Abstract
MicroRNAs (miRNAs) are short non-coding RNAs that have been recognized to regulate the expression of uncountable number of genes. Their aberrant expression has been found to be linked to the pathology of many diseases including cancer. There is a drive to develop miRNA targeted therapeutics for different diseases especially cancer. Nevertheless, reining in these short non-coding RNAs is not as straightforward as originally thought. This is in view of the recent discoveries that miRNAs are under epigenetic regulations at multiple levels. Exportin 5 protein (XPO5) nuclear export mediated regulation of miRNAs is one such important epigenetic mechanism. XPO5 is responsible for exporting precursor miRNAs through the nuclear membrane to the cytoplasm, and is thus a critical step in miRNA biogenesis. A number of studies have shown that variations in components of the miRNA biogenesis pathways, particularly the aberrant expression of XPO5, increase the risk of developing cancer. In addition to XPO5, the Exportin 1 protein (XPO1) or chromosome region maintenance 1 (CRM1) can also carry miRNA export function. These findings are supported by pathway analyses that reveal certain miRNAs as direct interaction partners of CRM1. An in depth understanding of miRNA export mediated regulatory mechanisms is important for the successful design of clinically viable therapeutics. In this review, we describe the current knowledge on the mechanisms of miRNA nuclear transport mediated regulation and propose strategies to selectively block this important mechanism in cancer.
Keywords: XPO5, XPO1, CRM1, Nuclear Export, Small Molecule Inhibitors, Selective Inhibitors of Nuclear Export
Introduction
The miRNAs are short non-coding RNAs that are approximately 18–25 nucleotides in length and are found in all animal and plant cells. The first miRNAs were recognized in Caenorhabditis elegans by Lee and colleagues [1]. In 2001, various small regulatory RNAs were discovered in plants and mammals and designated as the ‘microRNA’ [2-4]. At present there are more than 1,250 human miRNAs registered in the miRBase database (the miRBase sequence database is the major repository for published data on miRNA sequence and annotation) [5]. The miRNAs have been extensively studied for their involvement in RNA interference (RNAi) that results in regulation of different gene expression post-transcriptionally, and they are known to contribute to diverse physiological and pathophysiological functions, including regulation of tissue developmental [6], tissue differentiation [7], cell signaling regulation [8], stem cell regulation [9] and most importantly in carcinogenesis [10]. The biogenesis of miRNA and their RNAi functions have been intensively investigated (how miRNAs are synthesized and processed into a mature form, and how they influence gene expression) [11]. Furthermore, developments in miRNA-related technologies, such as miRNA expression profiling and synthetic oligoRNA, have contributed to the identification of miRNAs that are known to be involved in a number of physiological and pathological phenotypes. A Pubmed search (performed on May 14, 2013) for ‘microRNA’ returns >22,516 hits similarly key words ‘microRNA’ and ‘cancer’ return >9,133 research articles. Most interestingly, an evaluation of research publications from 2000 to 2013 shows an exponential increase in research indicating that the field is advancing rapidly (Fig. 1). These studies have enhanced the understanding of microRNA biogenesis, their regulatory control on different genes and strategies to target them for anti-cancer therapeutic benefits. However, some questions remain largely unanswered, such as how miRNA expression is controlled and which genes are regulated by each miRNA in a specific disease condition.
Fig. (1). MicroRNA Research in Numbers.
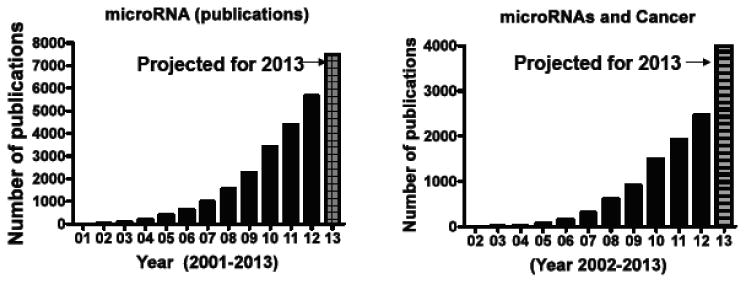
Graph showing Pubmed listings returned from key word search for [A] microRNAs and [B] microRNAs and Cancer (on May 14th 2013). The graph depicts research publications in the last 13 years. The 2013 publications are projected with a dashed line.
Nuclear Export is a Key Step in the MIRNA Biogenesis Process
Species such as ribosomal subunits, tRNAs, mRNAs, and other forms of RNA especially miRNAs are generated in the nucleus and exported to the cytoplasm [12, 13]. Nucleocytoplasmic shuttling of biomolecules (small RNAs, proteins) and drugs occurs through nuclear pore complexes, which penetrate the nuclear envelope and permit passage of material either by passive diffusion [14] or facilitated (carrier mediated) translocation [15]. Passive diffusion is rapid for small molecules, but limits to species with size <40 KDa [16]. In contrast, facilitated translocation allows passage of large structures with a molecular mass >40 KDa all the way up to several million Daltons. Facilitated translocation involves a receptor and is coupled to an input of metabolic energy (active transport), allowing accumulation of cargoes against gradients of chemical activity [17]. Extensive research in nuclear cytoplasmic shuttling mechanisms have lead to the consensus that all known RNA transport pathways are mediated by proteinaceous factors [18].
