Abstract
Fungi (Ascomycota and Basidiomycota) are prolific producers of structurally diverse terpenoid compounds. Classes of terpenoids identified in fungi include the sesqui-, di- and triterpenoids. Biosynthetic pathways and enzymes to terpenoids from each of these classes have been described. These typically involve the scaffold generating terpene synthases and cyclases, and scaffold tailoring enzymes such as e.g. cytochrome P450 monoxygenases, NAD(P)+ and flavin dependent oxidoreductases, and various group transferases that generate the final bioactive structures. The biosynthesis of several sesquiterpenoid mycotoxins and bioactive diterpenoids has been well-studied in Ascomycota (e.g. filamentous fungi). Little is known about the terpenoid biosynthetic pathways in Basidiomycota (e.g. mushroom forming fungi), although they produce a huge diversity of terpenoid natural products. Specifically, many trans-humulyl cation derived sesquiterpenoid natural products with potent bioactivities have been isolated. Biosynthetic gene clusters responsible for the production of trans-humulyl cation derived protoilludanes, and other sesquiterpenoids, can be rapidly identified by genome sequencing and bioinformatic methods. Genome mining combined with heterologous biosynthetic pathway refactoring has the potential to facilitate discovery and production of pharmaceutically relevant fungal terpenoids.
1 Introduction
Fungi are masters of evolution. With a history spanning at least 900 million years,1 fungi have successfully adapted to almost every habitat on earth.2 Conservative estimates place global fungal diversity beyond that of land plants by a ratio of 10:1.3 While there remains some uncertainty regarding the exact number of fungal species that exist,4 it is clear that fungi have been afforded with a unique evolutionary and environmental fitness. Fungi owe this inherent ability to survive to their extensive repertoire of natural product pathways. Many of the natural products from these pathways have antimicrobial, antifungal, immunosuppressive or cytotoxic effects.5–7 These bioactive properties enable fungi to successfully take hold of an ecological niche by conferring the ability to compete for nutrients, to deter predators, and to communicate with other organisms in the environment.8 These same fungal bioactive compounds can also be harnessed by humans to produce valuable medicines such as antimicrobial, anticancer and antiviral agents.9–16 Natural product pathway discovery and engineering from fungi therefore holds great promise for the pharmaceutical industry.
The importance of natural products for the survival of fungi is evidenced by the fact that they often have many different secondary metabolic pathways that are typically clustered within fungal genomes.17 One advantage that this feature presents for natural product discovery and engineering is that the physical clustering of genes facilitates genome mining approaches in the identification of biosynthetic pathways. If one gene in a secondary metabolic pathway can be identified, it is highly likely that the other pathway genes will be closely associated and therefore relatively easily identified by bioinformatic methods.18–20 The recent decrease in the cost of next-generation DNA sequencing technologies21 has led to a large increase in the number of sequenced fungal genomes that are publically available through databases such as the Joint Genome Institute (JGI). While some of the fungal species represented in this database are of interest due to their lignin degrading capabilities,22 recent trends have seen a shift towards genome sequencing and mining strategies to uncover natural product pathways in species that produce medicinally relevant compounds.23–25 With such a unique toolbox, those interested in fungal natural product discovery and biosynthesis can tap into the vast, yet largely unexplored contingent of bioactive natural product pathways.
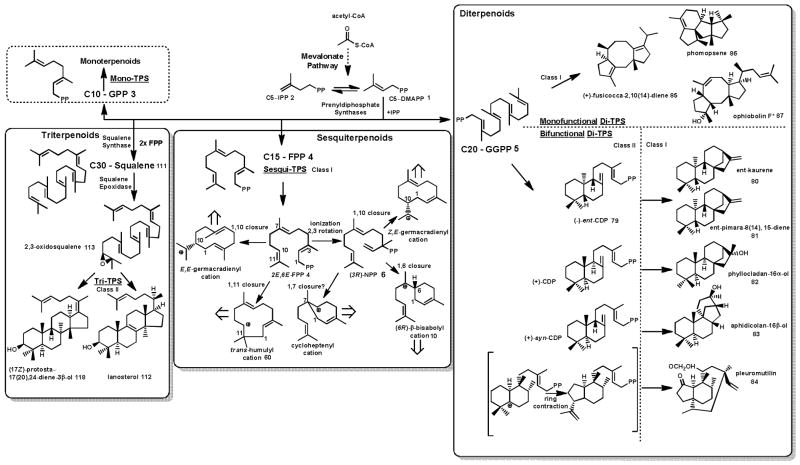
One of the largest groups of bioactive natural products that have been identified is the terpenoids. With over 55,000 initial terpene scaffolds so far described, terpenoids belong to an incredibly structurally diverse class of natural products.26 Despite this diversity, all terpenoids are derived from the simple five carbon precursor molecules dimethylallyl diphosphate (DMAPP) 1 and isopentenyl diphosphate (IPP) 2. In fungi, these two isomers are produced from acetyl-CoA via the mevalonate pathway.27 Condensation of IPP 2 and DMAPP 1 monomers results in linear hydrocarbons of varying length: C10 geranyl pyrophosphate (GPP) 3, C15 (2E, 6E)-farnesyl pyrophosphate ((2E, 6E)-FPP, or FPP) 4, and C20 geranylgeranyl pyrophosphate (GGPP) 5. These linear hydrocarbons undergo a dephosphorylation and cyclization cascade to produce terpenes. This highly complex reaction is catalyzed by enzymes known as terpene synthases.28 Two distinct classes of terpene synthase exist, defined according to substrate activation mechanism. Class I terpene synthases catalyze an ionization-dependent cyclization of substrate, while class II terpene synthases catalyze a protonation-dependent cascade.28, 29,30–32 Depending on the length of the precursor molecule, fungal terpene synthases are known to produce sesquiterpenes (C15), diterpenes (C20) and triterpenes (C30) (Scheme 1). Further biosynthetic pathway enzymes such as cytochrome P450 monooxygenases, oxidoreductases, and different group transferases modify this initial terpene scaffold, producing the final bioactive terpenoid natural product.
Scheme 1. Overview of fungal terpene biosynthesis.
The mevalonate pathway yields the C5 isoprene precursors dimethylallyl diphosphate (DMAPP) 1 and its isomer isopentenyl diphosphate (IPP) 2. Head-to-tail condensation of DMAPP 1 with one, two or three IPP 2 units catalyzed by prenyldiphosphate synthases yields geranyl diphosphate (GPP) 3, farnesyl diphosphate (FPP) 4 and geranylgeranyldiphosphate (GGPP) 5 that are cyclized by ionization-dependent class I or protonation-dependent class II terpene synthases into structurally diverse mono-, sesqui- or diterpenoids. Head-to-head condensation of two FPP 4 molecules yields squalene 111 as the precursor of triterpenoids. The cyclic scaffolds of diterpenoids are synthesized by either monofunctional terpene synthases that contain a class I catalytic domain fused to a prenyldiphosphate domain, or bifunctional terpene synthases that contain class I and class II catalytic domains (see also Scheme 9). Sesquiterpene synthases catalyze different ring-closures of 2E,6E-FPP 4 or upon isomerization, of (3R)-nerolidyl diphosphate (NPP) 6 to generate different sesquiterpenoids. At present, bona fide monoterpene synthases have not been isolated from fungi. CDP: Copalyldiphosphate. *: Ophiobolin F 87 is synthesized from the C25 diphosphate precursor farnesylgeranyl diphosphate.
This review will focus on the biosynthesis of the major fungal terpenoid natural product classes that have been described in the last three decades: the sesquiterpenoids, the diterpenoids and the triterpenoids. To date no bona fide fungal monoterpene synthases have been described. To keep this contribution focused, natural products of mixed isoprenoid biosynthetic origin (e.g. indole-diterpenoids, meroterpenoids) that involve different enzymatic steps to install the terpenoid moieties in these compounds will not be presented here.
Considering that terpenoids are the focus of a large body of literature, this article will discuss examples of the terpene synthases and cyclases responsible for the generation of the different fungal terpenoid scaffolds as well as examples of known terpenoid biosynthetic pathways. Comparisons will be drawn between the terpenomes of the two major fungal divisions: the Ascomycota which includes many well-known filamentous fungi like Aspergillus, Penicillium and Fusarium, and the Basidiomycota which includes the mushroom-forming fungi. Special emphasis will be placed upon the highly diverse and bioactive trans-humulyl cation derived sesquiterpenes produced by the Basidiomycota. Finally, to illustrate the promise of genome mining for the discovery of new bioactive terpenoids from fungi, a description of current bioinformatic approaches used in the identification of sesquiterpenoid biosynthetic pathways in Basidiomycota will be provided as example along with objectives for the development of tools to explore the largely untapped terpenome of this group of fungi.
2 Fungal sesquiterpenoids
2.1 Cyclization of FPP to produce the sesquiterpene scaffold
The sesquiterpenoids are a diverse group of cyclic hydrocarbons, with more than 300 sesquiterpene scaffolds described.33 All sesquiterpenes share in common a C15 backbone derived from the linear precursor FPP 4. Typically, FPP 4 is cyclized by class I terpene synthases known as sesquiterpene synthases, which are characterized by the signature active site motifs DDXXD and NSE.34–36 These amino acid residues play an important role in coordinating the catalytically essential divalent metal ions that stabilize the pyrophosphate group of FPP 4 within the active site cavity.35, 36 Cyclization is initiated by a metal ion induced ionization of the substrate and departure of inorganic pyrophosphate (PPi), which promotes structural displacements including closure of the active site lid.35 The resulting highly reactive carbocation undergoes an initial ring closure at the 1,10 or the 1,11 position. Some enzymes catalyze first the trans-cis isomerization of the 2,3-double-bond of (2E,6E)-FPP 4 to (3R)-nerolidylpyrophosphate 6 prior to generating a cisoid, allylic nerolidyl-carbocation upon PPi cleavage.37, 38 This nerolidyl-carbocation results in a known initial ring closure at either the 1,6 or the 1,10 position; 1,7 ring closure may be possible but has not yet been mechanistically shown (Scheme 1). Subsequent proton shifts, methyl shifts, and complex ring rearrangements are stabilized by the aromatic residues that line the active site cavity of all sesquiterpene synthases.28 The reaction cascade is quenched either by attack by a water molecule,39–41 or by deprotonation which, based on the fact that no active site base has yet been confirmed,42–44 is believed to be mediated by the leaving PPi group.45, 46 The final terpene product and divalent metal ions are released by the enzyme upon opening of the active site lid. Comprehensive mutational analyses and structural studies have provided detailed insights into the catalytic mechanisms used by fungal sesquiterpene synthases, therefore this class of terpene synthase is relatively well understood (reviewed in: 28, 30, 33).
Structures are known for several microbial and plant sesquiterpene synthases.34, 35, 39, 47–52 However, crystal structures have only been solved for two fungal sesquiterpene synthases; aristolochene 7 synthase from Penicillium roqueforti and Aspergillus terreus;46, 48, 52, 53 and trichodiene 8 synthase from Fusarium sporotrichioides.42, 51, 54, 55
Trichodiene 8 synthase (TS) from the plant pathogens Trichothecium roseum38 and Fusarium sporotrichioides was the first characterized fungal enzyme.56 Production of trichodiene 8 (Scheme 2) is the first committed step in the pathway to the trichothecene mycotoxins (see section 2.3) that cause the major cereal crop disease Fusarium head blight (FHB).57 The mechanistic details of TS have been deduced by site directed mutagenesis36 and structure solution of recombinant wild type and D100E enzyme in complex with PPi resulting from incubation with the substrate analogue 2-fluorofarnesyl diphosphate, and in complex with benzyl triethylammonium.42, 51, 54, 58 Removal of PPi from FPP 4 is initiated by three Mg2+ ions, two of which are coordinated by D100, and a third which is coordinated by the triad N225, S229 and E233.51, 54 The resulting cation is isomerized to (3R)-nerolidyl pyrophosphate (NPP) 6 upon recapture of PPi.59 Rotation about the C2–C3 bond allows the intermediate to adopt a cisoid conformation, and departure of PPi results in a 1,6 ring closure to yield a (6R)-β-bisabolyl cation 9. Residues K232, R304 and Y305 act to stabilize PPi, which remains bound at the active site throughout product generation.58 Subsequent hydride transfer and a 7,11 ring closure produces a secondary cuprenyl cation 10.60, 61 Finally, two methyl shifts and deprotonation, possibly mediated by the remaining PPi, results in trichodiene 8 as the major product (Scheme 2), and several minor products including α-cuprenene 11, α-barbatene 12, α- and β-bisabolene 13 and 14.38, 42, 54
Scheme 2. Examples of sesquiterpenoids generated by 1,6-cyclization of FPP upon isomerization to NPP.
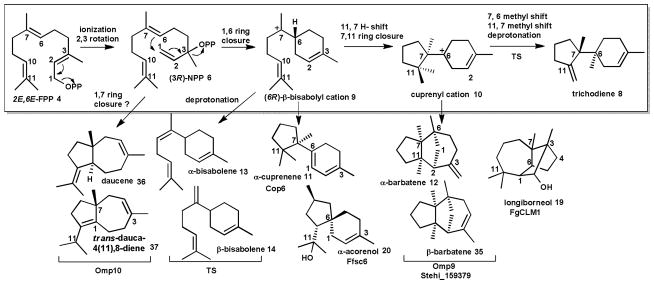
The cyclization of FPP 4 to trichodiene 8 by trichodiene synthase (TS) illustrates a well-studied cyclization mechanism. Other characterized fungal enzymes (see text) cyclize NPP into different major products.
The structure of aristolochene 7 synthase (AS) from Penicillium roqueforti (PR-AS)48 and Aspergillus terreus (AT-AS) is also known.46, 52, 53 AS belongs to the subclass of 1,10 cyclizing sesquiterpene synthases, catalyzing the ionization of FPP 4 and an initial ring closure to yield germacrene A 15 (Scheme 3). Protonation at C6 leads to the eudesmane carbocation 16,62 which following a methyl migration, hydride transfer, and deprotonation, yields (+)-aristolochene 7.45, 63 Aristolochene 7 is the sesquiterpenoid scaffold of several mycotoxins, including the PR-toxin 17, sporogen-AO1 and phomenone.64
Scheme 3. Examples of sesquiterpenoids generated by 1,10-cyclization of FPP.
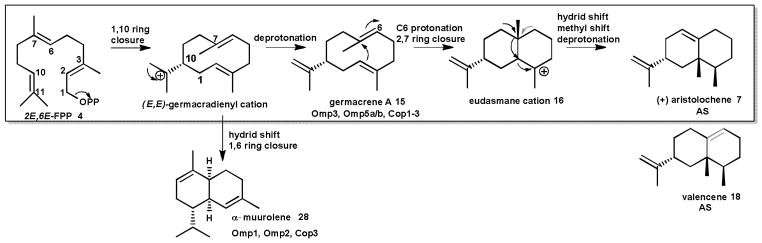
The cyclization of FPP to (+)-aristolochene 7 by aristolochene synthase (AS) illustrates a well-studied cyclization mechanism. Other characterized fungal enzymes (see text) cyclize FPP into different major products.
Detailed characterization of AS from the two distinct fungal sources has provided a unique perspective upon the evolution of sesquiterpene synthases. Both PR-AS and AT-AS follow the same cyclization mechanisms and display very similar three dimensional folds, indicating that the two enzymes share a common evolutionary ancestor. However, the underlying amino acid sequence of the two enzymes differs significantly, and this divergence in sequence has resulted in a divergence in fidelity. AT-AS produces (+)-aristolochene 7 (Scheme 3) as its sole product, while PR-AS is product promiscuous, producing (+)-aristolochene 7 as its major product and (-)-valencene 18 and germacrene A 15 as minor products (at a ratio of 94:2:4). Comparative site directed mutagenesis of the active site metal coordinating sites of both enzymes highlighted the importance of these conserved motifs in product fidelity, in particular the NSE motif. In PR-AS, the product ratio of mutants N244D, S248A and E252D shifted to 20:80 (+)-aristolochene 7: germacrene A 15, while E252Q produced only germacrene A 15. However, none of the mutants reported for PR-AS produced solely (+)-aristolochene 7. The product ratio of the corresponding AT-AS mutants N219D and E227D shifted from (+)-aristolochene 7 as the only product to 44:56 and 26:74 (+)-aristolochene 7: germacrene A 15, respectively.43 It would therefore appear that while the NSE metal coordinating motif serves as the modulator of product fidelity in AT-AS, other amino acids beyond these conserved residues play a role in determining the specific production of (+)-aristolochene 7 by PR-AS.
