Abstract
The rare AGG codon in Escherichia coli has been reassigned to code non-canonical amino acids using the pair. When Nε-alloc-lysine was used as a PylRS substrate, close to quantitative occupancy of Nε-alloc-lysine at an AGG codon site was achieved in minimal medium.
Keywords: Genetic code expansion, AGGA codon, AGG codon, sense codon reassignment, non-canonical amino acids
Pioneered by Schultz and others, using orthogonal aminoacyl-tRNA synthetase (aaRS)-suppressor tRNA pairs for the delivery of unique amino acids at nonsense and four-base codons have allowed the incorporation of more than 100 non-canonical amino acids (ncAAs) into proteins in living cells of Escherichia coli, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, and mammals.1 These ncAAs consist of diverse unique functionalities that can be applied for both basic studies and biotechnological development. Several broadly used orthogonal pairs have been constructed for this purpose, including one derived from the Methanocaldococcus jannaschii tyrosyl-tRNA synthetase (TyrRS)-tRNATyr pair for use in E. coli, two derived from the E. coli TyrRS-tRNATyr and leucyl-tRNA synthetase-tRNALeu pairs for use in eukaryotic cells, and the naturally existing pyrrolysyl-tRNA synthetase (PylRS)-tRNAPyl pair for use in both bacterial and eukaryotic cells.1e, 2 All four aaRS-tRNA pairs are primarily used for reassigning the amber UAG codon and two have been extended to reassign the opal UGA, ochre UAA, and four-base AGGA codons.3 Using all four codons to code ncAAs typically suffers low ncAA incorporation yields. For three nonsense codons, suppressor tRNAs have to compete against release factors (RFs) for their recognition during translation; for the four-base AGGA codon, two endogenous arginyl-tRNAPyl variants, and , recognize the first three nucleotides and potentially lead to misreading during translation. To improve amber suppression efficiency, cells with an orthogonal ribosome, a truncated ribosome with slow RF1 binding, and RF1 knockout have been engineered.4 In these cells, the fact that translation inclines to stop at amber codon still remains. Previously we reported that the PylRS-tRNAPyl pair could be reprogrammed for reassigning three nonsense codons.5 Here, we wish to show that the same pair can be engineered to reassign the rare AGG codon almost quantitatively to code ncAAs, resolving the low ncAA incorporation efficiency issue that is typically observed in codon reassignment experiments.
Flexible tRNA anticodon recognition of PylRS was previously reported.6 By simply mutating the anticodon of tRNAPyl, we showed that the pair could be successfully reprogrammed to reassign opal and ochre codons for coding ncAAs such as Nε-(t-butyloxycarbonyl)-lysine (BocK), Nε-(allyloxycarbonyl)-lysine (AllocK), and Nε-(t-propargyloxycarbonyl)-lysine (ProK) (Scheme 1).5 However, when we attempted to use the same strategy to reassign a four-base AGGA codon at the 134th amino acid site of a sequence-optimized superfolder green fluorescent protein (sfGFP) gene, a full-length protein with Arg134 was predominantly produced, indicating strong frameshift suppression from endogenous arginyl-tRNAArg. The presence of 5 mM BocK in the growth medium did not lead to a significant level of BocK incorporation at the AGGA codon site.6c However, when a different GFP strain, GFPUV with no sequence optimization and an AGGA mutation at its 149th amino acid site was used for a similar test, GFPUV with BocK149 was successfully produced in cells that expressed the pair and were grown in the presence of BocK (Figure 1A). The molecular weight of the purified protein determined by electrospray ionization mass spectrometry (ESI-MS) agreed well with its theoretical value (Figure 1B). In the absence of BocK, only a minimal amount of full-length GFPUV could be detected, indicating a very low basal level of frameshift suppression from endogenous arginyl-tRNAArg. To investigate effects of arginine on the AGGA suppression, we prepared an arginine-dropout auto-induction (AI) medium (see supplementary information for recipe) and used it to test BocK incorporation efficiencies at AGGA in the presence of different concentrations of arginine. As shown in Figure 1A, adding 0.1 mM arginine in the medium improved the expression yield of GFPUV, but the yield continued to drop when more arginine was provided. We suspect that the expression increase in the presence of 0.1 mM arginine was due to the relief of translation restriction at regular arginine codons and the decrease with further addition of arginine was caused by increasing misreading of the first three nucleotides of AGGA by endogenous arginyl-tRNAArg. These data strongly suggest that misreading at the first three nucleotides of AGGA is a competing process during the translational reassignment of AGGA to code a ncAA.
Scheme 1.
Three ncAAs used in the study.
Figure 1.