Earlier studies on RNA nuclear transport revealed that the mRNA export proceeds through a RanGTPase-independent mechanism, whereby NPC passage is facilitated by the yeast Mex67p/Mtr2p complex or its higher eukaryotic counterpart, the NXF (TAP)/p15 complex [19] in which the metabolic energy is fed in by RNA-helicases such as Dbp5p [20]. Other type of cellular RNAs, such as tRNA and U1 snRNA, are exported in a RanGTP-dependent manner [21-23]. Two exportins function in tRNA export, namely, exportin-t [24] and Exportin 5 (XPO5) [13]. This review focuses on miRNA nuclear export as discussed below.
The biogenesis of miRNAs is a multi-step process that involves multiple proteins [25]. MiRNA biogenesis begins with the expression of a primary ∼1000 nucleotide miRNA transcript designated as the pri-miRNA Pri-miRNA is cleaved to produce a ∼70 nucleotide precursor that is termed as pre-miRNA The pre-miRNAs are transported from the nucleus into the cytoplasm, where they are enzymatically processed into mature miRNAs [26]. The very critical step of nuclear export is performed by XPO5 [27]. XPO5 is a member of the importin-β family of proteins that comprise one major class of nucleocytoplasmic transporters and function as the major exporters of nuclear RNAs [28]. Okada and colleagues were the first group to capture the high resolution crystal structure of XPO5 bound to miRNA showing that XPO5 binds directly to its pre-miRNA cargo in a RanGTP-dependent manner [29]. Additionally, XPO5 can recognize and export structured RNAs that are unrelated to pre-miRNAs, including viral mini-helix RNA and tRNA, along with certain other proteins, [30, 31]. Earlier studies have verified that XPO5 can stabilize pre-miRNA leading to the assumption that XPO5 over-expression also increases the pool of pre-miRNAs that are available for nuclear export [32].
It has also been demonstrated that XPO5 plays a role in siRNA biogenesis and therefore is a key point of intersection between the siRNA and miRNA pathways [33]. Over-expression of XPO5 has been shown to result in enhanced miRNA activity that competes with Dicer which suggests that XPO5-mediated nuclear export of pre-miRNAs may be a rate-limiting step in miRNA biogenesis [34]. Conversely, genetic defects or loss of XPO5 binding results in reduced pre-miRNA expression and function [35]. The prognostic value of XPO5 in cancer is emerging and the importance of XPO5 in the miRNA pathway suggests that structural alterations in this transporter could potentially impact global miRNA expression, thereby altering an individual's risk of developing cancer.
Castanotto and colleagues have shown that miRNAs also use CRM1 for nuclear-cytoplasmic shuttling [36]. In their experiments, inhibition of CRM1 by the natural product inhibitor Leptomycin B resulted in nuclear accumulation of miRNA guide sequences. The authors also showed that nuclear to cytoplasmic transport could be inhibited by small interfering RNAs against CRM1, indicating that this pathway is shared by different classes of miRNAs. Additionally the authors also found that CRM1 co-immunoprecipitates with Ago-1, Ago-2, Topo2α, EzH2, and Mta, consistent with a role of Argonautes and small RNAs in chromatin remodeling. In another study Bussing and colleagues, identified the role of the Caenorhabditis elegans nuclear export receptor XPO1 and the cap-binding proteins CBP-20/NCBP-2 and CBP-80/NCBP-1 in miRNA maturation process [37]. The RNA interference of these genes retarded heterochronic phenotypes similar to those observed for animals with mutations in the let-7 miRNA or core miRNA machinery genes. Moreover, pre- and mature miRNAs become depleted, whereas primary miRNA transcripts accumulate. An involvement of XPO1 in miRNA biogenesis was found to be conserved in Drosophila, in which knockdown of XPO1 or its chemical inhibition through Leptomycin B (a CRM1 inhibitor) causes pri-miRNA accumulation. Their findings demonstrate that XPO-1/Emb promotes the pri-miRNA-to-pre-miRNA processing leading the authors to propose that this function involves intranuclear transport and/or nuclear export of primary miRNAs. The nuclear import of miRNA processed guide sequences by XPO1 and the interaction of XPO1 with different chromatin protein complexes raises important questions about the compartment dependent role of miRNA. There are a number of unexplored avenues such as localization and function of miRNAs in different organelles (nucleolus, mitochondria and exosomes, elles (nucleolus, mitochondria and exosomes, vesicles and microparticles) which add another level of complexity to the functions of these important small RNAs.
