Abstract
Cytosine methylation on CpG dinucleotides is an essential epigenetic modification in eukaryotes. How DNA methylation modulates nucleosome structure and dynamics has been a long standing question. We implemented a single molecule method to monitor effects of DNA methylation on the structure and dynamics of mononucleosomes. Our studies show that DNA methylation induces a more compact and rigid nucleosome structure, providing a physical basis for how DNA methylation might contribute to regulating chromatin structure.
DNA in eukaryotic cells is packaged into arrays of nucleosomes, each assembled from ~147bp of DNA wrapped around a histone octamer and folded into higher order chromatin structures. Enzyme mediated chemical modifications of DNA and histones regulate various genome transactions by modulating access to the DNA. One example of these epigenetic modifications is methylation of CpG dinucleotides, which is often associated with gene repression1. While CpG methylation is essential in development, inappropriate CpG methylation in tumor suppressor genes is associated with cancer2. A better understanding of how CpG methylation affects nucleosome structure will provide insights into the mechanism of gene repression and the development of novel drugs to treat methyl-CpG related diseases.
Two mechanisms have been proposed to explain how methylation might repress gene transcription. First, methylated CpGs may prevent transcriptional activators from binding its DNA target3. Second, transcriptional repressors with methyl-CpG binding domains associate with methylated CpGs and block transcription by modifying the surrounding chromatin or prevent interaction by activators4,5. Considering access to the underlying DNA is largely governed by DNA-histone interactions6,7, a third possibility is that the nucleosome structure is changed by methylation leading to a more closed state. To test this, we used a single molecule approach to monitor methylation induced conformational changes in mononucleosomes.
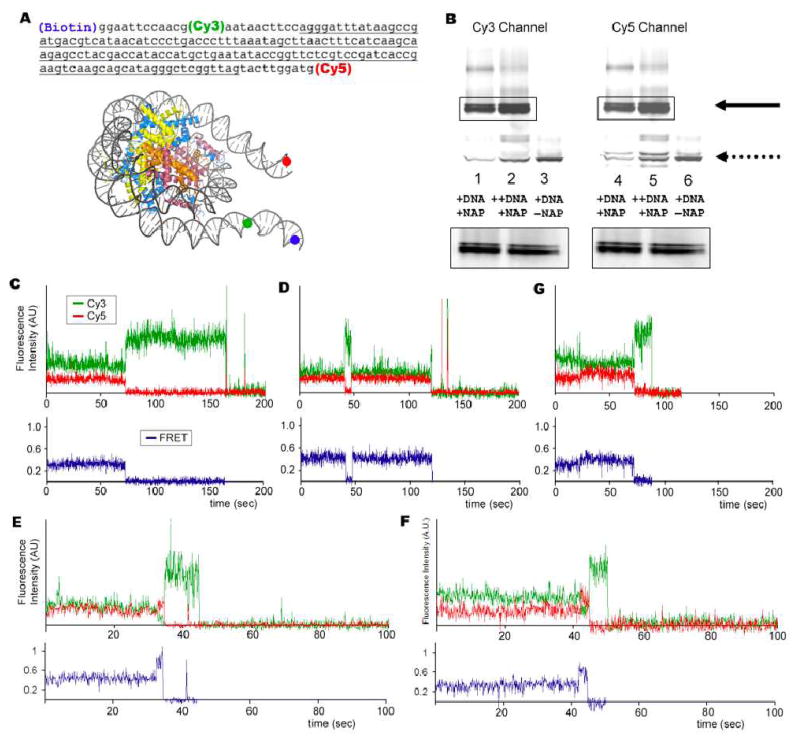
A single molecule system as shown in figure 1A was constructed that measures linker DNA end-to-end distance and flexibility by FRET and fluorescence polarization (methods in the Supporting Information). Two unevenly populated nucleosome bands were observed in a native gel analysis from reactions that contained 5S rDNA, histone octamers, and the NAP1 histone chaperone, but in the absence of NAP1 both products were absent (Fig. 1B). Consistent with the native gel analysis, we identified two unevenly distributed populations of nucleosomes with a low 0.29 (30%) or intermediate 0.4 (60%) FRET state (Figs. 1C, and 1D). No FRET signals were observed when the DNA is free of histones. A small population (8%) of nucleosomes made excursions between 0.29 to 0.4 and 0.4 to 0.29 FRET states (Fig. 1G). These excursions were observed mostly once in ~5 minute long FRET traces. These data are consistent with reports that the histone core can assemble nucleosomes at two translational positions on DNA – near the center of the DNA or biased toward one end8. Crossing the energetic barrier between these two states requires high heat treatment9. This is consistent with only a small population of complexes that make excursions between the 0.29 and 0.4 FRET states at a very low rate at room temperature (Fig. 1G).