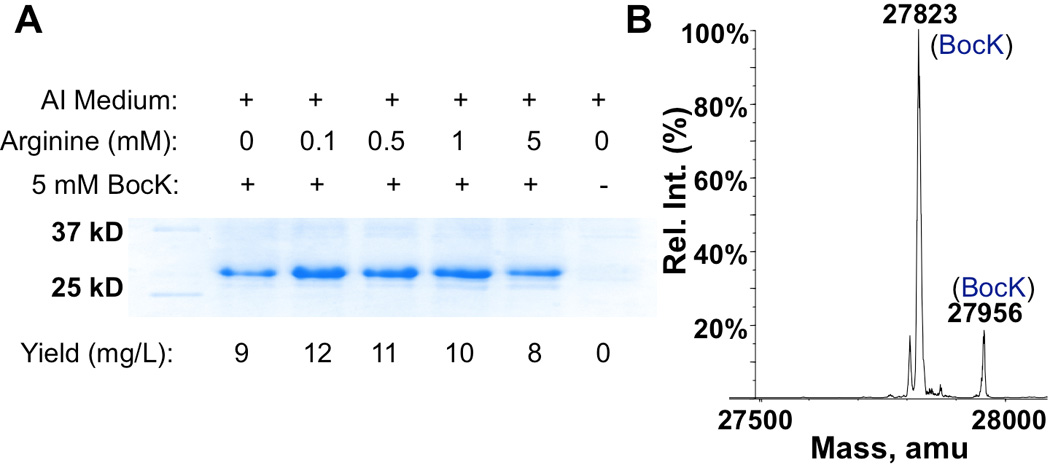
(A) Effects of arginine on the AGGA suppression. E. coli BL21(DE3) cells coding PylRS, , and GFPUV with an AGGA mutation at the 149 position and under control of an inducible T7 promoter were grown in AI medium supplemented with varying concentrations of arginine and BocK. Protein expression was induced by the addition of 0.5 mM IPTG. (B) The ESI-MS spectrum of purified GFPUV with BocK149. Theoretical molecular weights of full-length protein and full-length protein without Met1 are 27824 Da and 27955 Da, respectively.
Our AGGA suppression results with GFPUV prompted us to investigate the AGGA codon reassignment in sfGFP further. Since the sfGFP gene was codon-optimized, a potentially fast translation rate of its mRNA transcript might have favoured frameshift suppression at AGGA from arginyl-tRNAArg. To inquire this possibility, codons of five residues at the immediate N-terminal side of the 134th position of sfGFP were mutated to relatively rarely used codons. However, growing cells harbouring this gene, PylRS, and still led to the predominant production of sfGFP with Arg134 both in the absence and in the presence of BocK in LB medium (Figure 2). Using low temperature and GMML (M9 minimal medium with 1% glycerol and 0.3 mM leucine) for slow translation afforded similar results. At the current stage, we are still puzzled by conflicting AGGA suppression results for GFPUV and sfGFP. We suggest precautions when using the AGGA codon to code ncAAs. Although this further study did not unveil underlying reasons for frameshift suppression at AGGA from arginyl-tRNAArg, a close examination of the ESI-MS spectrum of sfGFP produced in the presence of BocK revealed two small peaks that did match theoretic molecular weights of sfGFP with BocK134 (Figure 2C). Therefore, was indeed able to deliver BocK at the AGGA mutation site but could not efficiently compete against endogenous arginyl-tRNAArg for binding to this particular site.
Figure 2.
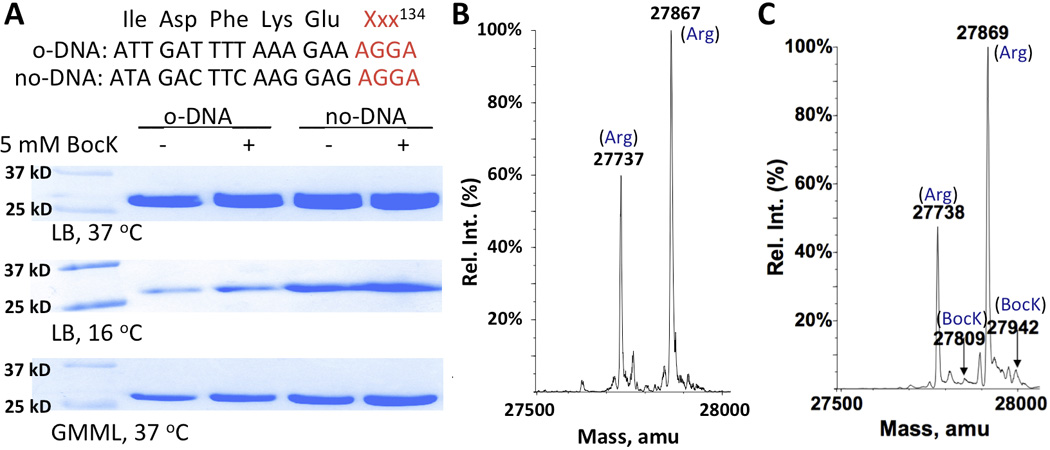
(A) Frameshift suppression at AGGA of sequence-optimized (opt) and sequence-unoptimized (unopt) sfGFP genes. E. coli BL21(DE3) cells coding PylRS, , and a sfGFP gene (opt or unopt) with an AGGA mutation at the 134th position and under control of an inducible T7 promoter were used for the test. Protein expression was induced with the addition of 0.5 mM IPTG. (B) The ESI-MS spectrum of sfGFP expressed in E. coli BL21(DE3) cells that coded PylRS, , and sequence-optimized sfGFP with an AGGA mutation at the 134th position and were grown at 37 °C in LB. Theoretic molecular weights are 27867 Da for sfGFP with Arg134 and 27735 Da for sfGFP with Arg134 but without Met1. (C) The ESI-MS spectrum of sfGFP expressed in E. coli BL21(DE3) cells that coded PylRS, , and sequence-unoptimized sfGFP with an AGGA mutation at the 134th position and were grown at 37 °C in LB with 5 mM BocK. Theoretic molecular weights are 27940 Da for sfGFP with BocK134 and 27809 Da for sfGFP with BocK134 but without Met1.