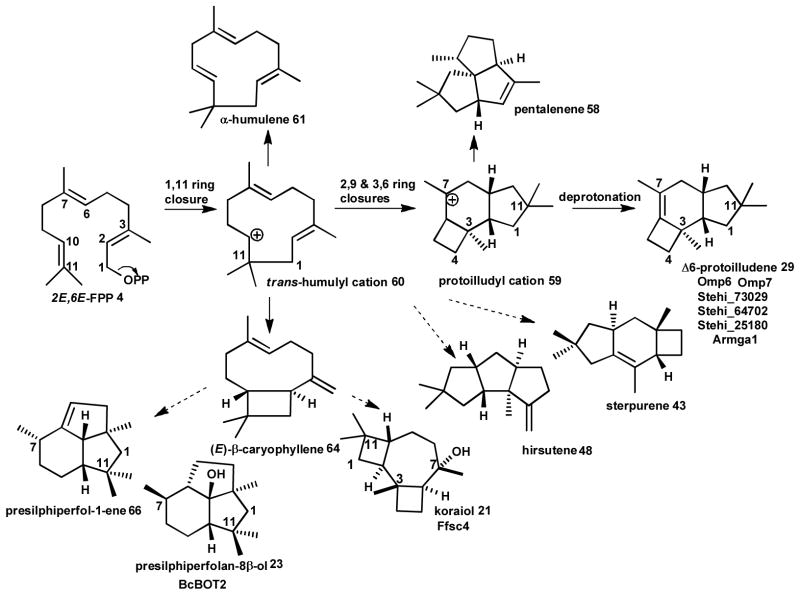
Three sesquiterpene synthases have recently been described from Fusarium strains. One terpene synthase (longiborneol 19 synthase FgCLM1, Scheme 2) from F. graminearum is presumed to catalyze a 1,6 cyclization to synthesize longiborneol 19 as the precursor of the antifungal compound culmorin.65 Gene deletion studies in the rice pathogen F. fujikuroi identified two terpene synthases involved in the production of α-acorenol 20 (acorenol synthase Ff_sc6, Scheme 2) and koraiol 21 (koraiol synthase Ff_sc4, Scheme 4) via a 1,6 and presumably a 1,11-cyclization reaction, respectively, followed by quenching of the final carbocation with water to yield the sesquiterpenoid alcohol products.66 The botrydial 22 biosynthetic gene cluster (see section 2.3) identified in the grey mold Botrytis cinerea encodes yet another 1,11-cyclizing enzyme (BcBOT2) which generates the tricyclic alcohol presilphiperfola-8β-ol 23 (Scheme 4) as precursor for this phytotoxin.67, 68
Scheme 4. Examples of sesquiterpenoids generated by 1,11-cyclization of FPP.
The initial trans-humulyl cation 60 cyclization product can be cyclized into different sesquiterpenoid scaffolds. Known fungal enzymes (see text) that carry out these cyclization reactions are shown.
Surprisingly, until a few years ago no sesquiterpene synthases were known from Basidiomycota, despite the fact that these fungi are prolific producers of bioactive sesquiterpenoids, many of which are derived from the trans-humulyl cation,9 which will be discussed in more detail in the next section. Mining of the first genome of a mushroom-forming Basidiomycota, C. cinerea, led to the cloning and biochemical characterization of six enzymes (Cop1–6) catalyzing 1,10 and 1,6 cyclization reactions of 3R-NPP 6 to α-cuprenene 11, germacrene D 24, γ-cadinene 25, δ-cadienene 26, cubebol 27 shown in Schemes 2 and 5, and 1,10 cyclization of FPP 4 to α-muurolene 28 and germacrene A 15 shown in Scheme 3. One of the enzymes, Cop6, catalyzes highly selective synthesis of α-cuprenene 11 as the precursor of the antimicrobial compound lagopodin.69–71
Scheme 5. Examples of sesquiterpenoids generated by 1,10-cyclization of FPP upon isomerization to NPP.
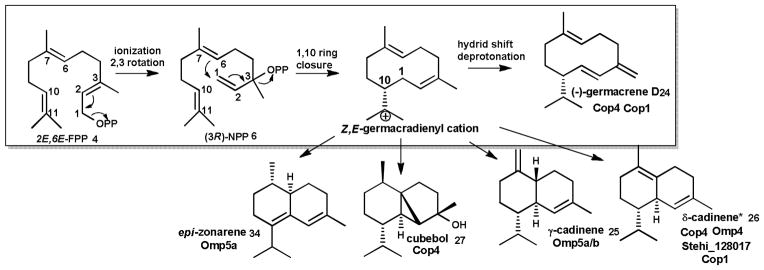
The initial Z,E-germacradienyl cation cyclization product can be cyclized into different sesquiterpenoid scaffolds Major products of known fungal enzymes (see text) are shown. Asterisk denotes that an alternative 1,10-cyclization pathway (Figure 10) is possible for δ−cadinene.
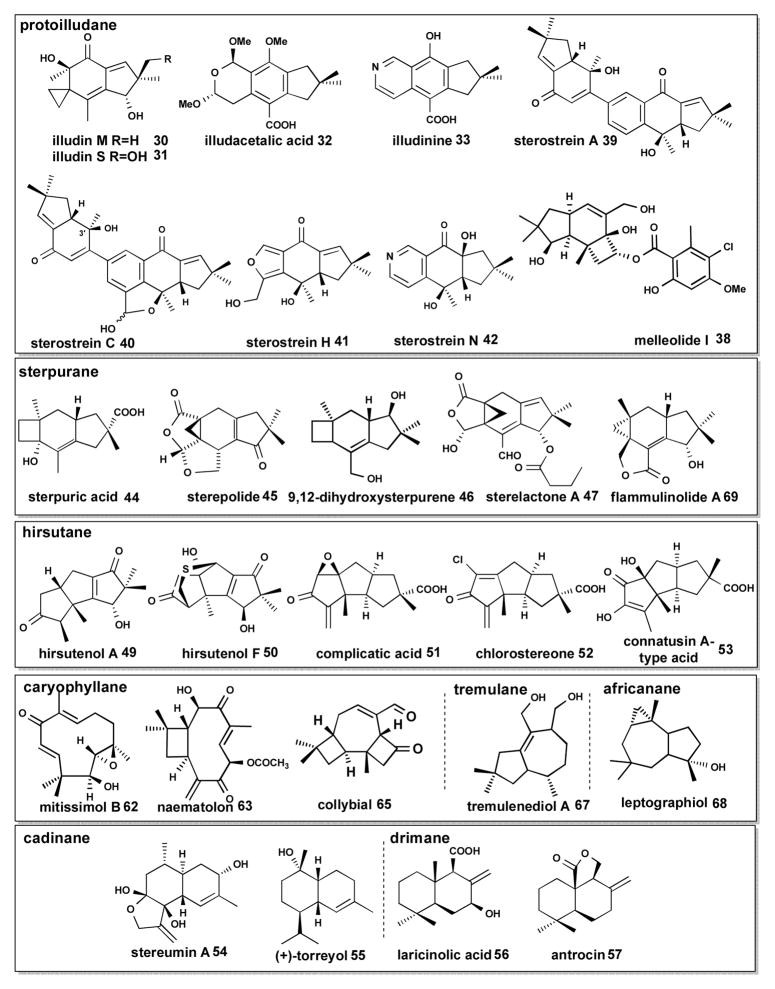
Of particular interest are enzymes that generate the 1,11-cyclization product Δ6-protoilludene 29 (Scheme 4), the precursor to many pharmaceutically relevant compounds,9 including the cytotoxic compounds illudin M 30 and S 31 (Scheme 6) that are being developed as anticancer therapeutics.15, 72, 73 With the objective of characterizing the biosynthesis of the anticancer illudins and their derivatives (compounds 30–33, Scheme 6) made by the Jack-O-Lantern mushroom Omphalotus olearius,74, 75 we sequenced its genome to apply a similar genome mining approach to find the Δ6-protoilludene 29 and presumably co-localized illudin biosynthetic genes. We discovered a surprisingly large complement of ten sesquiterpene synthases (Omp1–10) in this fungus. The recombinant enzymes catalyze all cyclization reactions of FPP shown in Scheme 1 (Schemes 2–5 show the major products for each Omp sesquiterpene synthase). The products made by of Omp1–5a/b are similar to those obtained with the C. cinerea enzymes Cop1–4, although Omp5a/b also makes as major products epi-zonarene 34 and γ-cadinene 25 (Scheme 5). Omp6 and Omp7 cyclize FPP highly selective into Δ6-protoilludene 29 (Scheme 4) while the other enzymes also display new cyclization activities, producing barbatenes 12, 35 (Omp9) and daucenes 36, 37 (Omp 10).23, 76 Another Δ6-protoilludene synthase was cloned from the honey mushroom Armillaria gallica (ArmGa1) which makes antimicrobial melleolide I 38 (Scheme 6) sesquiterpenoids.77
Scheme 6. Representative examples of modified sesquiterpenoids isolated from Basidiomycota.
Compounds are organized based on the sesquiterpenoid scaffolds that they are derived from (see text for details).
The biochemical characterization of the identified sesquiterpene synthases from Coprinus cinereus (Cop1–6)69–71 and of Omphalotus olearius (Omp1–10)23, 76 guided the subsequent development of in silico approaches for the directed discovery of new sesquiterpene synthases and their associated biosynthetic genes based upon cyclization mechanism of choice.20 Initiatives such as the 1000 Fungal Genomes Project78 have led to a substantial increase in the number of sequenced genomes that are publically available.79 The increase in the number of sequenced Basidiomycota genomes has been particularly steep – from less than a handful in 2008 to more than 100 genomes listed in the genome database of the Joint Genome Institute (JGI) at the time of writing. Our own BLAST searches for terpene synthases across Basidiomycota with sequenced genomes led to the identification of hundreds of putative sesquiterpene synthases, as well as associated biosynthetic gene clusters.20, 23, 69
The genome sequence of the wood-rotting mushroom Stereum hirsutum offers a unique opportunity to explore the terpenoid structural diversity of a fungal genus that is well known to produce a number of bioactive sesquiterpenoid natural products. 80–93 These include the sterostreins (e.g. 39–42), sterpurene 43 and its derivatives (e.g. 44–47), hirsutene 48 and its derivatives (e.g. 49–53), cadinane (25, 26) (e.g. 54–55) and drimane (e.g. 56, 57) compounds of which representative examples are shown in Scheme 6. We identified and predicted the cyclization mechanisms of 16 sesquiterpene synthases in this fungus. Presently, five of the predicted enzymes were cloned and biochemically characterized. Stehi1|25180, 64702, 73029 are highly product specific Δ6-protoilludene 29 synthases (Scheme 4) that are located in large biosynthetic gene clusters presumably involved in sterostrein (Scheme 6, e.g. compounds 39–42) biosynthesis.89, 91 Stehi1|159379 and Stehi1|128017 are 1,6- and 1,10-cyclizing enzymes, respectively (Schemes 2 and 5). Within the clade of not yet characterized 1,11-cyclizing sesquiterpene synthases sequences identified in S. hirsutum, there may be enzymes that synthesize hirsutene 48 and sterpurene 43 (Scheme 4) for the above mentioned modified sesquiterpenoid compounds isolated from this fungal genus.
Furthermore, pentalenene 58 (Scheme 4) has been observed as a minor volatile product in the culture headspace of S. hirsutum and O. olearius, and as a major volatile product of C. cinerea.20, 23, 69 However, no sesquiterpene synthase responsible for the production of pentalenene 58 has yet been identified from these fungi. Site directed mutagenesis and quantum-chemical modelling studies with bacterial pentalenene synthase indicate that the pathway to pentalenene 58 occurs through rearrangement of a protoilludyl cation 59.44, 94 It is therefore likely that one or more of the multiple Δ6-protoilludene synthases characterized from S. hirsutum and O. olearius are responsible for the production of pentalenene 58, under unidentified reaction conditions.20, 23 On the other hand, Cop5 is likely the sesquiterpene synthase responsible for the production of pentalenene 58 by C. cinerea, which does not produce Δ6-protoilludene 29. However, an active form of the predicted sesquiterpene synthase could not be cloned from C. cinerea cDNA,69 presumably due to incorrect prediction of gene borders, and/or the presence of frame shifting introns in all amplification products (see section 5 for more information).
2.2 The trans-humulyl cation derived sesquiterpenoids
While it is difficult to pin down the exact number of sesquiterpenoid-derived compounds characterized from the two major fungal divisions, searches in SciFinder™ and Web of Science™ return ten times more reports (~500) on sesquiterpenoids isolated from Basidiomycota than from Ascomycota. A diverse range of highly bioactive sesquiterpenoids with cytotoxic, antibacterial, nematocidal, and antiviral activities have been isolated from Basidiomycota (for reviews see:9, 95). Many of these compounds are thought to be made to protect fungal fruiting bodies from consumption.96, 97
Basidiomycota are known to synthesize sesquiterpenoids derived from 1,6 and 1,10 cyclized cations (Scheme 1), including germacranes,95 bisabolanes,96 cadinanes86, 90, 92 and drimanes,84, 98 including the mTOR signaling pathway inhibitor antrocin 57 produced by Antrodia camphorata90 (Scheme 6) However, the vast majority of sesquiterpenoids with medicinal or cytotoxic properties isolated from Basidiomycota are derived from the 1,11 cyclized trans-humulyl cation 60 as shown in Schemes 4 and 7, which will be the emphasis of this section.9 Of particular interest are derivatives of the tricyclic protoilludyl cation9, 97 which have potential applications as anticancer, antifungal and antibiotic agents.9
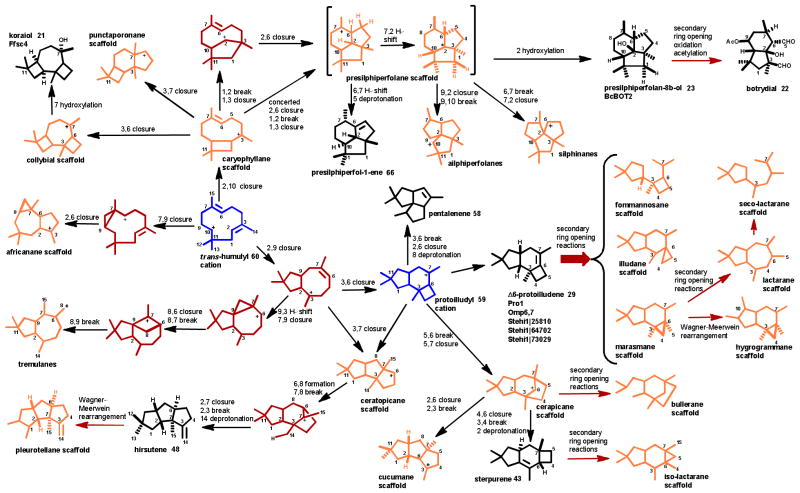
Scheme 7. Major fungal transformations of the trans-humulyl cation.
For consistency, carbon numbering is based on position in the linear FPP 4 precursor for all mechanism discussions. Where necessary for final products, published compound names are used. Enzymes that have been characterized as catalyzing transformations are listed beneath their major product. The key cation intermediates 59 and 60 are shown in blue, while minor intermediate cations are shown in red. Isolated final products are shown in black, and sesquiterpene scaffolds of known compound families are shown in orange. Rearrangements performed by sesquiterpene synthases are shown with black arrows, while reactions thought to be catalyzed by secondary enzymes are indicated by red arrows.
The trans-humulyl cation can be directly deprotonated to produce α-humulene 61 (Scheme 4), which has been observed in F. fujikorii and S. hirsutum.20, 66 Modified humulanes, such as the potential antitumor compound mitissimol B 62 (Scheme 6), have also been isolated from the fruiting bodies of several members of the genus Lactarius.99
Transformations of the trans-humulyl cation include a 2,10 ring closure, creating the distinctive dimethylated cyclobutyl ring of the caryophyllane scaffold. The mildly antibacterial sesquiterpenoid naematolon 63 (Scheme 6) is an oxidized, acetylated derivative of (E)-β-caryophyllene 64 produced by several members of the Naematoloma (Hypholoma) genus.96
In addition, the caryophyllane scaffold can be further cyclized to produce the tricyclic 4:7:4 collybial scaffold by a secondary 3,5 closure. The collybial scaffold is the precursor to the antibiotic collybial 65 (Scheme 6), produced by Collybia confluens.100 Hydroxylation at the C7 of the collybial scaffold results in koraiol 21. Alternatively, the caryophyllane scaffold can be cyclized to produce the triquinane presilphiperfolane scaffold, either via a stepwise cyclization,68 or via a concerted carbocation rearrangement.32 The resulting presilphiperfolane scaffold may then proceed through a 1,2 hydride shift and deprotonation to yield presilphiperfolan-1-ene 66, a minor volatile product of S. hirsutum.20 The presilphiperfolane scaffold may also undergo a 1,3 hydride shift and hydroxylated to produce 23,68 or be rearranged further to produce the silphiperfolane and silphinane families of triquinane sesquiterpenoids.101
Several tremulanes have been characterized from Phellinus tremulae, including tremulenediol A 67 (Scheme 6).102, 103 The potential route to the tremulane scaffold was deduced by 13C NMR analysis of tremulanediol A 67 formation by P. tremulae, and follows an initial 2,9 closure of the trans-humulyl cation 60. Following this, an additional cyclobutyl ring formation occurs at C6-C9. A secondary ring-opening rearrangement yields the fifth methyl group on C6 (Scheme 9).103
Scheme 9. Cyclization mechanism of the bifunctional CPS/KS type diterpene synthases.