Structure of Pre-MIRNA and XPO5 Interaction
Recently Lee and colleagues have reviewed in detail the x-ray structures and biology of the interaction between pre-miRNAs and XPO5 [38]. Being a Ran dependent process, the XPO5:RanGTP complex was shown to form a glove (mitt) like structure in which N-terminal HEAT repeats 6–13 form the outer side of the glove and C-terminal HEAT repeats 14–21 form the thumb region of the glove. HEAT repeats 6–19 of all 21 repeats are involved in binding pre-miRNA (schema in Fig. 2). The pre-miRNA aligns in the inside of the XPO5:RanGTP protein surface. The double-stranded (ds) stem region of the pre-miRNA is weakly caught in the glove and is stabilized in the XPO5:RanGTP complex by electrostatic interactions with a small number of residues within a radius of 3.5 A° to form a broad contact surface. Additionally, there are numerous secondary interactions that stabilize binding within a radius of ∼5 A°. The 3′ 2-nt overhang structure of the pre-miRNA in the tunnel is stabilized by additional hydrogen bonds and salt bridges with the basic residues in XPO5. It is speculated that XPO5 recognize a variety of 3′-nt sequences of pre-miRNAs as seven out of nine interactions involve atoms of the sugar-phosphate backbone, recognition of the 3′ 2-nt overhang by XPO5 is projected to be independent of RNA sequence and resembles the recognition interaction between the XPO5:RanGTP complex and the pre-miRNA stem. Ran contacts pre-miRNA using a limited surface area (83 A°2).
Fig. (2). Nuclear Export of pre-miRNAs a Key Step in Biogenesis Process.
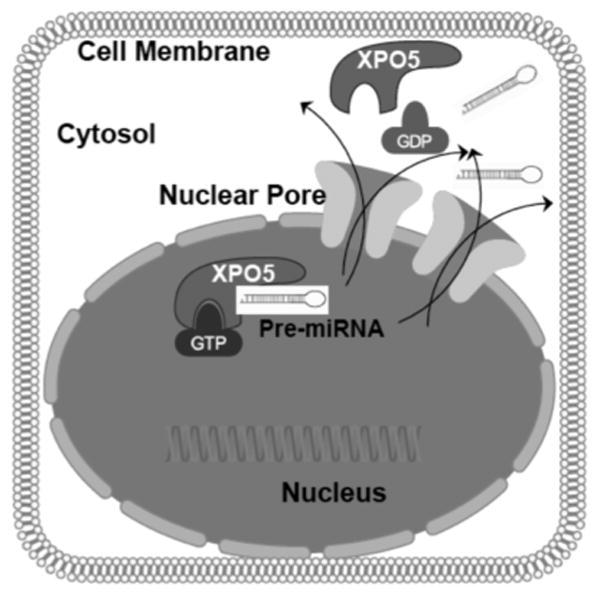
Precursor miRNAs (pre-miRNA) are exported from the nucleus with the aid of specialized protein exporters [primarily by exportin 5 (XPO5) and to a less extent exportin 1 (XPO1)]. Nuclear export begins with the binding of pre-miRNA to XPO5 in a RanGTP dependent manner. Once the pre-miRNA is exported to the cytoplasm, the RanGTP coverts to GDP bringing conformational change in the XPO5 resulting in the release of pre-miRNA from the XPO5 structure. (The color version of the figure is available in the electronic copy of the article).
Like the binding studies of XPO5-RanGTP, a number of studies have also shed light on the x-ray structure of the CRM1:RanGTP:RanBP1 complex [39]. In this case, the loop of the CRM1 HEAT 9 acts as an allosteric inhibitor of nuclear exclusion signal sequence (NES) binding because binding of RanBP1 displaces the HEAT 9 loop by C-terminal acidic residues of Ran. The displaced HEAT 9 loop interacts with the inner helix of HEATs 11–12, resulting in NES release by changing the conformation of the NES binding site in the outer helices of HEATs 11–12. Based on the simulation results, the authors suggested a different dissociation mechanism by RanBP1 binding to XPO5 or XPO-t (see review [38] for details). The model structure constructed from the RanGTP:RanBP1 and XPO-t:RanGTP:tRNA structures suggests that RanBP1 binding makes RanGTP C-terminus bump into XPO-t, thereby actively disassembling the tRNA export complex in the cytoplasm. This model agrees with previous conclusions that RanBP1 and RanGAP facilitate RanGTP hydrolysis by XPO-t and tRNA. However, XPO5 may have a different dissociation mechanism, and XPO5:RanGTP:dsRNAs may not be destabilized by RanBP1 because superimposition of the RanBP1:RanGTP structure onto the XPO5:RanGTP:pre-miRNA structure suggests that RanBP1 and the C-terminus of Ran would not sterically clash with XPO5 or with pre-miRNA. The rapid release of pre-miRNA from XPO5 upon translocation across the NPCs may not be favorable because free small RNAs are easily degraded (unlike stably folded tRNA). XPO5 might resist RanBP1 (or RanBP2)-mediated cargo release to protect pre-miRNA from degradation until the ternary export complex encounters the processing enzyme Dicer for the final maturation of miRNA in the cytoplasm.