Since ribosome is a catalytic machinery for translating three-nucleotide codons, its binding to may be disfavoured, making potentially disadvantageous in competing against arginyl-tRNAArg for binding to AGGA. Deleting the first nucleotide from the anticodon of to form will presumably improve its binding to ribosome. This new tRNAPyl mutant may be well charged by PylRS and efficiently compete against arginyl-tRNAArg in the recognition of the rare AGG codon, leading to its reassignment to code ncAAs. To examine this potential, a plasmid pET-AGG-GFPUV149AGG that carried genes coding PylRS, , and GFPUV with a single AGG codon at its 149th position was constructed. In this plasmid, PylRS and GFPUV149AGG are under control of an IPTG-inducible T7 promoter and is under control of a constitutive lpp promoter. Growing E. coli BL21(DE3) cells transformed with pET-AGG-GFPUV149AGG in 2×YT medium expressed GFPUV with Arg149 (Figure 3A). Providing 10 mM BocK in the medium led to the synthesis of two GFPUV variants, one with Arg149 and the other with BocK149 (Figure 3B). Integrating ESI-MS peaks of two variants indicated that GFPUV with BocK149 was roughly 24% of total GFPUV variants compared to 15% when 5 mM BocK was provided in the medium (Supplementary Figure 1). Therefore PylRS is active toward that does competitively recognize AGG during translation. Since 2×YT medium has a high concentration of arginine that promotes the reading of the AGG codon by endogenous arginyl-tRNAArg, we shifted to GMML to do further test. Growing E. coli BL21(DE3) cells transformed with pET-AGG-GFPUV149AGG in GMML led to a dominant production of GFPUV with BocK149 (Figure 3C). Among the total GFPUV variants, GFPUV with BocK149 accounts for 80%. To further increase the incorporation of BocK at the AGG codon site, an argW antisense RNA was coexpressed in E. coli BL21(DE3) cells that were transformed with pET-AGG-GFPUV149AGG. ArgW codes . Expression of its antisense RNA potentially reduces the recognition of AGG by endogenous . Shown in Figure 3D, coexpressing argW antisense RNA did increase the proportion of GFPUV with BocK149 in totally expressed GFPUV variants to 83% but was not significantly better than in cells with no argW antisense RNA expressed.
Figure 3.
ESI-MS spectra of GFPUV variants expressed in different conditions. (A) E. coli BL21(DE3) cells with pET-AGG-GFPUV149AGG were grown in 2×YT medium and induced with the addition of 0.5 mM IPTG. (B) E. coli BL21(DE3) cells with pET-AGG-GFPUV149AGG were grown in 2×YT medium and induced with the addition of 0.5 mM IPTG and 10 mM BocK. (C) E. coli BL21(DE3) cells with pET-AGG-GFPUV149AGG were grown in GMML and induced with the addition of 0.5 mM IPTG and 10 mM BocK. (D) E. coli BL21(DE3) cells co-transformed with pET-AGG-GFPUV149AGG and another plasmid containing an argW antisense RNA gene were grown in GMML and induced with the addition of 0.5 mM IPTG and 10 mM BocK.