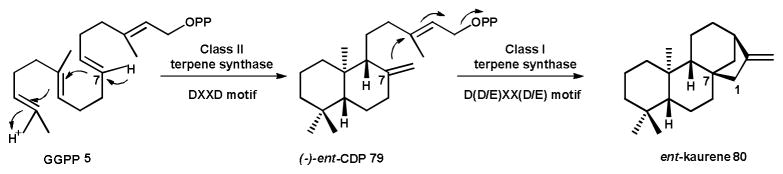
The precursor GGPP 5 undergoes an initial protonation dependent cyclization yielding (-)-ent-CDP 79 containing the typical bicyclic ring system of labdane-type diterpenoids. An ionization dependent dephosphorylation reaction and further cyclizations result in ent-kaurene 80.
The africananes are defined by a cyclopropyl group fused to a central cycloheptane ring, formed by a 7,9 cyclization of the trans-humulyl cation 60. Examples of modified africanane alcohols are leptographiol 68 (Scheme 6), isoleptographiol, and isoafricanol, isolated from Leptopgraphium lundbergii.97, 104 Interestingly, unmodified africanenes have been identified in the headspace of S. hirsutum and O. olearius liquid cultures.20, 23 However, the sesquiterpene synthase(s) responsible for their production have not yet been identified.
The largest and most diverse group of trans-humulyl cation 60 derived sesquiterpenoids produced by Basidiomycota are those derived from the 4:6:5 tricyclic protoilludyl cation 59.9 This scaffold is a key intermediate in the production of a large number of sesquiterpenes including e.g. sterpurene 43, hirsutene 48,105 Δ6-protoilludene 29 and thereof derived scaffolds,23 and of pentalenene 5894 as described below and shown in Scheme 7.
Δ6-protoilludene 29 is the precursor to diverse bioactive sesquiterpenoids, including the marasmanes, fommanosanes, (seco)lactaranes, hygrogammane and the illudanes (reviewed in: 9). These compounds are hypothesized to be produced by secondary ring opening and concurrent oxidations of Δ6-protoilludene 29, with the initial steps likely catalyzed by one or more cytochrome P450 monooxygenases and/or other oxygenases that are typically located in biosynthetic gene clusters of fungal sesquiterpenoid pathways.20, 23, 106, 107 In the case of the illudins 30 and 31, a ring opening and subsequent contraction of the Δ6-protoilludene 29 cyclobutyl ring yields the reactive cyclopropyl ring that defines the illudanes. Presently, no biosynthetic enzymes have been characterized that initiate these ring opening reactions.
Reopening of the protoilludyl cation 59 at C5-C6, and a subsequent 5,7 cyclization may afford the cerapicane scaffold which is the precursor to the rare cucumane and bullerane-type sesquiterpenes.9 The cerapicane scaffold is transformed to the sterpurane scaffold by an unusual ring rearrangement reaction that was deduced by 13C NMR analysis of 9,12-dihydroxysterpurene 46 synthesized by Stereum purpureum.81 Further ring opening modifications can then contract the cyclobutyl ring of sterpurene 43 into the defining dimethylated cyclopropyl ring of the iso-lactaranes, several of which have been identified from Lactarius rufus,108 S. purpureum,80 Phlebia uda,109 Flammulina velutipes,110 and Merulius tremillosus.111 The iso-lactaranes are often co-produced with sterpurenes, supporting the hypothesis of a shared biosynthetic pathway.80 Examples of iso-lactaranes include the cytotoxic flammulinolide A 69,110 sterepolides (e.g. 45),80 and the sterelactones (e.g. 47) (Scheme 6).87
Sterostrein A 39 produced by S. ostrea is a Δ6-protoilludene 29 dimer hypothesized to be formed by a Diels-Alder reaction that installs the central benzene ring.89 Biosynthesis of the melleolides (e.g. 38 in Scheme 6) by Armillaria gallica requires the esterification Δ6-protoilludene 29 with orsillinic acid.77, 112
The bioactive hirsutanes (e.g. compounds 49–53, Scheme 6) produced by S. hirsutum and other Basidiomycota20, 93,88, 113 may be synthesized either through 3,7 rearrangement of the protoilludyl cation 59 or directly via 3,7 cyclization of a bicyclic precursor shown in Scheme 7. While the intermediacy of a protoilludyl cation 59 has not been confirmed, a ceratopicane intermediate is consistent with the 13C labelling pattern observed in hirsutene 48 biosynthesis by Stereum complicatum.105 The ceratopicane intermediate undergoes multiple rearrangements, shifting the C3 (and its attached C14 methyl group) to its final position, and is then deprotonated to produce hirsutene 48.105 Hirsutene 48 may be converted by a Wagner-Meerwein rearrangement to produce the pleurotellane scaffold.9
2.3 Modifying the sesquiterpene scaffold
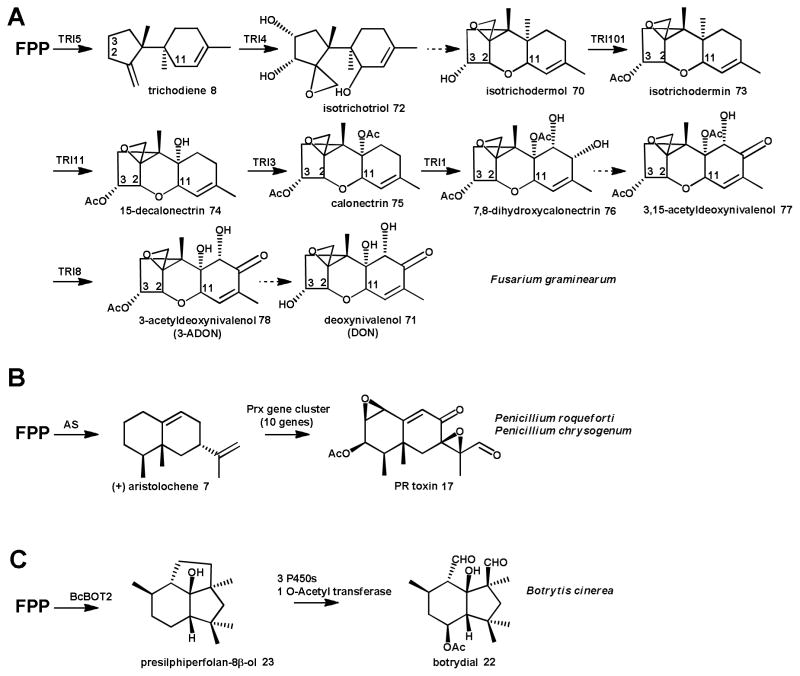
Compared to pathways responsible for the biosynthesis of diterpenoid natural products discussed in the next section, very few fungal sesquiterpenoid biosynthetic pathways have been characterized on a gene level. One of the most extensively characterized fungal sesquiterpenoid biosynthetic pathways is the route to the trichothecene mycotoxins (Scheme 8). Over 200 trichothecenes have been identified from diverse sources including the plant pathogens Fusarium and Myrothecium, the soil associated filamentous fungi Trichoderma and the mold Stachybotrys.114, 115 This particular pathway has been the focus of a great deal of research by the USDA during the past 30 years due to the significant economic impact that trichothecene related blight and ear rot has on global cereal crops each year, as well as the toxic effects of trichothecene contaminated feed on livestock.115–117 The trichothecene family of toxins is subclassified as Type A, B, C, and D depending on the chemical modifications to the common tricyclic core structure 12,13-epoxytrichothec-9-ene (EPT), exemplified by isotrichodermol 70 in Scheme 8. Of particular interest is deoxynivalenol (DON) 71, the Type B trichothecene that is most commonly associated with Fusarium head blight in cereal crops. DON 71 plays a vital role in activating fungal pathogenicity and virulence as part of a complex feedback exchange between plant, pathogen and environment.118–120
Scheme 8. Identified sesquiterpenoid biosynthetic pathways in Ascomycota.
Biosynthesis of (A) the trichothecene DON 71, (B) PR-toxin 17 and (C) botrydial 22 by the fungal strains shown.
The initial, common trichothecene biosynthetic steps in the pathway to DON 71 were established in the model fungus Fusarium sporotrichioides.56, 121 Later steps specific to the production of DON 71 were studied in a F. graminearum strain that was isolated from infected crops.122 Activation of biosynthesis of DON 71 is controlled by the global transcriptional regulator TRI6, which autoregulates in response to differences in nutrient levels.123 The first step on the biosynthetic pathway to all trichothecenes is the cyclization of FPP 4 to trichodiene 8 by the TRI5 encoded trichodiene 8 synthase,56 as described in section 2.1 and in Scheme 2. Trichodiene 8 is subjected to multiple oxidations by the cytochrome P450 monooxygenase TRI4, which installs oxygens at C2, C3, C11 and the C12-C13 epoxy group to give the intermediate isotrichotriol 72.124 Isotrichotriol 72 undergoes a spontaneous dual cyclization to yield isotrichodermol 70, a modified form of EPT.125 An acetyltransferase TRI101 acetylates the hydroxyl group at C3 to produce isotrichodermin 73, reducing toxicity of the intermediate to the host.126, 127 C15 of 73 is hydroxylated by the cytochrome P450 monoxygenase TRI11128 to form intermediate 74 and acetylated by TRI3129 yielding calonectrin 75, a key branch point on the pathway to biosynthesis of Type A and Type B trichothecenes by F. sporotrichioides and F. graminearum, respectively.115 For DON 71 production, calonectrin 75 is hydroxylated at positions C7 and C8 by TRI1 to give 7,8-dihydroxycalonectrin 76.130 The C8 hydroxyl group of 76 is then converted to a keto group by an unidentified enzyme, leading to 3,15-acetyldeoxynivalenol 77. Different variants of the deacetylase TRI8 remove the acetyl group from either the C3 or the C15 position, resulting in 15-acetyldeoxynivalenol (15-ADON) or 3-ADON 78, (the latter compound is shown in Scheme 8), respectively.131 The final biosynthetic step, deacetylation of ADON to DON 71, has yet to be elucidated (Scheme 8).
The ability of the fungal host to tolerate the toxic effects of intermediates and products of its own secondary metabolic pathways is key to survival. TRI12 is an integral membrane protein that acts as an efflux pump to export trichothecenes from hyphal cells, removing toxic products that could otherwise damage the cell.132 Subcellular localization of TRI12 to large motile vesicles133 indicated that F. graminearum could rely on an encapsulation mechanism to sequester trichothecene biosynthesis. The cytochrome P450 monooxygenases TRI1 and TRI4 co-localize with TRI12, suggesting that at least part of the trichothecene biosynthetic pathway is integrated within cellular targeting hubs, “toxisomes”, preventing toxic intermediates from causing unwanted effects.134 Whether this compartmentalization strategy is unique to F. graminearum, or whether it is a global mechanism employed by fungi producing bioactive secondary metabolites, remains to be established. Similarly, vesicles have been shown to also play an important role aflatoxin biosynthesis by Aspergillus parasiticus.135, 136
A much less complex gene cluster has been identified in the mold Botrytis cinerea that is responsible for the biosynthesis of the phytotoxin botrydial 22 (Scheme 8).67, 68 Presilphiperfolan-8β-ol 23, the product of terpene synthase BcBOT2 (Scheme 4), is converted by three cytochrome P450 monoxygenases and an acetyltransferase to the final phytotoxin. Most recently, screening of a genomic phage library and comparison with orthologous genes identified in the sequenced genome of a related strain, P. chysogenum, led to the identification of a 10-gene cluster encoding the biosynthetic pathway for the aristolochene 7-derived PR-toxin 17 in the blue cheese mold P. roqueforti (Scheme 8).137
As discussed in the previous section, multiple copies of Δ6-protoilludene 29 synthase are encoded in the genomes of O. olearius (Omp6 and Omp7) and S. hirsutum (Stehi1|25180, 64702, 73029) which are each located in predicted biosynthetic gene clusters responsible for the biosynthesis of illudin derived compounds (e.g. compounds 30–33) and potentially the sterostreins (e.g. compounds 39–42) isolated from O. olearius and Stereum ostrea BCC22955, respectively (Scheme 6).20, 23 The clusters share in common several cytochrome P450 monooxygenases, as well as further scaffold decorating enzymes including different types of oxygenases, oxidoreductases, group transferases and membrane transporters.
In O. olearius, Omp7 is located in a mini-gene cluster, including one P450 and an FAD-type oxidoreductase, while Omp6 is part of a large gene cluster.23 It appears that the two copies of protoilludene 29 synthase may have arisen from a gene duplication event, thereby boosting the production of illudin compounds.23 In the case of S. hirsutum, the three Δ6-protoilludene 29 synthases may have resulted from gene duplication, or they may have diverged from a common ancestor. Differences in catalytic efficiencies of the enzymes, as well as a diversity in product fidelity, could represent a spectrum of evolutionary time points on the course to a specific Δ-6 protoilludene 29 synthase.20 Furthermore, each of the three Δ–6 protoilludene 29 synthases is located in a distinct biosynthetic gene cluster which appear to be expressed at the same time under standard growth conditions.20 Heterologous refactoring of the entire gene clusters (see section 5) will be required to understand the role that each of these pathways plays in defining the natural product landscape of this fungus. It remains to be seen if all three biosynthetic gene clusters are responsible for the biosynthesis of the different Δ6-protoilludene 29 derived sterostreins (Scheme 8) isolated from Stereum, make yet uncharacterized compounds, or may even carry out a sophisticated combinatorial scheme among the three different clusters that each act on the same protoilludene scaffold.
3 Fungal diterpenoids
3.1 Cyclization of GGPP to produce the diterpene scaffold
Diterpenoids are a diverse class of natural products derived from the C20 precursor GGPP 5, with at least 12,000 compounds already described.138 The first committed step in diterpenoid biosynthesis is the cyclization of GGPP 5 to produce the diterpene scaffold, which occurs via a carbocation cascade. Classically, activation of the carbocation cascade by terpene synthases corresponds to the removal of the pyrophosphate group from the linear substrate (as described for the sesquiterpene synthases in section 2.1). This ionization-dependent reaction is catalyzed by class I terpene synthases.28
Notably, the initial generation of the reactive carbocation follows a different path in the biosynthesis of labdane-related diterpenes.139 Here, the carbocation cascade is preceded by a protonation-dependent bicyclization reaction to install the labdane bicycle. This reaction is catalyzed by class II diterpene synthases, more akin to the triterpene synthases (described in section 4.1).140 In this case, the pyrophosphate group of GGPP 5 remains intact and is later removed in a class I cyclization reaction (Scheme 9).139 Thus, this type of diterpene synthase appears to have evolved as an amalgamation of ancestral bacterial class I and class II terpene synthases.139
The first bifunctional fungal diterpene synthase was isolated and characterized from the gibberellin producer Phaeosphaeria sp. L487.141 The putative gene encoding the diterpene synthase was isolated using degenerate primers based upon conserved sequence motifs of plant copalyl diphosphate synthases. Characterization of the gene product revealed a conglomerate enzyme: that is, a bifunctional copalyl diphosphate synthase/ent-kaurene synthase (CPS/KS), which catalyzed a sequential class II followed by class I cyclization of GGPP 5 to produce (-)-ent-CDP 79 as an intermediate on the path to ent-kaurene 80 (Scheme 9).142 The class I activity is located in the N-terminal region, while the C-terminal region contains the class II activity. This differed from the previously characterized plant diterpene synthases, which appeared to involve separate mono-functional CPS and KS enzymes.143, 144
Several structures of plant diterpene synthases have recently been solved and shown to contain both an N-terminal α-helical domain (α-domain) typical of class I terpenoid synthases and a C-terminal α-barrel (or γβ-) domain typical of class II terpenoid synthases.145–147 Many of these modular γβα-domain plant diterpene synthases, however, are monofunctional in that only one of the cyclization activities (CPS or KS) is functional (reviewed in: 139). However, some plant enzymes (e.g. abietadiene synthase147) are bifunctional, like the fungal enzymes. The γβαdomain taxadiene synthase, however, converts GGPP 5 at its class I domain directly into the tricyclic taxadiene scaffold.146 Although protein sequences of diterpene synthases from plants and fungi share little similarity except for some conserved motifs, homology modelling suggests that they share the same domain organization.