Nuclear Transport Influences MRNA Regulation by MiRNAS
Not only has the nuclear transport dependent regulation of biogenesis of miRNAs been demonstrated, a number of studies have also shown that nuclear transport also affects the mRNA targets that are under miRNA control. Some of the earliest studies in this direction were performed by Rui and colleagues where they showed that the XPO5-mediated nuclear export of pre-miRNAs and the shRNAs can represent a rate-limiting step in miRNA and shRNA-derived siRNA biogenesis and function [40]. In this study the evidence in support of the hypothesis included the demonstration that Xpo5 over-expression specifically enhanced targeted inhibition of gene expression by an overexpressed miRNA or shRNA. This effect was specific, as XPO5 over-expression did not enhance RNAi mediated by an artificial siRNA duplex, which is predicted to gain direct access to the cytoplasm without requiring XPO-mediated nuclear export. Using an example of miR-30a miRNA and its precursor pre-miR-30 the authors showed that XPO5 over-expression enhanced miR-30a expression by ∼2 fold and enhanced the cytoplasmic expression of pre-miR-30, the target for nuclear export by XPO5, by ∼5 fold. Collectively their data demonstrated that XPO5 function is rate limiting for the biogenesis and function of over-expressed miRNAs and shRNAs in transfected human cancer cells. Their findings also suggested that XPO5 expression may also increase the pool of pre-miRNAs that are bound to be exported out of the nucleus.
Truesdell and colleagues have reported that microRNA-mediated enhancement of target mRNAs in oocytes is dependent on nuclear entry of the microRNAs [41]. In their study cytoplasmically-injected microRNAs was shown to repress target mRNAs. Components of the activated microRNP, AGO, FXR1 (FXR1-iso-a) and miR16 were found to be present in the nucleus and cytoplasm. Importantly, the authors found that microRNA target mRNAs for upregulation, Myt1, TNFα and a reporter bearing the TNFα AU-rich microRNA target sequence, are associated with AGO in immature oocyte nuclei and AGO2 in G0 human nuclei, respectively. mRNAs that are repressed or lack target sites are not associated with nuclear AGO. Cross linking-coupled immunopurification revealed greater association of AGO2 with FXR1 in the nucleus compared to cytoplasm. Consistently, overexpression of FXR1-iso-a rescues activation of cytoplasmically-injected RNAs and in low density, proliferating cells. These data indicate the importance of a compartmentalized AGO2-FXR1-iso-a complex for selective recruitment for microRNA-mediated upregulation.
Prognostic Value of XPO5-MIRNA Interrelationship in Cancer
To investigate the causes of breast cancer aggressiveness, Leaderer and colleagues performed genetic and epigenetic association studies of XPO5 in a case control study of breast cancer [42]. They first genotyped two missense SNPs in XPO5, rs34324334 (S241N) and rs11544382 (M1115T), and evaluated the methylation levels in the XPO5 promoter region for blood DNA samples from a breast cancer case-control study. Their primary findings included the capture of variant genotypes of rs11544382 that were associated with breast cancer risk compared to the homozygous commonly observed genotype. When stratified by menopausal status, the variant alleles of both rs11544382 and rs34324334 were found to be significantly associated with breast cancer risk in post-menopausal women. Their methylation analysis demonstrated that the “high” and combined “high/middle” tertiles of methylation index were associated with reduced risk of breast cancer. In conclusion the authors data corroborated by data from a publicly available tissue array, which showed lower levels of XPO5 expression in healthy controls relative to tumor or adjacent tissues from breast cancer patients with tumor tissue exhibiting the highest expression levels. These findings support the hypothesis that variations in components of the miRNA biogenesis pathway, most importantly the XPO5, may affect an individual's risk of developing breast cancer.
Very recently Iwasaki and colleagues demonstrated that that XPO5 mediated global elevation of miRNAs is critical for proper control of gene expression program during the key steps of cell cycle entry [43]. In their studies the authors showed that XPO5 is promptly induced during cell cycle entry by a PI3K-dependent post-transcriptional regulation. As a proof of concept, the Inhibition of XPO5 induction was shown to interfere with global miRNA elevation that results in a G1/S dependent proliferation defect. From these studies the authors concluded that during cell cycle entry, XPO5 plays a key role as a critical molecular hub controlling the gene expression program through global regulation of miRNAs. These data indicated that XPO5-mediated global miRNA elevation regulates a wide range of cellular events associated with cell cycle control. Similarly, Han and colleagues investigated in urothelial carcinoma model the effect of expression patterns of Dicer, Drosha, and XPO5 on the cell proliferation inhibition and apoptosis induced by silencing these genes [44]. In their studies the authors showed that all the three genes were up-regulated in bladder urothelial carcinoma compared to matched normal urothelium. Most importantly, Dicer, Drosha, and XPO5 expression levels were found to be directly correlated with high grade as well as invasive carcinomas. Additionally, silencing Dicer, Drosha, and more specifically XPO5 induced cell proliferation inhibition and apoptosis in bladder urothelial carcinoma T24 and 5637 cells. These studies indicate that XPO5 over expression regulates proliferative potential of bladder carcinomas and inhibition of this master exporter could become an attractive therapeutic strategy against different cancers.