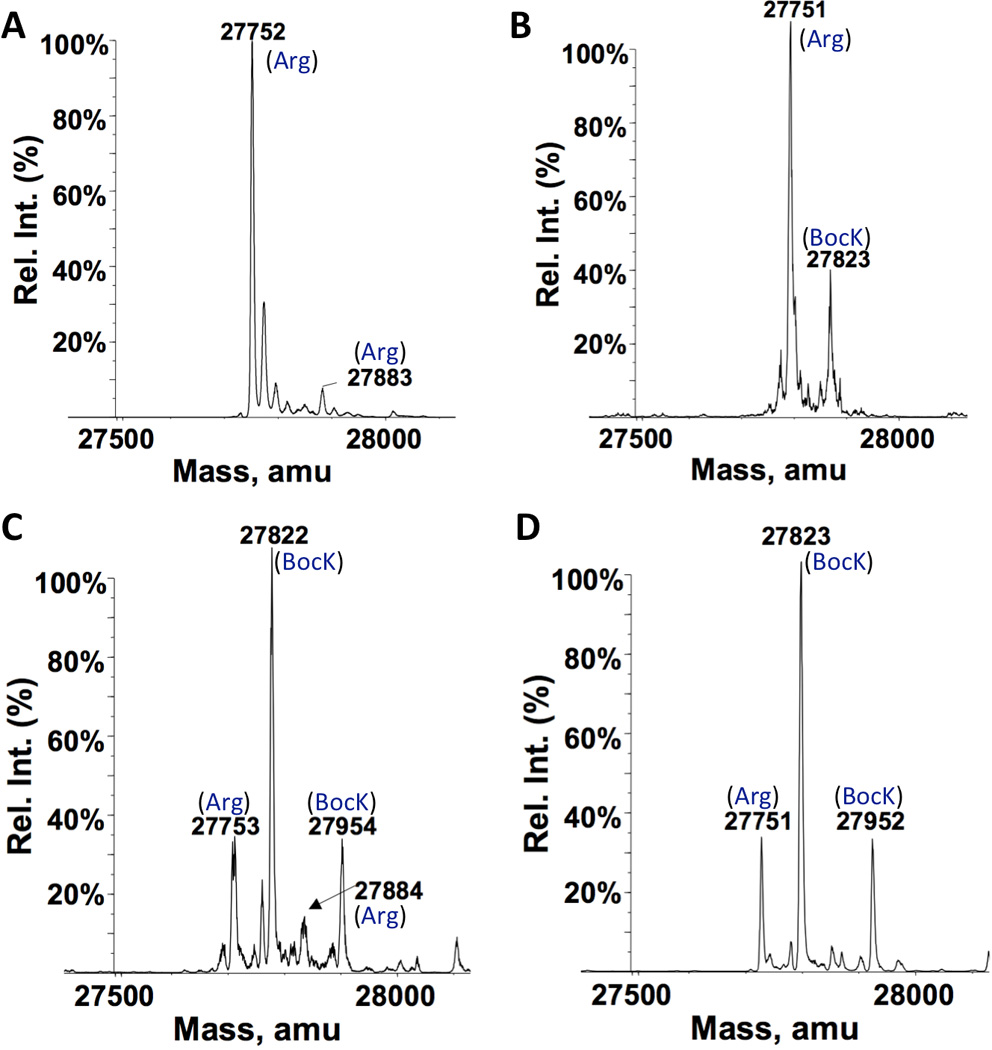
In experiments described in the previous paragraph, both IPTG and BocK were added into the medium at the same time to induce protein expression. Since PylRS was under control of an IPTG-inducible promoter, it surely took some time for PylRS to be expressed and achieve steady activity to charge with BocK after induction. During this time, some GFPUV with Arg149 had already been expressed since its expression was also induced by IPTG. One way to reduce this undesired expression of GFPUV with Arg149 is to achieve a stable level of before GFPUV expression is initiated. A separate plasmid pEVOL-AGG was constructed for this purpose. This plasmid contains genes coding PylRS under control of an arabinose-regulated pBAD promoter and under control of a constitutive proK promoter. The E. coli BL21-AI strain was chosen for the following protein expression studies due to the fact that this stain allows T7 RNA polymerase and a target gene to be expressed separately. E. coli BL21-AI has a T7 RNA polymerase gene under control of an arabinose-regulated pBAD promoter in its chromosome. As such, in this strain, T7 RNA polymerase can be expressed to a stable level by the addition of arabinose before IPTG is provided to induce expression of a target protein. Since AGG codon reassignment may potentially affect the translation of T7 RNA polymerase itself, stably expressed T7 RNA polymerase before the induction with IPTG and BocK will provide enough functional enzyme for expressing a target protein. E. coli BL21-AI cells were co-transformed with pEVOL-AGG and pET-AGG-GFPUV149AGG. A single transformant was then used to express GFPUV. Cells were grown in GMML to OD 0.5. arabinose, BocK, and IPTG were added sequentially in 60 min apart to induce the expression of GFPUV. ESI-MS analysis of purified GFPUV indicated a predominant GFPUV variant with BocK149 that accounted for 89% of total expressed GFPUV that had an expression level of 30 mg/L (Figure 4A). This represents a significant increase from 80% that was observed in the previous study with BL21(DE3) transformed with pET-AGG-GFPUV134AGG. We also tested another two PylRS substrates, AllocK and ProK. For AllocK, 92% of total expressed GFPUV with an expression level of 36 mg/L was the variant with AllocK149 (Figure 4B). This high incorporation efficiency at AGG is probably due to AllocK as a better substrate of PylRS than BocK. For ProK, a significant proportion of GFPUV with Arg149 (32%) was in the finally expressed GFPUV that had an expression level of 33 mg/L, indicating ProK is probably a weaker substrate of PylRS than BocK (Figure 4C). The GFPUV expression level in the absence of a ncAA was 24 mg/L. We also tested the toxic effects of the AGG reassignment. E. coli BL21-AI cells