A number of CPS/KS-type fungal diterpene synthases have been identified in addition to the one from Phaeosphaeria sp. These include the ent-kaurene 80 synthase from the gibberellic acid producers Fusarium fujikuroi (teleomorph: Gibberella fujikuroi),141, 142, 148, 149 the ent-primara-8(14),15-diene 81 synthase AN1495 from Aspergillus nidulans,150 phyllocladen-16 α-ol 82 synthase (PaDC1) and a CPS/KS synthase (PaDC2) with only a functional CPS domain151 from Phomopsis amygdali, and aphdicolan-16β-ol 83 synthase (PbACS) from Phoma betae.152 Only one labdane-type diterpene synthase has thus far been cloned from a Basidiomycota: pleuromutilin 84 synthase from Clitopilus passeckerianus.153 (see Scheme 1 for cyclization reactions).
Typically, these bifunctional diterpene synthases and their associated biosynthetic gene clusters (see section 3.2) have been identified by a sequence analysis strategy that first involved the identification of a GGPP 5 synthase (GGS) via genomic amplification followed by genome walking to identify flanking regions. GGPP 5 utilizing biosynthetic pathways require a dedicated GGS that extends the common isoprene precursor FPP 4 by one IPP 2 unit.148, 154 The gene encoding GGPP 5 synthase, which is essential for production of the precursor molecule is commonly clustered with other pathway genes.155 This gene finding strategy has led to the discovery two unusual monofunctional, non-labdane-type diterpene synthases from Phoma betae.156, 157 PaFS and PaPS, which synthesize fusicocca-2,10(14)-diene 85 and phomopsene 86 (Scheme 1), respectively, deviate from other CPS/KS-type fungal diterpene synthases in that they display multiple DDXXD motifs, as opposed to the prototypical N-terminal DXDD and C-terminal DEXXE motifs common to the dual-function diterpene synthases that carry out sequential type II and type I cyclization of GGPP 5 (Scheme 9).158 The DXDD motif is required for the class II type protonation dependent cyclization reaction, like that carried out by squalene hopene cyclase,159 while the DDXXD motif is associated with the class I type ionization dependent reaction, which has been described for terpene synthases and prenylchain synthases like GGPP 5 and FPP 4 synthases.34, 160
Both PaFS and PaPS appear to derive from a gene fusion, resulting in chimeric proteins with an N-terminal class I terpene synthase domain and another C-terminal class I domain that contains the GGPP 5 synthase activity. This physical co-localization of diterpene synthase and GGPP 5 synthase may have evolved to improve flux of precursor molecules towards downstream modifying enzymes. A chimeric sesterterpene (C25) synthase AcOS was recently identified in Aspergillus clavatus. Here a GFPP (geranylfarnesyl diphosphate) synthase domain supplies the substrate for subsequent cyclization by the terpene synthase domain into ophiobolin F 87 (Scheme 1).161 Structural analyses of plant and bacterial diterpene synthases indicate that modularity in domain architecture and diversity in mechanisms are a common theme for diterpene synthases.145–147
3.2 Modifying the diterpene scaffold
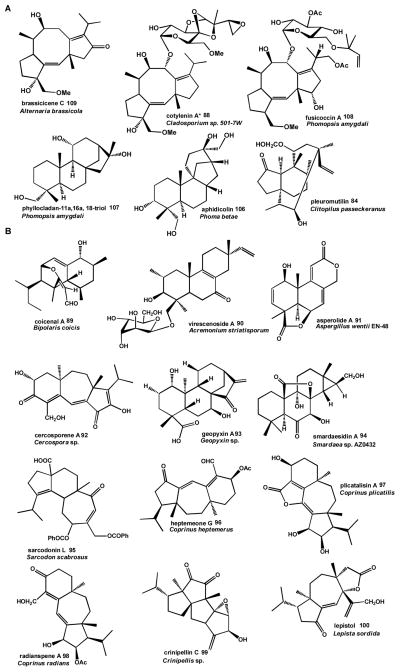
The diversity of diterpenoid compounds stems from the large number of modifications to the diterpene scaffold, typically catalyzed by decorating enzymes such as P450 monoxygenases, different types of oxidases and oxidoreductases as well as trasnferases. Diterpenoids are widely produced by plants, functioning as signaling compounds, as photosynthetic pigments, and in defense against infection.138 While a number of enzymes involved in the biosynthesis of diterpenoids in plants and bacteria have been characterized in detail, fewer diterpenoid biosynthetic enzymes have been characterized from fungi.138, 162 Except for pleuromutilin 84 biosynthesis by Clitopilus passeckeranus,153 all other characterized fungal diterpene pathways are from Ascomycota. Yet, diverse bioactive diterpenoids have been isolated from Basidiomycota, although their biosynthetic pathways remain to be identified (Table 1, see Scheme 10 for modified diterpenoid structures discussed below).163
Table 1.
Representive examples of fungal diterpenoids with characterized bioactivities.
| Biological activity | Fungal species | Compound name(s) | Reference |
|---|---|---|---|
|
Ascomycota
| |||
| Cytotoxic | Alternaria brassicola | Brassicicene C ( 109) † | 158, 173, 176 |
| Cytotoxic | Cladosporium sp. | Cotylenin A-E (88 ) | 234–236 |
| Phytotoxin | Phomopsis amygdali | Fusicoccins A ( 108)†, F | 157 |
| DNA-Pol. inhibitor | Phoma betae | Aphidicolin (106) † | 152, 237 |
| Anti-inflammatory | Bipolaris coicis* | Coicenals A( 89)-D | 238 |
| Cytotoxic | Acremonium striatisporum | Virescenosides A(90 ), B, C, M, N | 239 |
| Cytotoxic | Aspergillus wentii EN-48* | Asperolides A (91 )-C | 240 |
| Cytotoxic | Cercospora sp.* | Cercosporenes A ( 92)-F | 241 |
| Cytotoxic | Geopyxis sp.* | Geopyxins A (93 )-D | 242 |
| Cytotoxic | Smardaea sp.* | Smardaesidins A (94 ) -G | 243 |
| Growth hormone | Fusarium fujikuroi | Gibberellins GA14 (102),4 (103),3, 1 (104) | 148 |
| Growth hormone | Phaeosphaeria sp. L487 | Gibberellins GA12 (101),4 (103),9,20,1 (104) | 141, 171 |
| Growth hormone | Sphaceloma manihoticola | Gibberellins GA14 (102),4 (103) | 169 |
|
| |||
|
Basidiomycota
| |||
| Antibacterial | Clitopilus sp. & C. passeckeranus | Pleuromutilin (84)† | 153, 244 |
| Antibacterial | Sarcodon scabrosus | Sarcodonin L (95), M | 245 |
| Antifungal | Coprinus heptemerus | Heptemerones A-G (96) | 246 |
| Cytotoxic | Coprinus plicatilis | Plicatilisin A (97)-D | 247 |
| Cytotoxic | Coprinus radians | Radianspene A (98)-M | 248 |
| Cytotoxic | Crinipellis sp. | Crinipellins A-C (99), D | 249 |
| Cytotoxic | Lepista sordida | Lepistol (100) | 250 |
Scheme 10. Examples of fungal diterpenoid natural products and their producer organisms.
(A) Fuiscoccadiene (top) and labdane-type (bottom) diterpenoid compounds with known biosynthetic gene clusters. (B) Bioactive diterpenoids isolated from Ascomycota and Basidiomycota for which pathways are not known (see Table 1 for details and references)* Cotylenin A 88 biosynthesis is assumed to proceed via a pathway similar to fusiccocin A 108 174.
One of the first fungal species identified as a diterpenoid producer was the rice plant pathogen F. fujikuroi. This fungus causes stems of plants to become hyperelongated and etiolated, leading to what is commonly known as “foolish seedling” disease. The causative agent of this phenotype was discovered to be the gibberellin (GA) diterpenoids (GA compounds 101–104 Scheme 11) produced by F. fujikuroi. The biosynthetic pathway to the production of GAs by F. fujikuroi, which is clustered on the genome, has since been fully elucidated by Tudzynski and coworkers164 (Scheme 11). Detailed characterization of the enzymes involved in GA biosynthesis by F. fujikuroi has highlighted key differences in sequence and function from that of plant GA pathways,165 such that it has been suggested that GA biosynthesis in bacteria, plants and fungi evolved separately.155
Scheme 11. Proposed biosynthetic pathway to gibberellic acids (GA) in F. fujikuroi.
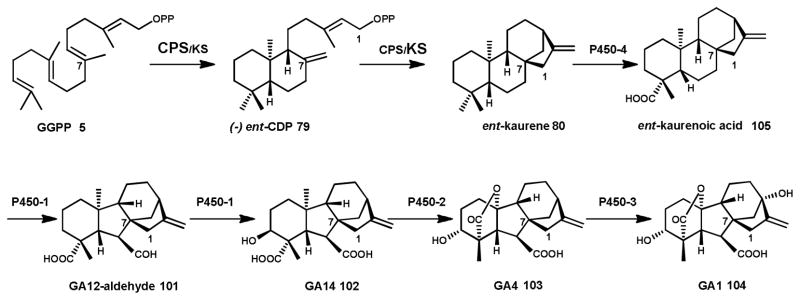
Production of the intermediate ent-kaurene 80 from GGPP 5 is catalyzed by a bifunctional CPS/KS type diterpene synthase (see Scheme 9). Ent-kaurene 80 is oxidized to the final product GA1 by four cytochrome P450s.
In GA biosynthesis, GGPP 5 is the substrate for a bifunctional diterpene synthase CPS/KS.149 The resulting tetracyclic ent-kaurene 80 diterpene is converted to ent-kaurenoic acid 105 through oxidation by the cytochrome P450 monooxygenase P450-4.148 Hydroxylation at the C7 position of ent-kaurenoic acid 105 is carried out by P450-1, resulting in aldehyde GA12 101 (note GA numbering is based on the order the compounds were identified), with a characteristic 6-5-6-5 ring structure. A further hydroxylation at the C3 position and an oxidation by the multifunctional P450-1 results in GA14 102, a key intermediate in the pathway. A closely related cytochrome P450, P450-2, catalyzes the essential oxidation of GA14 102 at C20 to yield the biologically active form, GA4 103.148, 166 Finally, P450-3 hydroxylates GA4 to GA1 104, another bioactive form of the growth hormone.167 Each of the steps catalyzed by the cytochrome P450s depends on a cytochrome P450 oxidoreductase (CPR) to transfer electrons from the cofactor NADPH via FAD and FMN.168
Subsequent identification and characterization of the GA biosynthetic pathway from S. manihoticola revealed that the organization of the gene cluster, the underlying gene sequences, and the function of the encoded GA enzymes, were similar to that of F. fujikuroi.169 Despite the fact that S. manihoticola lacked enzymes responsible for GA1 104, 3, and 7 production, complementation with its gene cluster was sufficient to restore GA biosynthesis in F. fujikuroi mutants. This indicates that the GA pathways of these two distantly related fungi may share a common evolutionary ancestor. By contrast, characterization of the GA biosynthetic pathway in Phaeosphaeria sp. L487 shows that it is more similar to that of plants.170, 171 Here, the GA pathway branch point is GA12 101 rather than GA14 102, which is the key intermediate linking the GA pathways of F. fujikuroi and S. manihoticola. Additionally, the C3 hydroxylation takes place later in the GA pathway of Phaeosphaeria sp. L487 than that of the other fungi.
Diversification of ancestral GA-pathways likely led to new diterpene pathways that produce different bioactive compounds. Several dozens of such putative diterpene biosynthetic gene clusters can be readily identified in published Ascomycota genome sequences. However, only very few diterpene biosynthetic gene clusters have been characterized so far. The only labdane-type diterpene pathway from a Basidiomycota has been disclosed in a patent application for pleuromutilin antibiotic biosynthesis in Clitopilus passeckeranus.153
Identification of the aphdicolan-16β-ol 83 synthase152 from Phoma betae (Scheme 1) facilitated subsequent identification and heterologous expression in Aspergillus oryzae of the aphidicolin 106 biosynthetic pathway (Scheme 6).172 In addition to the bifunctional diterpene synthase, this cluster encompasses two P450 enzymes that carry out three hydroxylations of the diterpenoid alcohol scaffold, as well as a transporter and the aforementioned GGPP 5 synthase. The two CPS/KS homologs PaDC1 phyllocladen-16 α-ol 82 synthase) and PaDC2 ((+)-CDP synthase) characterized from P. amygdali are each part of separate gene clusters that are involved in the biosynthesis of phyllocladan-11 α,16 α,18-triol 107.151 Interestingly, in addition to different types of oxygenases, each of two clusters includes its own GGPP synthase.
Genome mining recently led to the identification of an ent-primara-8(14),15-diene 81 synthase AN1495 in Aspergillus nidulans which is part of a gene cluster that has yet to be characterized.150 Encoded within this cluster are not only a GGPP synthase, but also a HMG-CoA synthase, a mevalonate pathway specific enzyme, which may boost precursor supply for diterpenoid biosynthesis by this pathway.
The monofunctional diterpene synthase, fusicocca-2,10(14)diene 85 synthase (PaFS), from P. amygdali is part of a five gene cluster,157 including three enzymes that catalyze oxidative modifications to the tricyclic diterpene scaffold.173 This cluster together with a later discovered second, nine-gene cluster are required to synthesize the bioactive fusiccocin A 108 diterpenoid.174 The presence of a prenyltransferase previously shown to catalyze the prenylation of glucose175 which is a moiety also present in fusiccocin A 108, led to the identification of this second fusiccocin cluster in the draft genome sequence of this fungus. Biochemical studies of recombinant enzymes together with gene deletions were carried to complete characterization of fusiccocin biosynthesis.174 The same group also identified and dissected another fusicoccadiene-type pathway for brassicicene C 109 biosynthesis in A. brassicola; as seen with the other diterpene pathways, a contingent of oxidizing enzymes (five cytochrome P450 enzymes, an oxidoreductase and a dioxygenase) are involved in modifying the terpene hydrocarbon scaffold.158, 173, 176
4 Fungal triterpenoids
4.1 Oxidosqualene/lanosterol cyclases
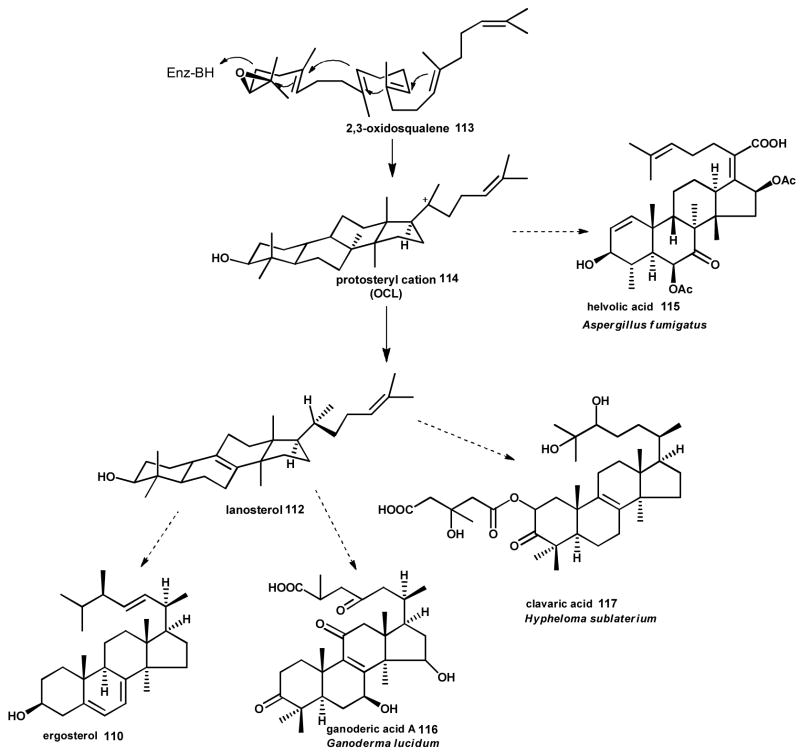
The triterpenoids are C30 prenyl chain derived compounds that are found widely in nature as steroids and sterols.177 The biosynthetic pathway to ergosterol 110, the major component of the plasma membrane in fungi, begins with the cyclization of squalene 111 to lanosterol 112.178 Epoxidation of squalene 111 yields 2,3-oxidosqualene 113 which adopts a chair-boat-chair conformation.179 Cyclization of the linear polyisoprene chain is catalyzed by a Class II type terpene synthase oxidosqualene/lanosterol cyclase (OSC) as part of a protonation-dependent reaction.159, 180 Activation of the epoxide in 113 is initiated by a conserved Asp, and a sequential ring-forming cascade results in a 6-6-6-5 tetracylic protosteryl cation 114. Two methyl migrations and hydride shifts, followed by a final deprotonation, results in lanosterol 112 (Scheme 12), the last common intermediate to the lanostane triterpenoids.140, 181, 182
Scheme 12. Triterpenoid biosynthesis in fungi.