Network Analysis Reveals MIR Targets of XPO5
Advancements in computational tools particularly systems biology and network analysis methods have helped the understanding of complex biological processes such as biological networks, drug interaction network and miRNAs [45-50]. The applications of computational tools range from microRNAs target identification to understanding the network of genes regulated by an individual microRNA. It is through systems levels investigations that researchers have been able to stratify the miRNA targets in a disease specific manner [51]. Pathway analysis tools have allowed the deeper evaluations of epigenetic regulatory mechanisms of miRNAs in different diseases particularly cancer [52]. Nevertheless, there is hardly any study that investigates, in a systems way, the effect nuclear transporter XPO5/XPO1-pre-miRNA or mRNA transport in a holistic manner (recently reviewed by Muqbil and colleagues [53]). The closest that any study has reached to systems level analysis are the investigations from Muralidhar and colleagues that showed that the most significantly up-regulated transcript following 5p gain was the microRNA (miRNA) processor Drosha, XPO5 and Dicer [54]. Drosha copy-number and expression were not elevated in pre-malignant cervical squamous intraepithelial lesions. Importantly, global miRNA profiling showed that Drosha over-expression in cervical SCC appears to be of functional significance. In their unsupervised principal component analysis of a mixed panel of cervical SCC cell lines and clinical specimens the authors showed statistically significant separation according to Drosha over-expression. miRNAs most significantly associated with Drosha over-expression were directly implicated in carcinogenesis in other distinct tissues, suggesting that they regulate fundamental processes in neo-plastic progression. Their evidence suggests that copy-number driven over-expression of Drosha and consequent changes in miRNAs are likely to be important contributors to the selective advantage provided by 5p gain in cervical neo-plastic progression. These studies indicate that more systems level work evaluating the XPO1/XPO5 on pre- and mature miRNAs in a global context is needed. Such investigations are expected to unwind the complexity of nuclear export regulatory mechanisms of miRNAs and may lead to the identification of novel agents that can interfere with the process in a controlled manner leading to some form of miRNA therapy.
Targeted Inhibition of XPO1 And XPO5 as a MIRNA Therapeutic Strategy
The importance of nuclear transport in drug resistance and disease pathology has been established [55]. Due to their central role in disease pathology, a number of agents have been designed to target the members of the nuclear transport family proteins [56, 57]. These drugs were designed with the idea that nuclear retention of major tumor suppressor proteins such as p53, FOXO, p27 and others can result in selective cancer cell death. Nevertheless the first generation compounds such as Leptomycin B (LMB) proved highly toxic in the clinic and were discontinued from use in patients [58]. Semi-synthetic derivatives of LMB have been developed, however, their clinical utility is yet to be tested [59]. Since the initial setback a number of newer and less toxic nuclear export inhibitors have been developed that have shown superior pre-clinical efficacy in both solid tumors and hematological malignancies [60, 61]. Such inhibitors have demonstrated broad efficacy against a spectrum of cancer types [62-68]. Our laboratory has demonstrated the pre-clinical efficacy of one such agent i.e. selective inhibitor of nuclear export (SINE) in pancreatic and non Hodgkin’s Lymphoma cell lines and xenograft models [69, 70]. The SINEs are undergoing Phase I clinical evaluation for both solid tumors and Non Hodgkin’s lymphomas. However, SINE and other inhibitors have been designed against XPO1 and not XPO5, the latter being the primary exporter of miRNA indicating that new class of XPO5 inhibitors are needed. It should be noted that XPO5 inhibition and the consequent nuclear retention of pre-miRNAs is bound to have adverse effects as well. This is simply because, a number of XPO5 target miRNAs have critical role in normal cell homeostasis. One can only speculate that global retention of miRNAs is cancer or diseased cells that carry aberrant genetic material will be subject to cell genome surveillance while normal cells (carrying normal genome) will be protected from the adverse effects (Fig. 3. summary diagram for exportin targeted drugs). Computational biology, particularly mathematical modeling and systems biology has been proposed to help understand these context dependent effects of XPO5 or CRM1 inhibition in cancer vs normal cells. Such unanswered questions make the nuclear retention of miRNAs through nuclear export inhibition an exciting yet unchartered territory worth exploring. Such a strategy may lead to development of newer therapeutic modality that target global nuclear retention of disease promoting miRNAs.
Fig. (3). Nuclear Export Inhibition as a Potential Therapeutic Strategy against miRNAs.