co-transformed with pEVOL-AGG and pET-AGG-GFPUV149AGG were grown with or without 10 mM ProK and induced with the addition of arabinose and IPTG. Cells in media with BocK grew faster than those in media without the addition of BocK, indicating a very low toxicity of the AGG reassignment to cells (Supplementary Figure 2). A faster growth pattern for cells in the presence of BocK might be due to that BocK could be metabolized to provide nutrients for cell growth. We also investigated the global incorporation of a ncAA at proteins whose genes naturally have AGG codons. Lysates of cells grown in the presence of 10 mM ProK were labelled with 3-azido-7-hydroxyl-coumarin using the copper-catalyzed click reaction. There were only a few proteins which could be fluorescently labelled (Supplementary Figure 3). This is not a surprise given that AGG is a rare codon and arginine has important structural roles in protein folding. When ProK replaces arginine, it may likely causes a protein not properly folded and then cleared out in cells. Since we previously observed strong arginine incorporation at the AGGA mutation site of the 134th amino acid position of sfGFP, we also tested whether a similar observation is associated with an AGG mutation at this site. A plasmid pET-AGG-sfGFP134AGG was constructed by swapping the GFPUV gene in pET-AGG-GFPUV149AGG with the sequence-optimized sfGFP gene with an AGG codon at its 134th amino acid position. This plasmid together with pEVOL-AGG was used to transform E. coli BL21-AI cells. A single transformant was then employed to produce sfGFP by following the same procedure for the production of GFPUV. BocK was used in this investigation. As presented in Figure 4D, sfGFP with BocK134 was the major sfGFP variant and accounted for 70% of total expressed sfGFP. Therefore, is competitive in recognizing this site. The low incorporation of BocK at the AGGA mutation of the same site by must be due to the weak binding of to ribosome, though we do not fully understand how arginyl-tRNAPyl could induce so strong frameshift suppression at an AGGA mutation site.
Figure 4.
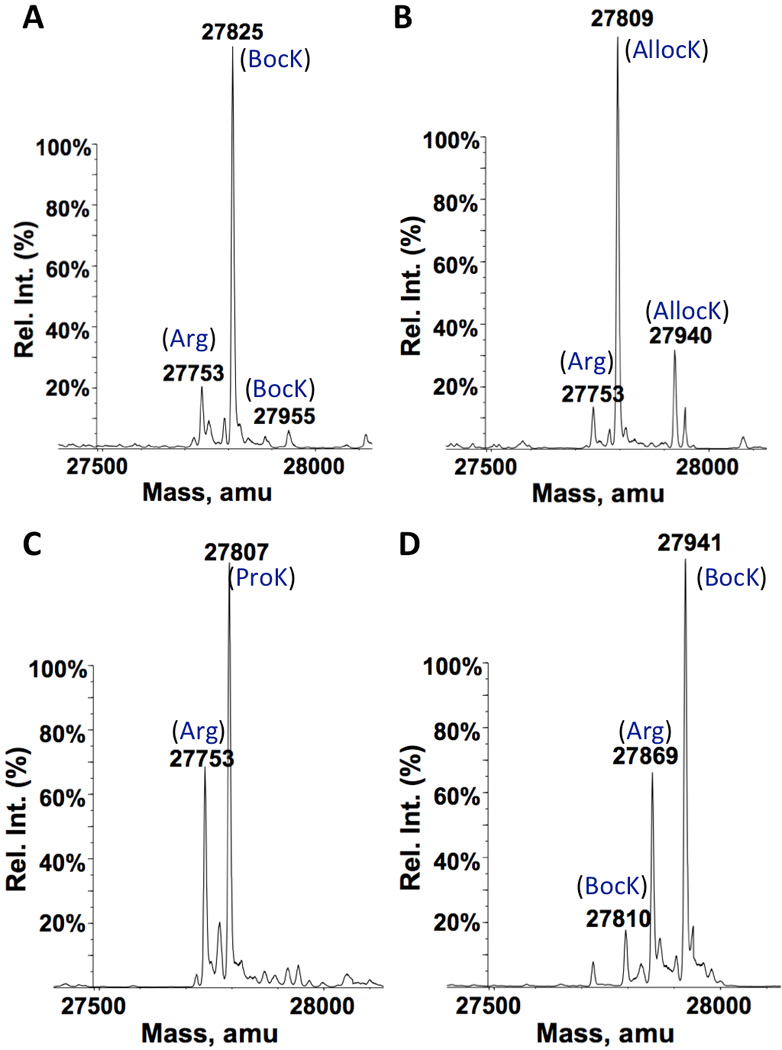
ESI-MS spectra of GFPUV and sfGFP variants expressed in different conditions. For A–C, E. coli BL21-AI cells transformed with pEVOL-AGG and pET-AGG-GFPUV149AGG were grown in GMML to OD 0.5. 0.2% arabinose, 10 mM ncAA (BocK for A, AllocK for B, and ProK for C), and 0.5 mM IPTG was provided in a sequential order with 1 h apart to induce the expression of GFPUV. For D, E. coli BL21-AI cells were transformed with pEVOL-AGG and pET-AGG-sfGFP134AGG. The expression condition of sfGFP was as same as in A. Theoretic molecular weights are 27867 Da for sfGFP with Arg134, 27940 Da for sfGFP with BocK134, and 27809 Da for sfGFP with BocK134 but without Met1.