Cyclization of 2,3-oxidosqualene 113 by an oxidosqualene cyclase (OSC) via a protonation dependent reaction results in a 6-6-6-5 tetracylic protosteryl cation that is the precursor to the membrane sterol ergosterol 110 and the bioactive triterpenoid examples shown.
Prokaryotic and eukaryotic triterpene cyclases share a similar overall protein fold, with two α-helical domains linked by a membrane spanning channel. The large aromatic-lined active site cavity spans the two domains, and the protonating group that initiates cyclization is located at the polar region at the top of the central channel.159, 180 Triterpene cyclases are also defined by conserved 16 amino acid long tandem repeats known as QW sequence motifs. These Gln and Trp residues provide a bonding network between the α-helices, which stabilizes the enzyme during the exergonic reaction, allowing stringent control over product formation.183 Beyond these structural similarities, bacterial squalene hopene cyclases (SHC) and eukaryotic OSC cyclases have diverged both in sequence and in mechanism. Notably, SHC is characterized by the presence of the protonating Asp in a conserved DXDD motif, while the protonating Asp of OSC is located in the motif XXDCX.182 This branching in the evolutionary tree of SHC and OSC has resulted in the mechanistically defining feature of the enzymes: SHC accepts squalene 111 as a substrate, while OSC accepts 2,3-oxidosqualene 113 as a substrate.182, 184
The first eukaryotic OSC to be cloned and characterized was the ergosterol 110 biosynthetic pathway enzyme ERG7 from Saccharomyces cerevisiae.185, 186 The catalytic mechanism of ERG7 has since been elucidated by mutational studies. Substrate activation is highly dependent upon the maintenance of a protonating environment, and aromatic residues play a key role in the stabilization of carbocation intermediates, which funnels the product trajectory of ERG7 specifically towards lanosterol 112.187–190 Further fungal OSCs have been characterized from diverse species, including the helvolic acid 115 antibiotic producers Cephalosporium caerulens191 and Aspergillus fumigatus,192 as well as the cytotoxic ganoderic acid (e.g. 116) producer Ganoderma lucidum.193, 194 Notably, characterization of an oxidosqualene-clavainone cyclase from Hypholoma sublateritium revealed the presence of two OSCs: one specifically for primary and one dedicated to secondary metabolism for clavaric acid 117 production.195
More recently, bioinformatic approaches led to a wider scale identification of putative OSCs from Ascomycota and Basidiomycota.196 This database represents a diversity of OSC sequences that could potentially be used to engineer a product promiscuous fungal OSC, offering biosynthetic routes to a greater diversity of fungal triterpenoids, like those produced by plant OSCs.197–199
4.2 Discovery of triterpenoid biosynthetic gene clusters
Triterpenoid biosynthesis is relatively well understood and characterized in plants.197, 200–202 Comparatively little, however, is known about fungal triterpenoid biosynthesis; despite the isolation of a large number of bioactive fungal triterpenoids mostly from Basidiomycota.203, 204
Recent genome sequencing and transcriptome projects have provided the first insights into the biosynthetic pathways to the triterpenoids.24, 205–207 G. lucidum is a prolific producer of bioactive ganoderic acid 116 triterpenoids, correspondingly it has an OSC and a large complement of cytochrome P450s encoded within its genome. Almost half of the genes encoding P450s are upregulated during the transition from primordia to fruiting bodies, which correlates with lanosterol 112 expression and triterpenoid production. Therefore, developmental stage of fungal growth plays an important role in regulation of secondary metabolite genes. Some of these same genes are physically clustered on the genome, albeit not with the gene encoding OSC.24 Likewise, transcriptome analysis of the medicinally relevant Wolfiporia cocos identified several P450s that were upregulated simultaneously with its OSC.205 Contrastingly, the genome of A. fumigatus shows that it contains multiple genes encoding OSC as opposed to a single copy. Furthermore, the putative enzymes related to helvolic acid 115 production are located in a gene cluster encoding protostadienol 118 synthase, multiple P450s, a reductase, acyltransferases and a dehydrogenase. Together, these enzymes are predicted to represent the full helvolic acid 115 biosynthetic pathway.192
5 Mining the fungal (sesqui)terpenome
5.1 Terpene synthase identification
Numerous databases exist to search the rapidly increasing number of fungal genomes for biosynthetic gene candidates, and for predicting putative coding sequences. For example, the Joint Genome Institute (JGI) listed 362 fungal genomes (117 Basidiomycota) at the time of writing, with a goal of 1000 sequenced genomes from all families in the fungal kingdom in the next few years.78 Users of the JGI web interface (http://genome.jgi-psf.org)208 benefit from standardized EST and/ or RNAseq data available for many of the fungal genomes that are that are displayed along with gene annotations.
Putative terpene synthases can be identified by performing a Basic Local Alignment Search Tool (BLAST)209 search of target genome(s) using characterized terpene synthases sequences as query sequences. We have used BLAST searches of fungal genomes, combined with biochemical knowledge of the initial FPP 4 cyclization reaction (see Scheme 1) catalyzed by the O. olearius enzymes (Omp1-10, section 2.1) to develop a predictive framework for further, targeted discovery of additional fungal sesquiterpene synthases and their associated biosynthetic gene clusters.20, 23
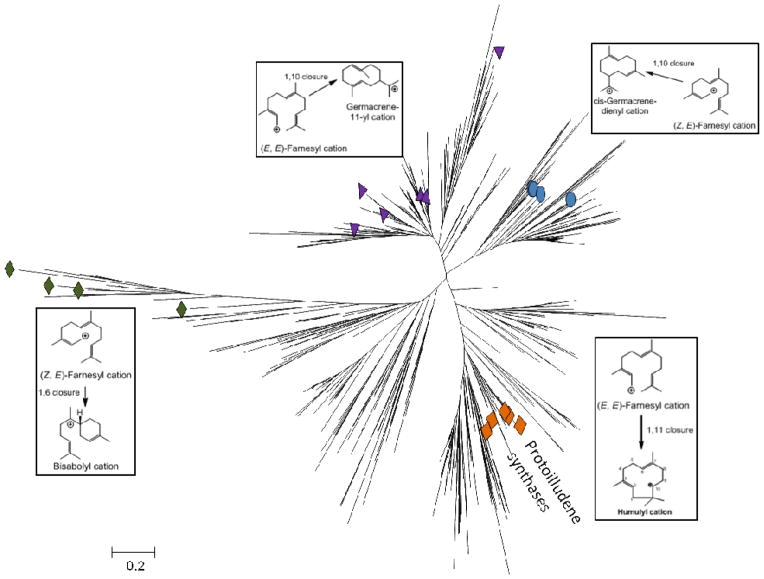
We found that sesquiterpene synthase sequences identified by BLAST analysis in the sequenced Basidiomycota genomes (presently more than 1000 sequences in ~100 genomes) cluster in distinct clades according to initial cyclization mechanism (Figure 1).23 This method led to the focused discovery and characterization of several 1,11-cyclizing terpene synthases, which produce trans-humulyl cation 60 derived sesquiterpenes (section 2.1.).20, 23 Notably, these characterized Δ6-protoilludene synthases form a single branch within the 1,11 cyclizing clade (Figure 1). Many other predicted trans-humulyl cation 60 producing sesquiterpene synthases remain yet to be discovered, potentially producing the diverse scaffolds shown in Scheme 7.
Figure 1. Putative Basidiomycota sesquiterpene synthases form clades consistent withtheir initial cyclization reaction.
117 Basidiomycota genomes listed in JGI were BLAST searched for gene catalogued protein homologues to Stehi1|73029, identifying 777 proteins. Of these, 577 were assembled in an unrooted neighbor-joining dendrogram with the 22 previously characterized Basidiomycota sesquiterpene synthases, Cop1–6 (Coprinus cinerea),69 Pro1 (A. gallica),77 Omp1–10 (O. olearius),23 Stehi1|159379, 128017, 25180, 73029, and 64702 (S. hirsutum).20 Enzymes cluster in clades according to initial cyclization mechanism. Characterized enzymes responsible for a: 1,10 cyclization of (E,E)-FPP are indicated by purple triangles; 1,10 cyclization of (3R)-NPP 6 are indicated by blue circles; 1,6 cyclization of (3R)-NPP 6 are indicated by green diamonds; 1,11 cyclization of (E,E)-FPP 4 are indicated by orange squares. The branch consisting of Δ6-protoilludene 29 synthases is also highlighted.
5.2 Identification of terpenoid biosynthetic gene clusters
Discovery of fungal terpenoid biosynthetic pathways by genome mining approaches is facilitated by the fact that fungi tend to cluster genes encoding biosynthetic pathways within their genome. Clusters can be identified by manually scanning the genome sequence ±20 kb upstream and downstream of predicted terpene synthase sequences e.g. identified using the strategy described in the previous section. BLAST searches of the NCBI protein database and the NCBI Conserved Domain Database (CDD) allows annotation of predicted genes surrounding the terpene synthase. Typical enzyme classes and proteins expected in such clusters are cytochrome P450 monooxygenases, oxidoreductases and oxygenases, group transferases, transporters, transcription factors and enzymes required for isoprenoid precursor biosynthesis. The likely boundaries of such biosynthetic clusters are predicted by the presence of large gaps between genes (>10 kb), or the presence of several consecutive genes that are not likely to be associated with natural product biosynthesis.18
However, manual identification of biosynthetic gene clusters is subject to user interpretation, is extremely time consuming, and as a result is a significant bottleneck in the identification of putative biosynthetic pathways. Therefore, automated search algorithms are required for the large scale identification of fungal natural product pathways and several web-based analysis tools have been developed for this purpose (reviewed in: 210, 211). When using these platforms, however, one has to keep in mind that these tools have often been trained for the identification of specific natural products gene clusters (e.g. non-ribosomal peptide (NRP) and polyketide synthase (PKS) pathways) common in certain groups of organisms (e.g. Ascomycota, bacteria). From our own experience we have found that these tools are not particularly adept at identifying terpenoid clusters in Basidiomycota. For example, antiSMASH 2.0212 identified four of the eleven manually identified putative terpene synthases in the genome of O. olearius.23 Similarly, five out of eighteen manually predicted terpene synthase sequences were identified in S. hirsutum.20 This and other tools also significantly underestimated biosynthetic gene cluster size compared to our manual annotations, probably due to the high degree of complexity and variability of fungal gene clusters, as well that fact that few Basidiomycota clusters have been fully characterized.213 As more natural product pathways from Basidiomycota are characterized, these tools can be trained for more accurate, automated cluster prediction in these types of fungi.
5.3 Characterization of terpenoid biosynthetic gene clusters
Characterization of fungal terpenoid gene clusters identified in Ascomycota described throughout this contribution has relied on a combined approach of biochemical characterization of recombinant enzymes and specific gene deletions in the native producer host, as well as transfer of biosynthetic cluster genes into closely related fungal hosts for which genetic tools are available.214–216 In order to access the incredible diversity of Basidiomycota terpenoid natural product pathways encoded in the large (and increasing) number of fungal genomes, different tools and methods need to be developed to go from genome mining to terpenoid biosynthesis. Some of the problems that need to be overcome for this to happen will be discussed briefly using the Basidiomycota as an example, but similar difficulties may also be encountered with other, less well-studied classes of fungi.
Several major challenges significantly impede efficient characterization of terpenoid biosynthetic gene clusters in Basidiomycota: 1. Correct cDNA prediction, 2. Therefore, necessity to clone genes from cDNA and thus the need to establish laboratory growth of strains under conditions where target genes/pathways are expressed, 3. Fungal strains are not genetically tractable, and therefore require heterologous gene expression and pathway assembly.
The first step following the identification of putative terpene synthases and the associated gene cluster in a genome requires a refined prediction of the correct coding sequences for each gene. However, this is in our experience not trivial for Basidiomycota, which tend to have have very intron-rich genomes.217 We have found several very small and unpredicted exons (sometimes only 11–18 bp in length 218 and introns/exons 27 and 28 bp in length23, 69) in genes cloned from cDNA. Genes synthesized based on cDNA predictions are therefore often non-functional. Figure 2 illustrates different structural gene predictions, alternative splice variants and the functional cDNAs of sesquiterpene synthases cloned from S. hirsutum. The gene prediction program Augustus (http://bioinf.uni-greifswald.de/augustus/)219, 220 can be used to identify the most likely cDNA sequences for a biosynthetic enzyme. Gene predictions created from the putative terpene synthase ±10 kb can then be used to manually identify potential gene start/stop codons by alignment in MEGA6.221 This allows the design of a small set of cloning primers to attempt amplification of the correct ORF from cDNA. In addition, the program FGENESH (http://linux1.softberry.com/) by SoftBerry (Mount Kisco, NY) offers multiple methods to predict gene models.222 However, these programs have been trained to a small set of fungal strains and often do not correctly predict the less common splicing pattern present in Basidiomycota.
Figure 2. Comparison of the intron architecture predicted by JGI and Augustus, and that found in cloned and characterized sesquiterpene synthases from S. hirsutum.
Included are the transcripts encoding the active Stehi1|25180, 73029, 64702, 128017 and 159379 enzymes. Stop codons are indicated with black arrowheads. Note that Stehi1|64702-short contained an additional splice site compared to the active transcript, while Stehi1|159379 isolate 6 was spliced to include a stop codon. An active form of Stehi1|113028 could not be obtained.2020
Collection of fungal tissue sufficient for cDNA isolation is often a significant barrier, as many fungi, and especially many Basidiomycota, are difficult or impossible to culture in the laboratory and may be not available from field collections. Also, the target genes for cloning must be expressed at the time of cDNA preparation. Fungal gene expression is highly complex, subject to RNAi silencing and influenced by trans-acting elements of the genome architecture.223, 224 Therefore, a large number of biosynthetic gene clusters remain silent.19 Strategies have been developed to activate fungal natural product pathways including modification of culture conditions, co-culture,225 and epigenetic manipulation, typically in high-throughput assays to produce novel fungal compounds for isolation.19, 226–229 However, the conditions required to express specific genes are generally unknown, and cannot be predicted with confidence.
Most Basidiomycota are not genetically tractable and no suitable tractable Basidiomycota model systems are available that could correctly splice and functionally express heterologous biosynthetic gene clusters cloned from other members of this fungal division. Hence, characterization of terpenoid natural product pathways identified in these fungi has to rely on the tedious assembly method of one gene at a time, in alternative hosts, such as yeast. Successful expression of pathways in these hosts can be challenging, as some enzymes e.g. cytochrome P450 monoxygenases, require a specific reductase for optimal activity230 and modified terpenoids may be toxic to the expression host, therefore complex cellular sequestration and secretion machinery could be required to prevent reduced cell growth.134 In the future, additional development of genetic platforms specifically designed for natural product pathway expression in Basidiomycota, such as Ustilago maydis, is needed,231, 232 similar to the advanced tools currently available for Ascomycota.233
6 Conclusions and outlook
Fungi have an enormous capacity for terpenoid natural product biosynthesis, and therefore represent an incredible resource for the discovery of new biosynthetic pathways. Identification and characterization of some of the pathways responsible for terpenoid production has revealed a diversity in structure, chemistry and function, including new classes of enzymes. The increasing availability of fungal genome sequences is streamlining the identification of novel pathways and enzymes to a wide range of bioactive terpenoids. The majority of such biosynthetic studies have focused on Ascomycota and only recently have we begun to look into the terpenome of Basidiomycota. Genome surveys clearly show that we have barely begun to unlock some of this biosynthetic potential encoded in the relatively small number of fungi with sequenced genomes that represent just a tiny fraction of the fungal diversity (Figure 1).