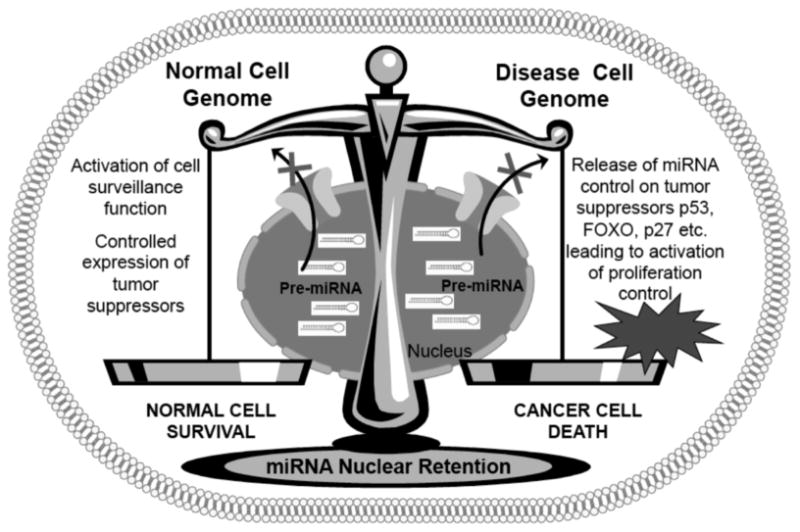
Targeted inhibition of nuclear export protein XPO5 and XPO1 can in principle, retain pre-miRNAs in cell nucleus. However, the impact of such global re-organization in normal cell is not known. We speculate that in diseased cells (carrying aberrant genome), the nuclear retention of pre-miRNA would suppress their negative control on major tumor suppressors such as p53, FOXO, p27 and other important cell surveillance molecules. This release from miRNA posttranslational control will allow the proper expression of different tumor suppressors resulting in activation of cell proliferation control pathways, apoptosis pathways and other surveillance mechanisms resulting in elimination of cancer cells. On the other hand in normal cells, the normal genome will block the over-activation of major tumor suppressors resulting in minimal toxicity to normal cells. (The color version of the figure is available in the electronic copy of the article).
Conclusions
miRNAs regulate uncountable number of genes and have been implicated for their roles in disease development, sustenance and drug resistance. A number of strategies have been suggested to tame in these important small RNAs. Nevertheless, we are still a long way from realizing their full potential as therapeutics against different diseases especially cancer. Even though a lot of work has been done on miRNA biogenesis mechanisms, more needs to be learned on their nuclear export regulation. Since nuclear export precede maturation of miRNAs indicating this important point as a viable therapeutic strategy against these small RNAs. The exportin family of transport proteins have been established to play a key role in the nuclear export of the pre-miRNAs transport. Nevertheless, there are no studies that have investigated the global impact of drugging these nuclear exporters. Additionally, there is less information on how mature RNAs are transported back in the nucleus. This is primarily due to the lack of understanding as to how the miRNA nuclear transport is regulated in normal and disease conditions. These critical points in miRNA biology are proposed to serve as a new form of therapeutic strategy. At present, more needs to be learned such as how the normal cells will react to inhibition of this critical nuclear export and related family members. Understanding the nature of export function in greater detail will lead to better miRNA therapeutic strategies that focus on their localization function within the tumor cell and may positively impact the treatment of cancer.
Acknowledgments
NIH R21 1R21CA16984801 and 1R21CA17597401 to RMM is acknowledged. We are thankful to Karyopharm Therapeutic for young investigator award to ASA.
Footnotes
Conflict Of Interest: The author(s) confirm that this article content has no conflicts of interest.
References
- 1.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 2.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9(2):175–9. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–9. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 4.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 5.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–38. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 6.Eulalio A, Mano M, Dal Ferro M, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492(7429):376–81. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 7.Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol. 2011;12(6):349–61. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- 8.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 9.Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10(2):116–25. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van KM, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11(9):644–56. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 11.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics. 2010;11(7):537–61. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grunwald D, Singer RH, Rout M. Nuclear export dynamics of RNA-protein complexes. Nature. 2011;475(7356):333–41. doi: 10.1038/nature10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leisegang MS, Martin R, Ramirez AS, Bohnsack MT. Exportin t and Exportin 5: tRNA and miRNA biogenesis - and beyond. Biol Chem. 2012;393(7):599–604. doi: 10.1515/hsz-2012-0146. [DOI] [PubMed] [Google Scholar]
- 14.Sugano K, Kansy M, Artursson P, et al. Coexistence of passive and carrier-mediated processes in drug transport. Nat Rev Drug Discov. 2010;9(8):597–614. doi: 10.1038/nrd3187. [DOI] [PubMed] [Google Scholar]
- 15.Dobson PD, Kell DB. Carrier-mediated cellular uptake of pharmaceutical drugs: an exception or the rule? Nat Rev Drug Discov. 2008;7(3):205–20. doi: 10.1038/nrd2438. [DOI] [PubMed] [Google Scholar]
