew the allelic origins of DNA fragments protected from cleavage by DNase I in human cells, suggesting that such single-nucleotide variants affect transcription factor occupancy. They also determined that such variants are not biased toward whether they result in synonymous or nonsynonymous changes in the protein sequence. Intriguingly, a large fraction of the variants that result in a nonsynonymous change are predicted not to alter protein function. This indicates that some variants within duons might primarily affect transcription factor binding instead. This supports the emerging idea that single-nucleotide variants within protein-coding regions can lead to disease without affecting protein structure or function (11, 12). Thus, the whole spectrum of “regulatory” codes within protein-coding regions should be considered when assessing the impact of single-nucleotide variants and interpreting disease mutation data from exome sequencing (only the protein-coding regions of the genome) and cancer genome studies.
Do the regulatory codes harmoniously coexist? Evidence is emerging that there can be conflicts. For example, in the fruit fly Drosophila melanogaster, there is a striking decrease in the use of codons that are optimal for translation, but a rise in codons that enhance RNA splicing, toward the end of exons (13). This may indicate that the requirement for accurate RNA splicing has superseded that for optimal translation. Likewise, Stergachis et al. observed that the binding motifs of transcription factors within protein-coding genomic regions are selectively devoid of sequences that contain a stop codon.
What features might permit synergistic coexistence of the regulatory and genetic codes? One major constraint of protein-coding genes is the requirement for the encoded polypeptide segment to fold into a defined tertiary structure. It is possible that in regions where folding constraints are not present, such as in intrinsically disordered regions (14), there might be increased tolerance for protein-coding genomic regions to harbor more regulatory elements that can be interpreted by different regulatory codes.
Stergachis et al. make a number of important genome-scale observations, but several mechanistic questions remain to be answered. For instance, although the authors report a weak tendency for transcription factors to preferentially bind to the protein-coding regions of highly expressed genes, it is unclear how the binding of a transcription factor within protein-coding regions mechanistically influences the expression of a gene. Perhaps this type of binding might result in alternative promoters with different transcriptional start sites or affect the expression of neighboring genes (by acting as a distal enhancer element, for example). It is also unclear whether binding of a transcription factor within a protein-coding region may not directly affect gene expression but instead determine the formation and maintenance of higher-order chromatin structure.
Future research will need to determine the number of overlapping codes that can be tolerated by the genetic code. There is also the question of possible trade-offs, in terms of maintaining regulation and functionality, that have been made to accommodate coexistence of codes and whether this can lead to nonoptimal or deleterious consequences. For instance, protein-coding regions that cannot tolerate mutations due to multiple overlapping codes may be exploited by pathogens during host infection. The investigation of overlapping codes opens new vistas on the functional interpretation of variation in coding regions and makes it clear that the story of the genetic code has not yet run its course.
Acknowledgments
We thank the Medical Research Council for support.
References
- 1.Nirenberg M. Trends Biochem Sci. 2004;29:46. doi: 10.1016/j.tibs.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Itzkovitz S, Hodis E, Segal E. Genome Res. 2010;20:1582. doi: 10.1101/gr.105072.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warnecke T, Weber CC, Hurst LD. Biochem Soc Trans. 2009;37:756. doi: 10.1042/BST0370756. [DOI] [PubMed] [Google Scholar]
- 4.Stergachis AB, et al. Science. 2013 In Press. [Google Scholar]
- 5.Birnbaum RY, et al. Genome Res. 2012;22:1059. doi: 10.1101/gr.133546.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin MF, et al. Genome Res. 2011;21:1916. doi: 10.1101/gr.108753.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shabalina SA, Ogurtsov AY, Spiridonov NA. Nucleic Acids Res. 2006;34:2428. doi: 10.1093/nar/gkl287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brest P, et al. Nat Genet. 2011;43:242. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 9.Fredrick K, Ibba M. Cell. 2010;141:227. doi: 10.1016/j.cell.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pechmann S, Frydman J. Nat Struct Mol Biol. 2013;20:237. doi: 10.1038/nsmb.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauna ZE, Kimchi-Sarfaty C. Nat Rev Genet. 2011;12:683. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 12.Plotkin JB, Kudla G. Nat Rev Genet. 2011;12:32. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warnecke T, Hurst LD. Mol Biol Evol. 2007;24:2755. doi: 10.1093/molbev/msm210. [DOI] [PubMed] [Google Scholar]
- 14.Babu MM, Kriwacki RW, Pappu RV. Science. 2012;337:1460. doi: 10.1126/science.1228775. [DOI] [PubMed] [Google Scholar]