ProK has a terminal alkyne. Its genetic incorporation followed by click labelling with azides have been used for site-selective protein labelling. This strategy was also applied to label GFPUV that was expressed in the conditions shown in Figure 4C. GFPUV expressed in the presence of ProK could be selectively labelled with a fluorescein azide (Figure 5).7 However, a same labelling reaction was not able to label GFPUV that was expressed in the absence of ProK. The labelling data and the ESI-MS analysis shown in Figure 4C collectively support the genetic incorporation of ProK at the AGG codon site.
Figure 5.
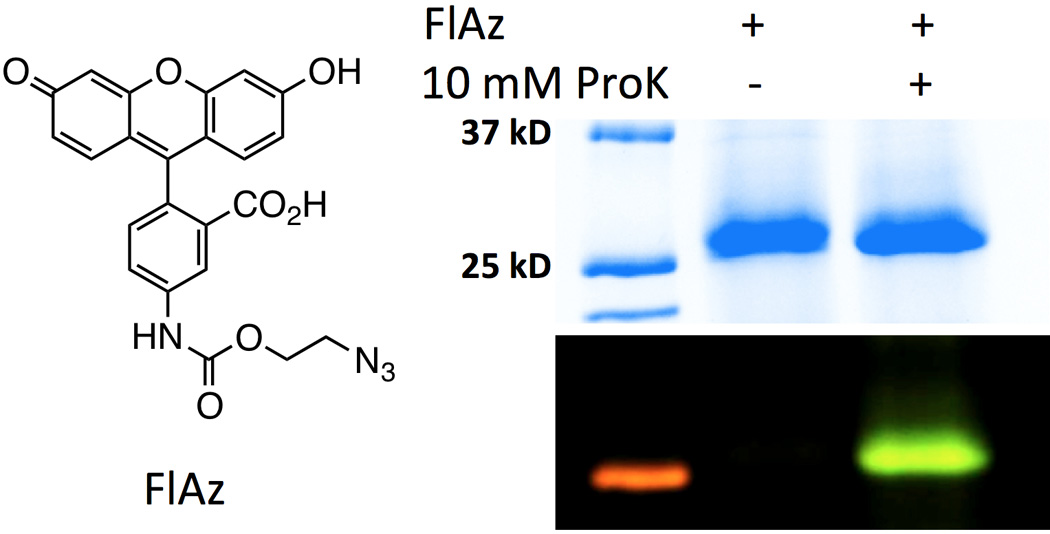
Labelling of GFPUV that were expressed in the absence and in the presence of ProK. Cells and expression conditions were as same as shown in Figure 4C except that GFPUV shown in the second lane was produced in the absence of ProK. To label GFPUV, Two purified GFPUV samples (1mg/mL) was incubated with 100 µM CuSO4, 1 mM NiCl, 0.5 mM tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine, and 2.5 mM FlAz, followed by 5 mM sodium ascorbate, RT for 3 h.