- 16.Keminer O, Peters R. Permeability of single nuclear pores. Biophys J. 1999;77(1):217–28. doi: 10.1016/S0006-3495(99)76883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guttler T, Gorlich D. Ran-dependent nuclear export mediators: a structural perspective. EMBO J. 2011;30(17):3457–74. doi: 10.1038/emboj.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misteli T. Physiological importance of RNA and protein mobility in the cell nucleus. Histochem Cell Biol. 2008;129(1):5–11. doi: 10.1007/s00418-007-0355-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segref A, Mattaj IW, Ohno M. The evolutionarily conserved region of the U snRNA export mediator PHAX is a novel RNA-binding domain that is essential for U snRNA export. RNA. 2001;7(3):351–60. doi: 10.1017/s1355838201002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17(9):2663–76. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izaurralde E, Mattaj IW. Transport of RNA between nucleus and cytoplasm. Semin Cell Biol. 1992;3(4):279–88. doi: 10.1016/1043-4682(92)90029-u. [DOI] [PubMed] [Google Scholar]
- 22.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78(4):657–68. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 23.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376(6542):709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 24.Kutay U, Lipowsky G, Izaurralde E, et al. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1(3):359–69. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- 25.Libri V, Miesen P, van Rij RP, Buck AH. Regulation of microRNA biogenesis and turnover by animals and their viruses. Cell Mol Life Sci. 2013 Jan 26; doi: 10.1007/s00018-012-1257-1. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 27.Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 2004;14(4):156–9. doi: 10.1016/j.tcb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Brownawell AM, Macara IG. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J Cell Biol. 2002;156(1):53–64. doi: 10.1083/jcb.200110082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okada C, Yamashita E, Lee SJ, et al. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326(5957):1275–9. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 30.Xu D, Farmer A, Chook YM. Recognition of nuclear targeting signals by Karyopherin-beta proteins. Curr Opin Struct Biol. 2010;20(6):782–90. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chook YM, Blobel G. Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature. 1999;399(6733):230–7. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- 32.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 33.Ohrt T, Muetze J, Svoboda P, Schwille P. Intracellular localization and routing of miRNA and RNAi pathway components. Curr Top Med Chem. 2012;12(2):79–88. doi: 10.2174/156802612798919132. [DOI] [PubMed] [Google Scholar]
- 34.Bennasser Y, Chable-Bessia C, Triboulet R, et al. Competition for XPO5 binding between Dicer mRNA, pre-miRNA and viral RNA regulates human Dicer levels. Nat Struct Mol Biol. 2011;18(3):323–7. doi: 10.1038/nsmb.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melo SA, Moutinho C, Ropero S, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18(4):303–15. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 36.Castanotto D, Lingeman R, Riggs AD, Rossi JJ. CRM1 mediates nuclear-cytoplasmic shuttling of mature microRNAs. Proc Natl Acad Sci USA. 2009;106(51):21655–9. doi: 10.1073/pnas.0912384106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bussing I, Yang JS, Lai EC, Grosshans H. The nuclear export receptor XPO-1 supports primary miRNA processing in C. elegans and Drosophila. EMBO J. 2010;29(11):1830–9. doi: 10.1038/emboj.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee SJ, Jiko C, Yamashita E, Tsukihara T. Selective nuclear export mechanism of small RNAs. Curr Opin Struct Biol. 2011;21(1):101–8. doi: 10.1016/j.sbi.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Koyama M, Matsuura Y. An allosteric mechanism to displace nuclear export cargo from CRM1 and RanGTP by RanBP1. EMBO J. 2010;29(12):2002–13. doi: 10.1038/emboj.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 2005;11(2):220–6. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Truesdell SS, Mortensen RD, Seo M, et al. MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci Rep. 2012;2:842. doi: 10.1038/srep00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leaderer D, Hoffman AE, Zheng T, et al. Genetic and epigenetic association studies suggest a role of microRNA biogenesis gene exportin-5 (XPO5) in breast tumorigenesis. Int J Mol Epidemiol Genet. 2011;2(1):9–18. [PMC free article] [PubMed] [Google Scholar]
- 43.Iwasaki YW, Kiga K, Kayo H, et al. Global microRNA elevation by inducible Exportin 5 regulates cell cycle entry. RNA. 2013;19(4):490–7. doi: 10.1261/rna.036608.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han Y, Liu Y, Gui Y, Cai Z. Inducing cell proliferation inhibition and apoptosis via silencing Dicer, Drosha, and Exportin 5 in urothelial carcinoma of the bladder. J Surg Oncol. 2013;107(2):201–5. doi: 10.1002/jso.23214. [DOI] [PubMed] [Google Scholar]
- 45.Bhattacharyya M, Das M, Bandyopadhyay S. A New Approach for Combining Knowledge from Multiple Co-expression Networks of MicroRNAs. IEEE Trans Biomed Eng. 2013 Mar 7; doi: 10.1109/TBME.2013.2250285. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 46.Azmi AS. Systems and Network Biology in Pharmaceutical Drug Discovery. Curr Pharm Des. 2013 Mar 19; doi: 10.2174/138161282001140113122054. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 47.Azmi AS. Adopting network pharmacology for cancer drug discovery. Curr Drug Discov Technol. 2013;10(2):95–105. doi: 10.2174/1570163811310020002. [DOI] [PubMed] [Google Scholar]