Our ability to characterize the fungal terpenome is dependent on, and currently severely limited by, our capability to rapidly identify and subsequently biochemically characterize terpenoid biosynthetic genes and their diverse functions. Current approaches of first finding potential pathways and then using molecular biology strategies to tease out the functions of individual enzymes in a biosynthetic pathway are slow, cumbersome and inadequate in the face of rapidly increasing genomic information. The development of better bioinformatic and genetic tools for the mining and heterologous assembly of large natural product pathways will be absolutely essential to enable high-throughput pathway discovery. For this to occur, it will be important for the natural products community to adopt synthetic biology and modular pathway assembly principles along with suitable platform organisms for process automation. The ultimate goal for the development of such discovery pipelines will be the ability to quickly move from in silico pathway prediction to high-throughput biosynthetic pathway assembly, heterologous expression, and analytical profiling of products followed by bioactivity screening. Revitalization of the natural products drug discovery pipeline will critically depend on the implementation of such strategies.
Acknowledgments
The authors apologize to any colleagues whose work was not discussed due to space limitations. The authors’ research in fungal terpenoid biosynthesis has been supported by the National Institutes of Health Grant GM080299 (to C.S-D.).
References
- 1.Galagan JE, Henn MR, Ma LJ, Cuomo CA, Birren B. Genome Res. 2005;15:1620–1631. doi: 10.1101/gr.3767105. [DOI] [PubMed] [Google Scholar]
- 2.Tedersoo L, May TW, Smith ME. Mycorrhiza. 2010;20:217–263. doi: 10.1007/s00572-009-0274-x. [DOI] [PubMed] [Google Scholar]
- 3.Blackwell M. Am J Bot. 2011;98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- 4.Bass D, Richards TA. Fungal Biol Rev. 2011;25:159–164. [Google Scholar]
- 5.Alves MJ, Ferreira IC, Dias J, Teixeira V, Martins A, Pintado M. Planta Med. 2012;78:1707–1718. doi: 10.1055/s-0032-1315370. [DOI] [PubMed] [Google Scholar]
- 6.Alves MJ, Ferreira IC, Martins A, Pintado M. J Appl Microbiol. 2012;113:466–475. doi: 10.1111/j.1365-2672.2012.05347.x. [DOI] [PubMed] [Google Scholar]
- 7.Fraga BM. Nat Prod Rep. 2012;29:1334–1366. doi: 10.1039/c2np20074k. [DOI] [PubMed] [Google Scholar]
- 8.Gershenzon J, Dudareva N. Nat Chem Biol. 2007;3:408–414. doi: 10.1038/nchembio.2007.5. [DOI] [PubMed] [Google Scholar]
- 9.Abraham WR. Curr Med Chem. 2001;8:583–606. doi: 10.2174/0929867013373147. [DOI] [PubMed] [Google Scholar]
- 10.Elisashvili V. Int J Med Mushrooms. 2012;14:211–239. doi: 10.1615/intjmedmushr.v14.i3.10. [DOI] [PubMed] [Google Scholar]
- 11.Evidente A, Kornienko A, Cimmino A, Andolfi A, Lefranc F, Mathieu V, Kiss R. Nat Prod Rep. 2014;31:617–627. doi: 10.1039/c3np70078j. [DOI] [PubMed] [Google Scholar]
- 12.Misiek M, Hoffmeister D. Planta Med. 2007;73:103–115. doi: 10.1055/s-2007-967104. [DOI] [PubMed] [Google Scholar]
- 13.Wasser SP. Appl Microbiol Biotechnol. 2011;89:1323–1332. doi: 10.1007/s00253-010-3067-4. [DOI] [PubMed] [Google Scholar]
- 14.Zaidman BZ, Yassin M, Mahajna J, Wasser SP. Appl Microbiol Biotechnol. 2005;67:453–468. doi: 10.1007/s00253-004-1787-z. [DOI] [PubMed] [Google Scholar]
- 15.Schobert R, Knauer S, Seibt S, Biersack B. Curr Med Chem. 2011;18:790–807. doi: 10.2174/092986711794927766. [DOI] [PubMed] [Google Scholar]
- 16.Brase S, Encinas A, Keck J, Nising CF. Chem Rev. 2009;109:3903–3990. doi: 10.1021/cr050001f. [DOI] [PubMed] [Google Scholar]
- 17.Walton JD. Fungal Genet Biol. 2000;30:167–171. doi: 10.1006/fgbi.2000.1224. [DOI] [PubMed] [Google Scholar]
- 18.Wawrzyn GT, Bloch SE, Schmidt-Dannert C. Methods Enzymol. 2012;515:83–105. doi: 10.1016/B978-0-12-394290-6.00005-7. [DOI] [PubMed] [Google Scholar]
- 19.Wiemann P, Keller NP. J Ind Microbiol Biotechnol. 2014;41:301–313. doi: 10.1007/s10295-013-1366-3. [DOI] [PubMed] [Google Scholar]
- 20.Quin MB, Flynn CM, Wawrzyn GT, Choudhary S, Schmidt-Dannert C. ChemBioChem. 2013;14:2480–2491. doi: 10.1002/cbic.201300349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shendure J, Lieberman Aiden E. Nat Biotechnol. 2012;30:1084–1094. doi: 10.1038/nbt.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martinez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Gorecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kues U, Kumar TK, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, Rokas A, Ruiz-Duenas FJ, Sabat G, Salamov A, Samejima M, Schmutz J, Slot JC, St John F, Stenlid J, Sun H, Sun S, Syed K, Tsang A, Wiebenga A, Young D, Pisabarro A, Eastwood DC, Martin F, Cullen D, Grigoriev IV, Hibbett DS. Science. 2012;336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 23.Wawrzyn GT, Quin MB, Choudhary S, Lopez-Gallego F, Schmidt-Dannert C. Chem Biol. 2012;19:772–783. doi: 10.1016/j.chembiol.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S, Xu J, Liu C, Zhu Y, Nelson DR, Zhou S, Li C, Wang L, Guo X, Sun Y, Luo H, Li Y, Song J, Henrissat B, Levasseur A, Qian J, Li J, Luo X, Shi L, He L, Xiang L, Xu X, Niu Y, Li Q, Han MV, Yan H, Zhang J, Chen H, Lv A, Wang Z, Liu M, Schwartz DC, Sun C. Nature Communications. 2012;3:913. doi: 10.1038/ncomms1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bushley KE, Raja R, Jaiswal P, Cumbie JS, Nonogaki M, Boyd AE, Owensby CA, Knaus BJ, Elser J, Miller D, Di Y, McPhail KL, Spatafora JW. PLoS Genet. 2013;9:e1003496. doi: 10.1371/journal.pgen.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christianson DW. Curr Opin Chem Biol. 2008;12:141–150. doi: 10.1016/j.cbpa.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miziorko HM. Arch Biochem Biophys. 2011;505:131–143. doi: 10.1016/j.abb.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christianson DW. Chem Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 29.Wendt KU, Schulz GE. Structure. 1998;6:127–133. doi: 10.1016/s0969-2126(98)00015-x. [DOI] [PubMed] [Google Scholar]
- 30.Oldfield E, Lin FY. Angew Chem Int Ed Engl. 2012;51:1124–1137. doi: 10.1002/anie.201103110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dickschat JS. Nat Prod Rep. 2011;28:1917–1936. doi: 10.1039/c1np00063b. [DOI] [PubMed] [Google Scholar]
- 32.Tantillo DJ. Nat Prod Rep. 2011;28:1956–1956. doi: 10.1039/c1np00006c. [DOI] [PubMed] [Google Scholar]
- 33.Miller DJ, Allemann RK. Nat Prod Rep. 2012;29:60–71. doi: 10.1039/c1np00060h. [DOI] [PubMed] [Google Scholar]
- 34.Lesburg CA, Zhai G, Cane DE, Christianson DW. Science. 1997;277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- 35.Starks CM, Back KW, Chappell J, Noel JP. Science. 1997;277:1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 36.Cane DE, Xue Q, Fitzsimmons BC. Biochemistry. 1996;35:12369–12376. doi: 10.1021/bi961344y. [DOI] [PubMed] [Google Scholar]
- 37.Cane DE, Ha HJ, Pargellis C, Waldmeier F, Swanson S, Murthy PPN. Bioorg Chem. 1985;13:246–265. [Google Scholar]
- 38.Cane DE, Swanson S, Murthy PPN. J Am Chem Soc. 1981;103:2136–2138. [Google Scholar]
- 39.Baer P, Rabe P, Citron CA, de Oliveira Mann CC, Kaufmann N, Groll M, Dickschat JS. ChemBioChem. 2014;15:213–216. doi: 10.1002/cbic.201300708. [DOI] [PubMed] [Google Scholar]
- 40.Rabe P, Citron CA, Dickschat JS. ChemBioChem. 2013;14:2345–2354. doi: 10.1002/cbic.201300329. [DOI] [PubMed] [Google Scholar]
- 41.Garms S, Kollner TG, Boland W. J Org Chem. 2010;75:5590–5600. doi: 10.1021/jo100917c. [DOI] [PubMed] [Google Scholar]
- 42.Vedula LS, Jiang J, Zakharian T, Cane DE, Christianson DW. Arch Biochem Biophys. 2008;469:184–194. doi: 10.1016/j.abb.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Felicetti B, Cane DE. J Am Chem Soc. 2004;126:7212–7221. doi: 10.1021/ja0499593. [DOI] [PubMed] [Google Scholar]
- 44.Seemann M, Zhai G, de Kraker JW, Paschall CM, Christianson DW, Cane DE. J Am Chem Soc. 2002;124:7681–7689. doi: 10.1021/ja026058q. [DOI] [PubMed] [Google Scholar]
- 45.Faraldos JA, Gonzalez V, Allemann RK. Chem Commun. 2012;48:3230–3232. doi: 10.1039/c2cc17588f. [DOI] [PubMed] [Google Scholar]
- 46.Chen M, Al-lami N, Janvier M, D'Antonio EL, Faraldos JA, Cane DE, Allemann RK, Christianson DW. Biochemistry. 2013;52:5441–5453. doi: 10.1021/bi400691v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aaron JA, Lin X, Cane DE, Christianson DW. Biochemistry. 2010;49:1787–1797. doi: 10.1021/bi902088z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caruthers JM, Kang I, Rynkiewicz MJ, Cane DE, Christianson DW. J Biol Chem. 2000;275:25533–25539. doi: 10.1074/jbc.M000433200. [DOI] [PubMed] [Google Scholar]
- 49.Gennadios HA, Gonzalez V, Di Costanzo L, Li A, Yu F, Miller DJ, Allemann RK, Christianson DW. Biochemistry. 2009;48:6175–6183. doi: 10.1021/bi900483b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McAndrew RP, Peralta-Yahya PP, DeGiovanni A, Pereira JH, Hadi MZ, Keasling JD, Adams PD. Structure. 2011;19:1876–1884. doi: 10.1016/j.str.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 51.Rynkiewicz MJ, Cane DE, Christianson DW. Proc Natl Acad Sci USA. 2001;98:13543–13548. doi: 10.1073/pnas.231313098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shishova EY, Di Costanzo L, Cane DE, Christianson DW. Biochemistry. 2007;46:1941–1951. doi: 10.1021/bi0622524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shishova EY, Yu F, Miller DJ, Faraldos JA, Zhao Y, Coates RM, Allemann RK, Cane DE, Christianson DW. J Biol Chem. 2008;283:15431–15439. doi: 10.1074/jbc.M800659200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rynkiewicz MJ, Cane DE, Christianson DW. Biochemistry. 2002;41:1731–1741. doi: 10.1021/bi011960g. [DOI] [PubMed] [Google Scholar]
- 55.Vedula LS, Cane DE, Christianson DW. Biochemistry. 2005;44:12719–12727. doi: 10.1021/bi0510476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hohn TM, Vanmiddlesworth F. Arch Biochem Biophys. 1986;251:756–761. doi: 10.1016/0003-9861(86)90386-3. [DOI] [PubMed] [Google Scholar]
- 57.Osborne LE, Stein JM. Int J Food Microbiol. 2007;119:103–108. doi: 10.1016/j.ijfoodmicro.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 58.Vedula LS, Zhao Y, Coates RM, Koyama T, Cane DE, Christianson DW. Arch Biochem Biophys. 2007;466:260–266. doi: 10.1016/j.abb.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cane DE, Ha HJ. J Am Chem Soc. 1988;110:6865–6870. [Google Scholar]
- 60.Dickschat JS, Brock NL, Citron CA, Tudzynski B. ChemBioChem. 2011;12:2088–2095. doi: 10.1002/cbic.201100268. [DOI] [PubMed] [Google Scholar]
- 61.Hong YJ, Tantillo DJ. Org Lett. 2006;8:4601–4604. doi: 10.1021/ol061884f. [DOI] [PubMed] [Google Scholar]
- 62.Miller DJ, Gao J, Truhlar DG, Young NJ, Gonzalez V, Allemann RK. Org Biomol Chem. 2008;6:2346–2354. doi: 10.1039/b804198a. [DOI] [PubMed] [Google Scholar]
- 63.Faraldos JA, Gonzalez V, Senske M, Allemann RK. Org Biomol Chem. 2011;9:6920–6923. doi: 10.1039/c1ob06184d. [DOI] [PubMed] [Google Scholar]
- 64.Motohashi K, Hashimoto J, Inaba S, Khan ST, Komaki H, Nagai A, Takagi M, Shin-ya K. J Antibiot (Tokyo) 2009;62:247–250. doi: 10.1038/ja.2009.21. [DOI] [PubMed] [Google Scholar]
- 65.McCormick SP, Alexander NJ, Harris LJ. Appl Environ Microbiol. 2010;76:136–141. doi: 10.1128/AEM.02017-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brock NL, Huss K, Tudzynski B, Dickschat JS. ChemBioChem. 2013;14:311–315. doi: 10.1002/cbic.201200695. [DOI] [PubMed] [Google Scholar]
- 67.Pinedo C, Wang CM, Pradier JM, Dalmais B, Choquer M, Le Pecheur P, Morgant G, Collado IG, Cane DE, Viaud M. ACS Chem Biol. 2008;3:791–801. doi: 10.1021/cb800225v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang CM, Hopson R, Lin X, Cane DE. J Am Chem Soc. 2009;131:8360–8361. doi: 10.1021/ja9021649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Agger S, Lopez-Gallego F, Schmidt-Dannert C. Mol Microbiol. 2009;72:1181–1195. doi: 10.1111/j.1365-2958.2009.06717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lopez-Gallego F, Agger SA, Abate-Pella D, Distefano MD, Schmidt-Dannert C. ChemBioChem. 2010;11:1093–1106. doi: 10.1002/cbic.200900671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lopez-Gallego F, Wawrzyn GT, Schmidt-Dannert C. Appl Environ Microbiol. 2010;76:7723–7733. doi: 10.1128/AEM.01811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schobert R, Seibt S, Mahal K, Ahmad A, Biersack B, Effenberger-Neidnicht K, Padhye S, Sarkar FH, Mueller T. J Med Chem. 2011;54:6177–6182. doi: 10.1021/jm200359n. [DOI] [PubMed] [Google Scholar]
- 73.Kelner MJ, McMorris TC, Beck WT, Zamora JM, Taetle R. Cancer Res. 1987;47:3186–3189. [PubMed] [Google Scholar]
- 74.McMorris TC, Anchel M. J Am Chem Soc. 1965;87:1594–1600. doi: 10.1021/ja01085a031. [DOI] [PubMed] [Google Scholar]
- 75.Nair MS, Anchel M. Tetrahedron Lett. 1972;13:2753–2754. [Google Scholar]