Figure 1.
Experimental system to study effects of DNA methylation on nucleosome structure and sample data. (A) Solid circles in blue, green and red indicate the positions of biotin, Cy3 and Cy5, respectively on the DNA. (B) Native polyacrylimide gel analysis of nucleosome. Solid arrow indicates assembled nucleosomes. Dotted arrow indicates DNA. (C) to (F) Sample FRET traces.
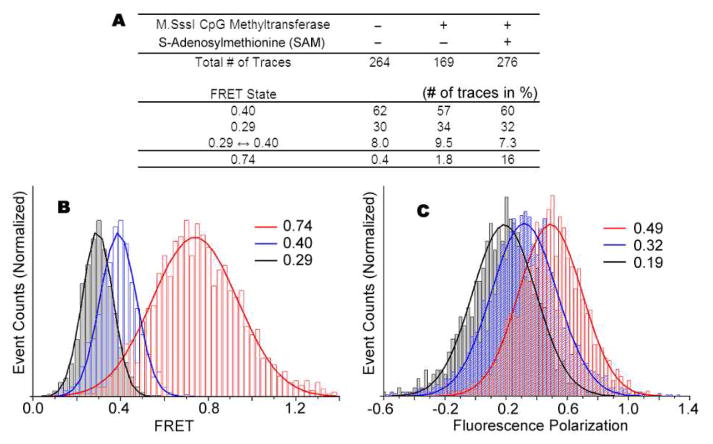
Next, we addressed the question of how DNA methylation might modulate nucleosomal structure. Changes in DNA-histone interactions as a result of CpG methylation would likely alter the linker DNA end-to-end distance. We predict that a decrease in DNA-histone contacts leads to a more open structure and a longer linker DNA end-to-end distance. Conversely, an increase in DNA-histone contacts leads to a more closed structure and a shorter DNA linker end-to-end distance. Nucleosomes were immobilized as in the previous experiment followed by addition of M.SssI CpG methyltransferase. Strikingly, after a 90 minute incubation, the population of nucleosome complexes that made excursions to a high 0.74 FRET state increased from 0.4% to 16% (Figs. 1E, 1F and Fig. 2A). Importantly, the appearance of nucleosomes with excursions to 0.74 FRET states depended on the methyl donor S-adenonsylmethionine (SAM), consistent with methylation of DNA (Fig. 2A). The 40-fold increase in complexes with a high 0.74 FRET state suggests that upon CpG methylation the DNA end-to-end distance shortens, possibly through more DNA-histone contacts, forming a more compact nucleosome structure (Fig. 3 top). However, we cannot completely rule out the possibility that the DNA may take an alternative path to shorten the end-to-end distance. This would require unwrapping more internal regions of DNA to shorten the end-to-end distance without forming a more compact nucleosome structure. Previously, FRET studies showed that DNA ends wrapping and unwrapping around the histone core were responsible for end-to-end distance changes10,11. Thus, we favor the former interpretation that a more compact nucleosome structure is induced by CpG methylation. A higher polarization of the high FRET state also supports our interpretation (see discussion below).
Figure 2.
Effects of DNA methylation on nucleosome structure. (A) Statistics on the FRET states of nucleosomes. (B) FRET efficiency distributions of the three FRET states. (C) Cy5 fluorescence polarization of the three FRET states.