- 48.Azmi AS. Network pharmacology: an emerging field in cancer drug discovery. Curr Drug Discov Technol. 2013;10(2):93–94. doi: 10.2174/1570163811310020001. [DOI] [PubMed] [Google Scholar]
- 49.Azmi AS. Network pharmacology for cancer drug discovery: are we there yet? Future Med Chem. 2012;4(8):939–41. doi: 10.4155/fmc.12.44. [DOI] [PubMed] [Google Scholar]
- 50.Azmi AS, Wang Z, Philip PA, Mohammad RM, Sarkar FH. Proof of concept: network and systems biology approaches aid in the discovery of potent anticancer drug combinations. Mol Cancer Ther. 2010;9(12):3137–44. doi: 10.1158/1535-7163.MCT-10-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Satoh J, Tabunoki H. Comprehensive analysis of human microRNA target networks. BioData Min. 2011;4:17. doi: 10.1186/1756-0381-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Azmi AS, Beck FW, Bao B, Mohammad RM, Sarkar FH. Aberrant epigenetic grooming of miRNAs in pancreatic cancer: a systems biology perspective. Epigenomics. 2011;3(6):747–59. doi: 10.2217/epi.11.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muqbil I, Kauffman M, Shacham S, Mohammad RM, Azmi AS. Understanding XPO1 target networks using systems biology and mathematical modeling. Curr Pharm Des. 2013 Mar 19; doi: 10.2174/13816128113199990611. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 54.Muralidhar B, Winder D, Murray M, et al. Functional evidence that Drosha overexpression in cervical squamous cell carcinoma affects cell phenotype and microRNA profiles. J Pathol. 2011;224(4):496–507. doi: 10.1002/path.2898. [DOI] [PubMed] [Google Scholar]
- 55.Turner JG, Dawson J, Sullivan DM. Nuclear export of proteins and drug resistance in cancer. Biochem Pharmacol. 2012;83(8):1021–32. doi: 10.1016/j.bcp.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishi K, Yoshida M, Nishimura M, et al. A leptomycin B resistance gene of Schizosaccharomyces pombe encodes a protein similar to the mammalian P-glycoproteins. Mol Microbiol. 1992;6(6):761–9. doi: 10.1111/j.1365-2958.1992.tb01526.x. [DOI] [PubMed] [Google Scholar]
- 57.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269(9):6320–4. [PubMed] [Google Scholar]
- 58.Newlands ES, Rustin GJ, Brampton MH. Phase I trial of elactocin. Br J Cancer. 1996;74(4):648–9. doi: 10.1038/bjc.1996.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mutka SC, Yang WQ, Dong SD, et al. Identification of nuclear export inhibitors with potent anticancer activity in vivo. Cancer Res. 2009;69(2):510–7. doi: 10.1158/0008-5472.CAN-08-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sakakibara K, Saito N, Sato T, et al. CBS9106 is a novel reversible oral CRM1 inhibitor with CRM1 degrading activity. Blood. 2011;118(14):3922–31. doi: 10.1182/blood-2011-01-333138. [DOI] [PubMed] [Google Scholar]
- 61.Kalid O, Toledo WD, Shechter S, Sherman W, Shacham S. Consensus Induced Fit Docking (cIFD): methodology, validation, and application to the discovery of novel Crm1 inhibitors. J Comput Aided Mol Des. 2012;26(11):1217–28. doi: 10.1007/s10822-012-9611-9. [DOI] [PubMed] [Google Scholar]
- 62.Tai YT, Landesman Y, Acharya C, et al. CRM1 inhibition induces tumor cell cytotoxicity and impairs osteoclastogenesis in multiple myeloma: molecular mechanisms and therapeutic implications. 2013 Apr 16; doi: 10.1038/leu.2013.115. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Etchin J, Sanda T, Mansour MR, et al. KPT-330 inhibitor of CRM1 (XPO1)-mediated nuclear export has selective anti-leukaemic activity in preclinical models of T-cell acute lymphoblastic leukaemia and acute myeloid leukaemia. Br J Haematol. 2013;161(1):117–27. doi: 10.1111/bjh.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Inoue H, Kauffman M, Shacham S, et al. CRM1 Blockade by Selective Inhibitors of Nuclear Export Attenuates Kidney Cancer Growth. J Urol. 2013;189(6):2317–26. doi: 10.1016/j.juro.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lapalombella R, Sun Q, Williams K, et al. Selective inhibitors of nuclear export show that CRM1/XPO1 is a target in chronic lymphocytic leukemia. Blood. 2012;120(23):4621–34. doi: 10.1182/blood-2012-05-429506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang K, Wang M, Tamayo AT, et al. Novel selective inhibitors of nuclear export CRM1 antagonists for therapy in mantle cell lymphoma. Exp Hematol. 2013;41(1):67–78. doi: 10.1016/j.exphem.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 67.Etchin J, Sun Q, Kentsis A, et al. Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells. Leukemia. 2013;27(1):66–74. doi: 10.1038/leu.2012.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ranganathan P, Yu X, Na C, et al. Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood. 2012;120(9):1765–1773. doi: 10.1182/blood-2012-04-423160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Azmi AS, Al-Katib A, Aboukameel A, et al. Selective inhibitors of nuclear export for the treatment of non-Hodgkin's Lymphomas. Haematologica. 2013;98(7):1098–106. doi: 10.3324/haematol.2012.074781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Azmi AS, Aboukameel A, Bao B, et al. Selective inhibitors of nuclear export block pancreatic cancer cell proliferation and reduce tumor growth in mice. Gastroenterology. 2013;144(2):447–56. doi: 10.1053/j.gastro.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]