In summary, we have shown that the rare AGG codon can be reassigned to code ncAAs using the pair. When AllocK was used as a PylRS substrate, close to quantitative occupancy at the AGG codon was achieved in minimal medium. The other two PylRS substrates, BocK and ProK were also successfully incorporated at the AGG codon with high efficiencies. So far, PylRS and its engineered mutants have been shown to recognize close to more than 90 ncAAs.1i, 2d, 8 These ncAAs can be potentially incorporated at the AGG codon with varying efficiencies depending on their activities toward corresponding enzymes. Since AGG is a sense codon, the reported approach resolves the typical low ncAA incorporation issue that has been pegged to the ncAA mutagenesis and therefore allow bulk preparation of proteins with site-selectively incorporated ncAAs for applications such as therapeutic protein production. One unresolved obstacle of the current method is the production of a protein mixture with either a ncAA or an arginine at a designated AGG site. However, a ncAA such as ProK can be easily modified such as with polyethylene glycol. The modified protein can then be separated from the arginine version. Church et al. recently reported an E. coli cell strain in which RF1 was removed and all 321 TAG codons in the E. coli genome were replaced with TAA codons.9 The same technique can be applied to construct an E. coli cell strain that has all AGG codons replaced with other arginine codons and removed. This new cell strain will allow the exclusive incorporation of a ncAA at an AGG codon site. A cell strain that has both TAG and AGG reassigned can also be potentially constructed for the easy production of proteins with two different ncAAs.3a, 10
Experimental Section
Details of plasmid construction, protein expression, ESI-MS analysis, and protein labelling are presented in the supplementary materials.
Supplementary Material
Acknowledgements
We thank Dr. Yohannes H. Reznom from the Laboratory for Biological Mass Spectrometry at Texas A&M University for characterizing proteins with electrospray-ionization mass spectrometry. This work was supported in part by the National Institutes of Health (grant 1R01CA161158), the National Science Foundation (grant CHEM-1148684), and the Welch Foundation (grant A-1715).
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.20xxxxxxx.
References
- 1.a) Furter R. Protein Sci. 1998;7:419–426. doi: 10.1002/pro.5560070223. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang L, Brock A, Herberich B, Schultz PG. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]; c) Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]; d) Liu W, Brock A, Chen S, Chen S, Schultz PG. Nat. Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]; e) Wu N, Deiters A, Cropp TA, King D, Schultz PG. J. Am. Chem. Soc. 2004;126:14306–14307. doi: 10.1021/ja040175z. [DOI] [PubMed] [Google Scholar]; f) Hancock SM, Uprety R, Deiters A, Chin JW. J. Am. Chem. Soc. 2010;132:14819–14824. doi: 10.1021/ja104609m. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Bianco A, Townsley FM, Greiss S, Lang K, Chin JW. Nat. Chem. Biol. 2012;8:748–750. doi: 10.1038/nchembio.1043. [DOI] [PubMed] [Google Scholar]; h) Greiss S, Chin JW. J. Am. Chem. Soc. 2011;133:14196–14199. doi: 10.1021/ja2054034. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Mukai T, Kobayashi T, Hino N, Yanagisawa T, Sakamoto K, Yokoyama S. Biochem. Biophys. Res. Commun. 2008;371:818–822. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]; j) Liu CC, Schultz PG. Annu. Rev. Biochem. 2010;79:413–414. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]; k) Wan W, Tharp JM, Liu WR. Biochim. Biophys. Acta. 2014;1844:1059–1070. doi: 10.1016/j.bbapap.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Wang L, Schultz PG. Chem. Biol. 2001;8:883–890. doi: 10.1016/s1074-5521(01)00063-1. [DOI] [PubMed] [Google Scholar]; b) Chin JW, Cropp TA, Chu S, Meggers E, Schultz PG. Chem. Biol. 2003;10:511–519. doi: 10.1016/s1074-5521(03)00123-6. [DOI] [PubMed] [Google Scholar]; c) Blight SK, Larue RC, Mahapatra A, Longstaff DG, Chang E, Zhao G, Kang PT, Green-Church KB, Chan MK, Krzycki JA. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]; d) Neumann H, Peak-Chew SY, Chin JW. Nat. Chem. Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 3.a) Wan W, Huang Y, Wang Z, Russell WK, Pai PJ, Russell DH, Liu WR. Angew. Chem. Int. Ed. Engl. 2010;49:3211–3214. doi: 10.1002/anie.201000465. [DOI] [PubMed] [Google Scholar]; b) Neumann H, Slusarczyk AL, Chin JW. J. Am. Chem. Soc. 2010;132:2142–2144. doi: 10.1021/ja9068722. [DOI] [PubMed] [Google Scholar]; c) Niu W, Schultz PG, Guo J. ACS Chem. Biol. 2013;8:1640–1645. doi: 10.1021/cb4001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Rackham O, Chin JW. J. Am. Chem. Soc. 2005;127:17584–17585. doi: 10.1021/ja055338d. [DOI] [PubMed] [Google Scholar]; b) Huang Y, Russell WK, Wan W, Pai PJ, Russell DH, Liu W. Mol. Biosyst. 2010;6:683–686. doi: 10.1039/b920120c. [DOI] [PubMed] [Google Scholar]; c) Johnson DB, Xu J, Shen Z, Takimoto JK, Schultz MD, Schmitz RJ, Xiang Z, Ecker JR, Briggs SP, Wang L. Nat. Chem. Biol. 2011;7:779–786. doi: 10.1038/nchembio.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Odoi KA, Huang Y, Rezenom YH, Liu WR. PLoS One. 2013;8:357035. doi: 10.1371/journal.pone.0057035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Ambrogelly A, Gundllapalli S, Herring S, Polycarpo C, Frauer C, Soll D. Proc. Natl. Acad. Sci. USA. 2007;104:3141–3146. doi: 10.1073/pnas.0611634104. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. J. Mol. Biol. 2008;378:634–652. doi: 10.1016/j.jmb.2008.02.045. [DOI] [PubMed] [Google Scholar]; c) O'Donoghue P, Prat L, Heinemann IU, Ling J, Odoi K, Liu WR, Soll D. FEBS Lett. 2012;586:3931–3937. doi: 10.1016/j.febslet.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu B, Wang Z, Huang Y, Liu WR. Chembiochem. 2012;13:1405–1408. doi: 10.1002/cbic.201200281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Polycarpo CR, Herring S, Berube A, Wood JL, Soll D, Ambrogelly A. FEBS Lett. 2006;580:6695–6700. doi: 10.1016/j.febslet.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. Chem. Biol. 2008;15:1187–1197. doi: 10.1016/j.chembiol.2008.10.004. [DOI] [PubMed] [Google Scholar]; c) Chen PR, Groff D, Guo J, Ou W, Cellitti S, Geierstanger BH, Schultz PG. Angew. Chem. Int. Ed. Engl. 2009;48:4052–4055. doi: 10.1002/anie.200900683. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Fekner T, Li X, Lee MM, Chan MK. Angew. Chem. Int. Ed. Engl. 2009;48:1633–1635. doi: 10.1002/anie.200805420. [DOI] [PubMed] [Google Scholar]; e) Li WT, Mahapatra A, Longstaff DG, Bechtel J, Zhao G, Kang PT, Chan MK, Krzycki JA. J. Mol. Biol. 2009;385:1156–1164. doi: 10.1016/j.jmb.2008.11.032. [DOI] [PubMed] [Google Scholar]; f) Li X, Fekner T, Ottesen JJ, Chan MK. Angew. Chem. Int. Ed. Engl. 2009;48:9184–9187. doi: 10.1002/anie.200904472. [DOI] [PubMed] [Google Scholar]; g) Nguyen DP, Lusic H, Neumann H, Kapadnis PB, Deiters A, Chin JW. J. Am. Chem. Soc. 2009;131:8720–8721. doi: 10.1021/ja900553w. [DOI] [PubMed] [Google Scholar]; h) Ai HW, Lee JW, Schultz PG. Chem. Commun. (Camb) 2010;46:5506–5508. doi: 10.1039/c0cc00108b. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Gautier A, Nguyen DP, Lusic H, An W, Deiters A, Chin JW. J. Am. Chem. Soc. 2010;132:4086–4088. doi: 10.1021/ja910688s. [DOI] [PubMed] [Google Scholar]; j) Groff D, Chen PR, Peters FB, Schultz PG. Chembiochem. 2010;11:1066–1068. doi: 10.1002/cbic.200900690. [DOI] [PMC free article] [PubMed] [Google Scholar]; k) Huang Y, Wan W, Russell WK, Pai PJ, Wang Z, Russell DH, Liu W. Bioorg. Med. Chem. Lett. 2010;20:878–880. doi: 10.1016/j.bmcl.2009.12.077. [DOI] [PubMed] [Google Scholar]; l) Li X, Fekner T, Chan MK. Chem. Asian. J. 2010;5:1765–1769. doi: 10.1002/asia.201000205. [DOI] [PMC free article] [PubMed] [Google Scholar]; m) Wang YS, Wu B, Wang Z, Huang Y, Wan W, Russell WK, Pai PJ, Moe YN, Russell DH, Liu WR. Mol. Biosyst. 2010;6:1557–1560. doi: 10.1039/c002155e. [DOI] [PubMed] [Google Scholar]; n) Chou CJ, Uprety R, Davis L, Chin JW, Deiters A. Chemical Science. 2011;2:480–483. [Google Scholar]; o) Gautier A, Deiters A, Chin JW. J. Am. Chem. Soc. 2011;133:2124–2127. doi: 10.1021/ja1109979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lajoie MJ, Rovner AJ, Goodman DB, Aerni HR, Haimovich AD, Kuznetsov G, Mercer JA, Wang HH, Carr PA, Mosberg JA, Rohland N, Schultz PG, Jacobson JM, Rinehart J, Church GM, Isaacs FJ. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]; b) Xiao H, Chatterjee A, Choi SH, Bajjuri KM, Sinha SC, Schultz PG. Angew. Chem. Int. Ed. Engl. 2013;52:14080–14083. doi: 10.1002/anie.201308137. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chatterjee A, Xiao H, Bollong M, Ai HW, Schultz PG. Proc. Natl. Acad. Sci. USA. 2013;110:11803–11808. doi: 10.1073/pnas.1309584110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