- 76.Quin MB, Wawrzyn G, Schmidt-Dannert C. Acta Crystallogr Sect F. 2013;69:574–577. doi: 10.1107/S1744309113010749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Engels B, Heinig U, Grothe T, Stadler M, Jennewein S. J Biol Chem. 2011;286:6871–6878. doi: 10.1074/jbc.M110.165845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martin F, Cullen D, Hibbett D, Pisabarro A, Spatafora JW, Baker SE, Grigoriev IV. New Phytol. 2011;190:818–821. doi: 10.1111/j.1469-8137.2011.03688.x. [DOI] [PubMed] [Google Scholar]
- 79.Cuomo CA, Birren BW. Methods Enzymol. 2010;470:833–855. doi: 10.1016/S0076-6879(10)70034-3. [DOI] [PubMed] [Google Scholar]
- 80.Ayer WA, Saeedighomi MH. Tetrahedron Lett. 1981;22:2071–2074. [Google Scholar]
- 81.Abell C, Leech AP. Tetrahedron Lett. 1988;29:4337–4340. [Google Scholar]
- 82.Ainsworth AM, Rayner ADM, Broxholme SJ, Beeching JR, Pryke JA, Scard PR, Berriman J, Powell KA, Floyd AJ, Branch SK. Mycol Res. 1990;94:799–809. [Google Scholar]
- 83.Yun BS, Lee IK, Cho Y, Cho SM, Yoo ID. J Nat Prod. 2002;65:786–788. doi: 10.1021/np010602b. [DOI] [PubMed] [Google Scholar]
- 84.Kim YH, Yun BS, Ryoo IJ, Kim JP, Koshino H, Yoo ID. J Antibiot. 2006;59:432–434. doi: 10.1038/ja.2006.61. [DOI] [PubMed] [Google Scholar]
- 85.Yoo N-H, Kim J-P, Yun B-S, Ryoo I-J, Lee I-K, Yoon E-S, Koshino H, Yoo I-D. J Antibiot. 2006;59:110–113. doi: 10.1038/ja.2006.16. [DOI] [PubMed] [Google Scholar]
- 86.Li GH, Duan M, Yu ZF, Li L, Dong JY, Wang XB, Guo JW, Huang R, Wang M, Zhang KQ. Phytochemistry. 2008;69:1439–1445. doi: 10.1016/j.phytochem.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 87.Opatz T, Kolshorn H, Anke H. J Antibiot. 2008;61:563–567. doi: 10.1038/ja.2008.75. [DOI] [PubMed] [Google Scholar]
- 88.Liermann JC, Schuffler A, Wollinsky B, Birnbacher J, Kolshorn H, Anke T, Opatz T. J Org Chem. 2010;75:2955–2961. doi: 10.1021/jo100202b. [DOI] [PubMed] [Google Scholar]
- 89.Isaka M, Srisanoh U, Choowong W, Boonpratuang T. Org Lett. 2011;13:4886–4889. doi: 10.1021/ol2019778. [DOI] [PubMed] [Google Scholar]
- 90.Rao YK, Wu AT, Geethangili M, Huang MT, Chao WJ, Wu CH, Deng WP, Yeh CT, Tzeng YM. Chem Res Toxicol. 2011;24:238–245. doi: 10.1021/tx100318m. [DOI] [PubMed] [Google Scholar]
- 91.Isaka M, Srisanoh U, Sappan M, Supothina S, Boonpratuang T. Phytochemistry. 2012;79:116–120. doi: 10.1016/j.phytochem.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 92.Zheng X, Li GH, Xie MJ, Wang X, Sun R, Lu H, Zhang KQ. Phytochemistry. 2013;86:144–150. doi: 10.1016/j.phytochem.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 93.Ma K, Bao L, Han J, Jin T, Yang X, Zhao F, Li S, Song F, Liu M, Liu H. Food Chem. 2014;143:239–245. doi: 10.1016/j.foodchem.2013.07.124. [DOI] [PubMed] [Google Scholar]
- 94.Zu L, Xu M, Lodewyk MW, Cane DE, Peters RJ, Tantillo DJ. J Am Chem Soc. 2012;134:11369–11371. doi: 10.1021/ja3043245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fraga BM. Nat Prod Rep. 2013;30:1226–1264. doi: 10.1039/c3np70047j. [DOI] [PubMed] [Google Scholar]
- 96.Erkel G, Anke T. In: Biotechnology: Products of Secondary Metabolism. 2. Rehm H-J, Reed G, editors. Wiley; 1997. pp. 490–533. [Google Scholar]
- 97.Kramer R, Abraham W-R. Phytochem Rev. 2011;11:15–37. [Google Scholar]
- 98.Jansen BJM, de Groot A. Nat Prod Rep. 2004;21:449–477. doi: 10.1039/b311170a. [DOI] [PubMed] [Google Scholar]
- 99.Luo D-Q, Gao Y, Gao J-M, Wang F, Yang X-L, Liu J-K. J Nat Prod. 2006;69:1354–1357. doi: 10.1021/np060153l. [DOI] [PubMed] [Google Scholar]
- 100.Simon B, Anke T, Anders U, Neuhaus M, Hansske E. Zeitschrift Fur Naturforschung C-a Journal of Biosciences. 1995;50:173–180. doi: 10.1515/znc-1995-3-403. [DOI] [PubMed] [Google Scholar]
- 101.Coates RM, Ho JZ, Klobus M, Zhu LJ. J Org Chem. 1998;63:9166–9176. [Google Scholar]
- 102.Ayer WA, Cruz ER. J Org Chem. 1993;58:7529–7534. [Google Scholar]
- 103.Cruz ER. Can J Chem. 1997;75:834–839. [Google Scholar]
- 104.Abraham WR, Ernst L, Witte L, Hanssen HP, Sprecher E. Tetrahedron. 1986;42:4475–4480. [Google Scholar]
- 105.Feline TC, Mellows G, Jones RB, Phillips L. J Chem Soc, Chem Commun. 1974;2:63–64. [PubMed] [Google Scholar]
- 106.Covello PS, Teoh KH, Polichuk DR, Reed DW, Nowak G. Phytochemistry. 2007;68:1864–1871. doi: 10.1016/j.phytochem.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 107.Tokai T, Koshino H, Takahashi-Ando N, Sato M, Fujimura M, Kimura M. Biochem Biophys Res Commun. 2007;353:412–417. doi: 10.1016/j.bbrc.2006.12.033. [DOI] [PubMed] [Google Scholar]
- 108.Daniewski WM, Vidari G. Constituents of Lactarius (mushrooms) Springer-Verlag Wien; 1999. pp. 69–171. [Google Scholar]
- 109.Schuffler A, Wollinsky B, Anke T, Liermann JC, Opatz T. J Nat Prod. 2012;75:1405–1408. doi: 10.1021/np3000552. [DOI] [PubMed] [Google Scholar]
- 110.Wang Y, Bao L, Liu D, Yang X, Li S, Gao H, Yao X, Wen H, Liu H. Tetrahedron. 2012;68:3012–3018. [Google Scholar]
- 111.Sterner O, Anke T, Sheldrick WS, Steglich W. Tetrahedron. 1990;46:2389–2400. [Google Scholar]
- 112.Lackner G, Bohnert M, Wick J, Hoffmeister D. Chem Biol. 2013;20:1101–1106. doi: 10.1016/j.chembiol.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 113.Opatz T, Kolshorn H, Birnbacher J, Schuffler A, Deininger F, Anke T. Eur J Org Chem. 2007:5546–5550. [Google Scholar]
- 114.Grove JF. Progr Chem Org Nat Prod. 2007;88:63–130. [PubMed] [Google Scholar]
- 115.McCormick SP, Stanley AM, Stover NA, Alexander NJ. Toxins (Basel) 2011;3:802–814. doi: 10.3390/toxins3070802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Desjardins AE. J Agric Food Chem. 2009;57:4478–4484. doi: 10.1021/jf9003847. [DOI] [PubMed] [Google Scholar]
- 117.Kimura M, Tokai T, Takahashi-Ando N, Ohsato S, Fujimura M. Biosci, Biotechnol, Biochem. 2007;71:2105–2123. doi: 10.1271/bbb.70183. [DOI] [PubMed] [Google Scholar]
- 118.Kazan K, Gardiner DM, Manners JM. Mol Plant Pathol. 2012;13:399–413. doi: 10.1111/j.1364-3703.2011.00762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Audenaert K, Vanheule A, Hofte M, Haesaert G. Toxins (Basel) 2014;6:1–19. doi: 10.3390/toxins6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gu Q, Zhang C, Liu X, Ma Z. Mol Plant Pathol. 2014 doi: 10.1111/mpp.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hohn TM, Beremand PD. Gene. 1989;79:131–138. doi: 10.1016/0378-1119(89)90098-x. [DOI] [PubMed] [Google Scholar]
- 122.Brown DW, McCormick SP, Alexander NJ, Proctor RH, Desjardins AE. Fungal Genet Biol. 2002;36:224–233. doi: 10.1016/s1087-1845(02)00021-x. [DOI] [PubMed] [Google Scholar]
- 123.Nasmith CG, Walkowiak S, Wang L, Leung WW, Gong Y, Johnston A, Harris LJ, Guttman DS, Subramaniam R. PLoS Path. 2011;7:e1002266. doi: 10.1371/journal.ppat.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McCormick SP, Alexander NJ, Proctor RH. Can J Microbiol. 2006;52:636–642. doi: 10.1139/w06-011. [DOI] [PubMed] [Google Scholar]
- 125.Hesketh AR, Bycroft BW, Dewick PM, Gilbert J. Phytochemistry. 1993;32:105–116. [Google Scholar]
- 126.McCormick SP, Alexander NJ, Trapp SE, Hohn TM. Appl Environ Microbiol. 1999;65:5252–5256. doi: 10.1128/aem.65.12.5252-5256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ohsato S, Ochiai-Fukuda T, Nishiuchi T, Takahashi-Ando N, Koizumi S, Hamamoto H, Kudo T, Yamaguchi I, Kimura M. Plant Cell Rep. 2007;26:531–538. doi: 10.1007/s00299-006-0251-1. [DOI] [PubMed] [Google Scholar]
- 128.Alexander NJ, Hohn TM, McCormick SP. Appl Environ Microbiol. 1998;64:221–225. doi: 10.1128/aem.64.1.221-225.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McCormick SP, Hohn TM, Desjardins AE. Appl Environ Microbiol. 1996;62:353–359. doi: 10.1128/aem.62.2.353-359.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brown DW, Dyer RB, McCormick SP, Kendra DF, Plattner RD. Fungal Genet Biol. 2004;41:454–462. doi: 10.1016/j.fgb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 131.Alexander NJ, McCormick SP, Waalwijk C, van der Lee T, Proctor RH. Fungal Genet Biol. 2011;48:485–495. doi: 10.1016/j.fgb.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 132.Alexander NJ, McCormick SP, Hohn TM. Mol Genet Genomics. 1999;261:977–984. doi: 10.1007/s004380051046. [DOI] [PubMed] [Google Scholar]
- 133.Menke J, Dong Y, Kistler HC. Mol Plant-Microbe Interact. 2012;25:1408–1418. doi: 10.1094/MPMI-04-12-0081-R. [DOI] [PubMed] [Google Scholar]
- 134.Menke J, Weber J, Broz K, Kistler HC. PLoS One. 2013;8:e63077. doi: 10.1371/journal.pone.0063077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chanda A, Roze LV, Kang S, Artymovich KA, Hicks GR, Raikhel NV, Calvo AM, Linz JE. Proc Natl Acad Sci U S A. 2009;106:19533–19538. doi: 10.1073/pnas.0907416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chanda A, Roze LV, Linz JE. Eukaryot Cell. 2010;9:1724–1727. doi: 10.1128/EC.00118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hidalgo PI, Ullan RV, Albillos SM, Montero O, Fernandez-Bodega MA, Garcia-Estrada C, Fernandez-Aguado M, Martin JF. Fungal Genet Biol. 2014;62:11–24. doi: 10.1016/j.fgb.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 138.Zi J, Mafu S, Peters RJ. Annu Rev Plant Biol. 2014;65 doi: 10.1146/annurev-arplant-050213-035705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Peters RJ. Nat Prod Rep. 2010;27:1521–1530. doi: 10.1039/c0np00019a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Abe I. Nat Prod Rep. 2007;24:1311–1331. doi: 10.1039/b616857b. [DOI] [PubMed] [Google Scholar]
- 141.Kawaide H, Imai R, Sassa T, Kamiya Y. J Biol Chem. 1997;272:21706–21712. doi: 10.1074/jbc.272.35.21706. [DOI] [PubMed] [Google Scholar]
- 142.Kawaide H, Sassa T, Kamiya Y. J Biol Chem. 2000;275:2276–2280. doi: 10.1074/jbc.275.4.2276. [DOI] [PubMed] [Google Scholar]
- 143.Sun TP, Kamiya Y. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yamaguchi S, Saito T, Abe H, Yamane H, Murofushi N, Kamiya Y. Plant J. 1996;10:203–213. doi: 10.1046/j.1365-313x.1996.10020203.x. [DOI] [PubMed] [Google Scholar]
- 145.Koksal M, Hu H, Coates RM, Peters RJ, Christianson DW. Nat Chem Biol. 2011;7:431–433. doi: 10.1038/nchembio.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Koksal M, Jin Y, Coates RM, Croteau R, Christianson DW. Nature. 2011;469:116–120. doi: 10.1038/nature09628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhou K, Gao Y, Hoy JA, Mann FM, Honzatko RB, Peters RJ. J Biol Chem. 2012;287:6840–6850. doi: 10.1074/jbc.M111.337592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tudzynski B, Holter K. Fungal Genet Biol. 1998;25:157–170. doi: 10.1006/fgbi.1998.1095. [DOI] [PubMed] [Google Scholar]
- 149.Tudzynski B, Kawaide H, Kamiya Y. Curr Genet. 1998;34:234–240. doi: 10.1007/s002940050392. [DOI] [PubMed] [Google Scholar]
- 150.Bromann K, Toivari M, Viljanen K, Vuoristo A, Ruohonen L, Nakari-Setala T. PLoS One. 2012;7:e35450. doi: 10.1371/journal.pone.0035450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Toyomasu T, Niida R, Kenmoku H, Kanno Y, Miura S, Nakano C, Shiono Y, Mitsuhashi W, Toshima H, Oikawa H, Hoshino T, Dairi T, Kato N, Sassa T. Biosci Biotechnol Biochem. 2008;72:1038–1047. doi: 10.1271/bbb.70790. [DOI] [PubMed] [Google Scholar]
- 152.Oikawa H, Toyomasu T, Toshima H, Ohashi S, Kawaide H, Kamiya Y, Ohtsuka M, Shinoda S, Mitsuhashi W, Sassa T. J Am Chem Soc. 2001;123:5154–5155. doi: 10.1021/ja015747j. [DOI] [PubMed] [Google Scholar]
- 153.Mitterbauer R, Specht T. WO/2011/110610. 2011;EP2011/053571:WO2011110610A2011110611. [Google Scholar]
- 154.Mende K, Homann V, Tudzynski B. Mol Genet Genomics. 1997;255:96–105. doi: 10.1007/s004380050477. [DOI] [PubMed] [Google Scholar]
- 155.Bomke C, Tudzynski B. Phytochemistry. 2009;70:1876–1893. doi: 10.1016/j.phytochem.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 156.Toyomasu T, Kaneko A, Tokiwano T, Kanno Y, Kanno Y, Niida R, Miura S, Nishioka T, Ikeda C, Mitsuhashi W, Dairi T, Kawano T, Oikawa H, Kato N, Sassa T. J Org Chem. 2009;74:1541–1548. doi: 10.1021/jo802319e. [DOI] [PubMed] [Google Scholar]
- 157.Toyomasu T, Tsukahara M, Kaneko A, Niida R, Mitsuhashi W, Dairi T, Kato N, Sassa T. Proc Natl Acad Sci USA. 2007;104:3084–3088. doi: 10.1073/pnas.0608426104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Minami A, Tajima N, Higuchi Y, Toyomasu T, Sassa T, Kato N, Dairi T. Bioorg Med Chem Lett. 2009;19:870–874. doi: 10.1016/j.bmcl.2008.11.108. [DOI] [PubMed] [Google Scholar]
- 159.Wendt KU, Poralla K, Schulz GE. Science. 1997;277:1811–1815. doi: 10.1126/science.277.5333.1811. [DOI] [PubMed] [Google Scholar]
- 160.Tarshis LC, Yan M, Poulter CD, Sacchettini JC. Biochemistry. 1994;33:10871–10877. doi: 10.1021/bi00202a004. [DOI] [PubMed] [Google Scholar]
- 161.Chiba R, Minami A, Gomi K, Oikawa H. Org Lett. 2013;15:594–597. doi: 10.1021/ol303408a. [DOI] [PubMed] [Google Scholar]
- 162.Smanski MJ, Peterson RM, Huang SX, Shen B. Curr Opin Chem Biol. 2012;16:132–141. doi: 10.1016/j.cbpa.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shen JW, Ruan Y, Ma BJ. J Basic Microbiol. 2009;49:242–255. doi: 10.1002/jobm.200800102. [DOI] [PubMed] [Google Scholar]
- 164.Tudzynski B. Appl Microbiol Biotechnol. 2005;66:597–611. doi: 10.1007/s00253-004-1805-1. [DOI] [PubMed] [Google Scholar]
- 165.Hedden P, Phillips AL, Rojas MC, Carrera E, Tudzynski B. J Plant Growth Regul. 2001;20:319–331. doi: 10.1007/s003440010037. [DOI] [PubMed] [Google Scholar]
- 166.Tudzynski B, Rojas MC, Gaskin P, Hedden P. J Biol Chem. 2002;277:21246–21253. doi: 10.1074/jbc.M201651200. [DOI] [PubMed] [Google Scholar]
- 167.Tudzynski B, Mihlan M, Rojas MC, Linnemannstons P, Gaskin P, Hedden P. J Biol Chem. 2003;278:28635–28643. doi: 10.1074/jbc.M301927200. [DOI] [PubMed] [Google Scholar]
- 168.Malonek S, Rojas MC, Hedden P, Gaskin P, Hopkins P, Tudzynski B. J Biol Chem. 2004;279:25075–25084. doi: 10.1074/jbc.M308517200. [DOI] [PubMed] [Google Scholar]
- 169.Bomke C, Rojas MC, Gong F, Hedden P, Tudzynski B. Appl Environ Microbiol. 2008;74:5325–5339. doi: 10.1128/AEM.00694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kawaide H. Biosci, Biotechnol, Biochem. 2006;70:583–590. doi: 10.1271/bbb.70.583. [DOI] [PubMed] [Google Scholar]
- 171.Kawaide H, Sassa T, Kamiya Y. Phytochemistry. 1995;39:305–310. [Google Scholar]
- 172.Fujii R, Minami A, Tsukagoshi T, Sato N, Sahara T, Ohgiya S, Gomi K, Oikawa H. Biosci Biotechnol Biochem. 2011;75:1813–1817. doi: 10.1271/bbb.110366. [DOI] [PubMed] [Google Scholar]
- 173.Ono Y, Minami A, Noike M, Higuchi Y, Toyomasu T, Sassa T, Kato N, Dairi T. J Am Chem Soc. 2011;133:2548–2555. doi: 10.1021/ja107785u. [DOI] [PubMed] [Google Scholar]
- 174.Noike M, Ono Y, Araki Y, Tanio R, Higuchi Y, Nitta H, Hamano Y, Toyomasu T, Sassa T, Kato N, Dairi T. PLoS One. 2012;7:e42090. doi: 10.1371/journal.pone.0042090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Noike M, Liu C, Ono Y, Hamano Y, Toyomasu T, Sassa T, Kato N, Dairi T. ChemBioChem. 2012;13:566–573. doi: 10.1002/cbic.201100725. [DOI] [PubMed] [Google Scholar]
- 176.Hashimoto M, Higuchi Y, Takahashi S, Osada H, Sakaki T, Toyomasu T, Sassa T, Kato N, Dairi T. Bioorg Med Chem Lett. 2009;19:5640–5643. doi: 10.1016/j.bmcl.2009.08.026. [DOI] [PubMed] [Google Scholar]
- 177.Hill RA, Connolly JD. Nat Prod Rep. 2013;30:1028–1065. doi: 10.1039/c3np70032a. [DOI] [PubMed] [Google Scholar]
- 178.Kristan K, Rizner TL. J Steroidal Biochem Mol Biol. 2012;129:79–91. doi: 10.1016/j.jsbmb.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 179.Garaiova M, Zambojova V, Simova Z, Griac P, Hapala I. FEMS Yeast Res. 2014;14:310–323. doi: 10.1111/1567-1364.12107. [DOI] [PubMed] [Google Scholar]
- 180.Thoma R, Schulz-Gascg T, D'Arcy B, Benz J, Aebi J, Dehmlow H, Hennig M, Stihle M, Ruf A. Nature. 2004;432:118–122. doi: 10.1038/nature02993. [DOI] [PubMed] [Google Scholar]
- 181.Corey EJ, Virgil SC. J Am Chem Soc. 1991;113:4025–4026. [Google Scholar]
- 182.Siedenburg G, Jendrossek D. Appl Environ Microbiol. 2011;77:3905–3915. doi: 10.1128/AEM.00300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Poralla K, Hewelt A, Prestwich GD, Abe I, Reipen I, Sprenger G. Trends Biochem Sci. 1994;19:157–158. doi: 10.1016/0968-0004(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 184.Frickey T, Kannenberg E. Environ Microbiol. 2009;11:1224–1241. doi: 10.1111/j.1462-2920.2008.01851.x. [DOI] [PubMed] [Google Scholar]
- 185.Corey EJ, Matsuda SPT, Bartel B. Proc Natl Acad Sci USA. 1994;91:2211–2215. doi: 10.1073/pnas.91.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shi Z, Buntel CJ, Griffin JH. Proc Natl Acad Sci USA. 1994;91:7370–7374. doi: 10.1073/pnas.91.15.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Corey EJ, Cheng H, Hunter Baker C, Matsuda SPT, Li D, Song XD. J Am Chem Soc. 1997;119:1289–1296. [Google Scholar]
- 188.Wu TK, Wen HY, Chang CH, Liu YT. Org Lett. 2008;10:2529–2532. doi: 10.1021/ol800799n. [DOI] [PubMed] [Google Scholar]
- 189.Chang CH, Wen HY, Shie WS, Lu CT, Li ME, Liu YT, Li WH, Wu TK. Org Biomol Chem. 2013;11:4214–4219. doi: 10.1039/c3ob40493e. [DOI] [PubMed] [Google Scholar]
- 190.Wu TK, Yu MT, Liu YT, Chang CH, Wang HJ, Diau EWG. Org Lett. 2006;8:1319–1322. doi: 10.1021/ol053134w. [DOI] [PubMed] [Google Scholar]
- 191.Abe I, Naito K, Takagi Y, Noguchi H. Biochim Biophys Acta. 2001;1522:67–73. doi: 10.1016/s0167-4781(01)00307-4. [DOI] [PubMed] [Google Scholar]
- 192.Mitsuguchi H, Seshime Y, Fujii I, Shibuya M, Ebizuka Y, Kushiro T. J Am Chem Soc. 2009;131:6402–6411. doi: 10.1021/ja8095976. [DOI] [PubMed] [Google Scholar]
- 193.Shang C-H, Shi L, Ren A, Qin L, M-Zhao W. Biosci, Biotechnol, Biochem. 2010;74:974–978. doi: 10.1271/bbb.90833. [DOI] [PubMed] [Google Scholar]
- 194.Cheng CR, Yue QX, Wu ZY, Song XY, Tao SJ, Wu XH, Xu PP, Liu X, Guan SH, Guo DA. Phytochemistry. 2010;71:1579–1585. doi: 10.1016/j.phytochem.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 195.Godio RP, Martin JF. Fungal Genet Biol. 2009;46:232–242. doi: 10.1016/j.fgb.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 196.Racolta S, Juhl PB, Sirim D, Pleiss J. Proteins. 2012;80:2009–2019. doi: 10.1002/prot.24089. [DOI] [PubMed] [Google Scholar]
- 197.Phillips DR, Rasbery JM, Bartel B, Matsuda SP. Curr Opin Plant Biol. 2006;9:305–314. doi: 10.1016/j.pbi.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 198.Liu YT, Hu TC, Chang CH, Shie WS, Wu TK. Org Lett. 2012;14:5222–5225. doi: 10.1021/ol302341h. [DOI] [PubMed] [Google Scholar]
- 199.Lodeiro S, Xiong Q, Wilson WK, Kolesnikova MD, Onak CS, Matsuda SP. J Am Chem Soc. 2007;129:11213–11222. doi: 10.1021/ja073133u. [DOI] [PubMed] [Google Scholar]
- 200.Moses T, Pollier J, Thevelein JM, Goossens A. New Phytol. 2013;200:27–43. doi: 10.1111/nph.12325. [DOI] [PubMed] [Google Scholar]
- 201.Sawai S, Saito K. Front Plant Sci. 2011;2:25. doi: 10.3389/fpls.2011.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Xue Z, Duan L, Liu D, Guo J, Ge S, Dicks J, Osbourn OMPA, Qi X. New Phytol. 2012;193:1022–1038. doi: 10.1111/j.1469-8137.2011.03997.x. [DOI] [PubMed] [Google Scholar]
- 203.Rios JL, Andujar I, Recio MC, Giner RM. J Nat Prod. 2012;75:2016–2044. doi: 10.1021/np300412h. [DOI] [PubMed] [Google Scholar]
- 204.Zhao M, Godecke T, Gunn J, Duan JA, Che CT. Molecules. 2013;18:4054–4080. doi: 10.3390/molecules18044054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Shu S, Chen B, Zhou M, Zhao X, Xia H, Wang M. PLoS One. 2013;8:e71350. doi: 10.1371/journal.pone.0071350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Liu D, Gong J, Dai W, Kang X, Huang Z, Zhang HM, Liu W, Liu L, Ma J, Xia Z, Chen Y, Chen Y, Wang D, Ni P, Guo AY, Xiong X. PLoS One. 2012;7:e36146. doi: 10.1371/journal.pone.0036146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Yu GJ, Wang M, Huang J, Yin YL, Chen YJ, Jiang S, Jin YX, Lan XQ, Wong BH, Liang Y, Sun H. PLoS One. 2012;7:e44031. doi: 10.1371/journal.pone.0044031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, Shabalov I, Smirnova T, Grigoriev IV, Dubchak I. Nucleic Acids Res. 2014;42:D26–31. doi: 10.1093/nar/gkt1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 210.Fedorova N, Moktali V, Medema MH. Fungal Secondary Metabolism: Methods and Protocols. Springer Science; 2012. [Google Scholar]
- 211.Weber T. Int J Med Microbiol. 2014;304:230–235. doi: 10.1016/j.ijmm.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 212.Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. Nucleic Acids Res. 2013;41:W204–212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wiemann P, Guo CJ, Palmer JM, Sekonyela R, Wang CC, Keller NP. Proc Natl Acad Sci USA. 2013;110:17065–17070. doi: 10.1073/pnas.1313258110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Pahirulzaman KA, Williams K, Lazarus CM. Methods Enzymol. 2012;517:241–260. doi: 10.1016/B978-0-12-404634-4.00012-7. [DOI] [PubMed] [Google Scholar]
- 215.Tagami K, Minami A, Fujii R, Liu C, Tanaka M, Gomi K, Dairi T, Oikawa H. ChemBioChem. 2014 doi: 10.1002/cbic.201402195. [DOI] [PubMed] [Google Scholar]
- 216.Itoh T, Tokunaga K, Matsuda Y, Fujii I, Abe I, Ebizuka Y, Kushiro T. Nat Chem. 2010;2:858–864. doi: 10.1038/nchem.764. [DOI] [PubMed] [Google Scholar]
- 217.Da Lage JL, Binder M, Hua-Van A, Janecek S, Casane D. BMC Evol Biol. 2013;13:40. doi: 10.1186/1471-2148-13-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.C. M. Flynn, 2014.
- 219.Hoff KJ, Stanke M. Nucleic Acids Res. 2013;41:W123–128. doi: 10.1093/nar/gkt418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Stanke M, Steinkamp R, Waack S, Morgenstern B. Nucleic Acids Res. 2004;32:W309–312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Solovyev V, Kosarev P, Seledsov I, Vorobyev D. Genome Biol. 2006;7(Suppl 1):S10.11–12. doi: 10.1186/gb-2006-7-s1-s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Nakayashiki H. FEBS Lett. 2005;579:5950–5957. doi: 10.1016/j.febslet.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 224.Noble LM, Andrianopoulos A. Genome Biol Evol. 2013;5:1336–1352. doi: 10.1093/gbe/evt077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Bertrand S, Bohni N, Schnee S, Schumpp O, Gindro K, Wolfender JL. Biotechnol Adv. 2014 doi: 10.1016/j.biotechadv.2014.03.001. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 226.Chiang YM, Chang SL, Oakley BR, Wang CC. Curr Opin Chem Biol. 2011;15:137–143. doi: 10.1016/j.cbpa.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Cichewicz RH. Nat Prod Rep. 2010;27:11–22. doi: 10.1039/b920860g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Du L, King JB, Morrow BH, Shen JK, Miller AN, Cichewicz RH. J Nat Prod. 2012;75:1819–1823. doi: 10.1021/np300473h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Brakhage AA, Schroeckh V. Fungal Genet Biol. 2011;48:15–22. doi: 10.1016/j.fgb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 230.Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, Polichuk DR, Teoh KH, Reed DW, Treynor T, Lenihan J, Fleck M, Bajad S, Dang G, Dengrove D, Diola D, Dorin G, Ellens KW, Fickes S, Galazzo J, Gaucher SP, Geistlinger T, Henry R, Hepp M, Horning T, Iqbal T, Jiang H, Kizer L, Lieu B, Melis D, Moss N, Regentin R, Secrest S, Tsuruta H, Vazquez R, Westblade LF, Xu L, Yu M, Zhang Y, Zhao L, Lievense J, Covello PS, Keasling JD, Reiling KK, Renninger NS, Newman JD. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 231.Steinberg G, Perez-Martin J. Trends Cell Biol. 2008;18:61–67. doi: 10.1016/j.tcb.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 232.Feldbrugge M, Kellner R, Schipper K. Appl Microbiol Biotechnol. 2013;97:3253–3265. doi: 10.1007/s00253-013-4777-1. [DOI] [PubMed] [Google Scholar]
- 233.Yaegashi J, Oakley BR, Wang CC. J Ind Microbiol Biotechnol. 2014;41:433–442. doi: 10.1007/s10295-013-1386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Asahi K, Honma Y, Hazeki K, Sassa T, Kubohara Y, Sakurai A, Takahashi N. Biochem Biophys Res Commun. 1997;238:758–763. doi: 10.1006/bbrc.1997.7385. [DOI] [PubMed] [Google Scholar]
- 235.Sassa T, Ooi T, Nukina M, Ikeda M, Kato N. Biosci, Biotechnol, Biochem. 1998;62:1815–1818. doi: 10.1271/bbb.62.1815. [DOI] [PubMed] [Google Scholar]
- 236.Sassa T, Togashi M, Kitaguchi T. Agric Biol Chem. 1975;39:1735–1744. [Google Scholar]
- 237.Toyomasu T, Nakaminami K, Toshima H, Mie T, Watanabe K, Ito H, Matsui H, Mitsuhashi W, Sassa T, Oikawa H. Biosci, Biotechnol, Biochem. 2004;68:146–152. doi: 10.1271/bbb.68.146. [DOI] [PubMed] [Google Scholar]
- 238.Wang Q-X, Qi Q-Y, Wang K, Li L, Bao L, Han J-J, Liu M-M, Zhang L-X, Cai L, Liu H-W. Org Lett. 2013;15:3982–3985. doi: 10.1021/ol401736z. [DOI] [PubMed] [Google Scholar]
- 239.Afiyatullov S, Kuznetsova TA, Isakov VV, Pivkin MV, Prokofeva NG, Elyakov GB. J Nat Prod. 2000;63:848–850. doi: 10.1021/np9904004. [DOI] [PubMed] [Google Scholar]
- 240.Sun HF, Li XM, Meng L, Cui CM, Gao SS, Li CS, Huang CG, Wang BG. J Nat Prod. 2012;75:148–152. doi: 10.1021/np2006742. [DOI] [PubMed] [Google Scholar]
- 241.Feng Y, Ren F, Niu S, Wang L, Li L, Liu X, Che Y. J Nat Prod. 2014 doi: 10.1021/np4009688. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 242.Wijeratne EM, Bashyal BP, Liu MX, Rocha DD, Gunaherath GM, U'Ren JM, Gunatilaka MK, Arnold AE, Whitesell L, Gunatilaka AA. J Nat Prod. 2012;75:361–369. doi: 10.1021/np200769q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Wang XN, Bashyal BP, Wijeratne EM, U'Ren JM, Liu MX, Gunatilaka MK, Arnold AE, Gunatilaka AA. J Nat Prod. 2011;74:2052–2061. doi: 10.1021/np2000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hartley AJ, de Mattos-Shipley K, Collins CM, Kilaru S, Foster GD, Bailey AM. FEMS Microbiol Lett. 2009;297:24–30. doi: 10.1111/j.1574-6968.2009.01656.x. [DOI] [PubMed] [Google Scholar]
- 245.Shibata H, Irie A, Morita Y. Biosci, Biotechnol, Biochem. 1998;62:2450–2452. doi: 10.1271/bbb.62.2450. [DOI] [PubMed] [Google Scholar]
- 246.Valdivia C, Kettering M, Anke H, Thines E, Sterner O. Tetrahedron. 2005;61:9527–9532. [Google Scholar]
- 247.Liu Y, Li Y, Ou Y, Xiao S, Lu C, Zheng Z, Shen Y. Bioorg Med Chem Lett. 2012;22:5059–5062. doi: 10.1016/j.bmcl.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 248.Ou YX, Li YY, Qian XM, Shen YM. Phytochemistry. 2012;78:190–196. doi: 10.1016/j.phytochem.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 249.Li Y-Y, Shen Y-M. Helv Chim Acta. 2010;93:2151–2157. [Google Scholar]
- 250.Mazur X, Becker U, Anke T, Sterner O. Phytochemistry. 1996;43:405–407. doi: 10.1016/0031-9422(96)00327-5. [DOI] [PubMed] [Google Scholar]