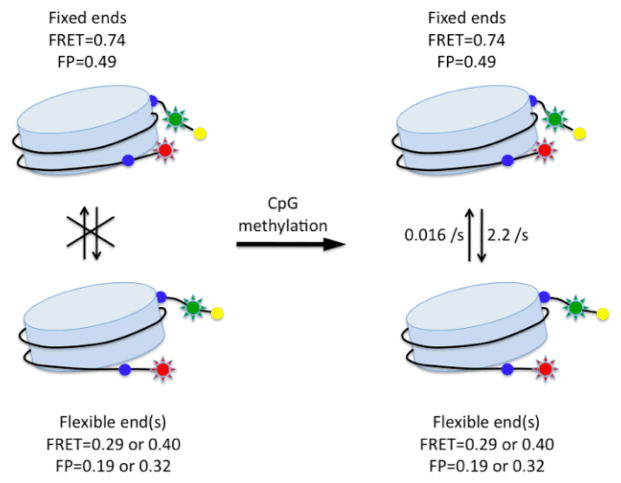
Figure 3.
Summary of CpG methylation effect on nucleosomal structure. Data analysis is included in the Supporting Information.
Finally, we examined if a more compact structure may be correlated with a more rigid DNA wrapped around the histone core. Fluorescence anisotropy has long been utilized to determine the rotational diffusion time scale of a biological molecule12. A heavier, restricted or rigid molecule yields higher anisotropy of fluorescence emission than a lighter, unrestricted, or flexible molecule. The fluorescence polarization, a measure of anisotropy in the fluorescence emission, from the FRET acceptor Cy5 represents the structural flexibility of the Cy5 labeled end with respect to the Cy3 labeled region of the nucleosomal DNA. Consequently, a higher Cy5 emission polarization suggests a higher rigidity of the DNA wrapped around the histone core (further details in the Supporting Information).
As shown in figure 2, the high FRET state (0.74 FRET) has higher polarization (0.49) than the two lower FRET states (0.32 and 0.19 for the 0.40 and 0.29 FRET states, respectively). The polarization difference for 0.40 and 0.29 FRET states may reflect the difference in the structural flexibility caused by different translational positioning – i.e. one end of DNA in one translational positioning may be longer and more flexible than in the other, where the ends are equal and less flexible. The two bands in the native gel also suggest different flexibilities of the two states (Fig 1B). The highly polarized emission from 0.74 FRET state suggests that nucleosomal DNA motion around the histone core is more restricted when the DNA is methylated. Based on the highly polarized FRET acceptor emission, we conclude that DNA methylation enhances the interaction between DNA and histone core, resulting in a more rigid nucleosomal structure.
Based on our findings, we propose that CpG methylation induces excursions to a closed and rigid nucleosomal conformation, resulting on average in a more tightly wrapped nucleosome structure (Fig. 3). Therefore, in addition to acting as “molecular handles” recognized by methyl-CpG binding proteins or as a means to prevent association of transcription factors by masking the underlying sequence1,13,14, a third mechanism of function of CpG methylation involves directly altering the structural dynamics of nucleosomes.
To our knowledge this is the first report to show how DNA methylation changes the structure and dynamics of mononucleosomes leading to a more compact and rigid structure. Our single molecule system allowed us to observe changes in structure and dynamics that otherwise would be challenging to detect with traditional biochemical methods. In conclusion, this report strongly suggests that CpG methylation may contribute to the repression of gene transcription and other genome processes by inducing a more compact and rigid nucleosome conformation.
Supplementary Material
Acknowledgments
The authors thank J. Huh, R. Dutnall, M. Mattson and A. Islas for NAP1 expression and advice, and J. Cotes for 5S rDNA. JSC was an American Cancer Society postdoctoral fellow. THL is supported by Searle Scholar Award and the Camillie and Henry Dreyfus New Faculty Award.
Footnotes
Supporting Information Available: Methods and two figures with captions are provided as Supporting Information.
References
- 1.Bird A. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Esteller M. Nat Rev Genet. 2007;8:286–98. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 3.Tate PH, Bird AP. Curr Opin Genet Dev. 1993;3:226–31. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 4.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Nat Genet. 1998;19:187–91. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 5.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Nature. 1998;393:386–9. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 6.Razin A, Cedar H. Proc Natl Acad Sci U S A. 1977;74:2725–8. doi: 10.1073/pnas.74.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solage A, Cedar H. Biochemistry. 1978;17:2934–8. doi: 10.1021/bi00607a036. [DOI] [PubMed] [Google Scholar]
- 8.Luger K. Current Opinion in Genetics & Development. 2003;13:127–135. doi: 10.1016/s0959-437x(03)00026-1. [DOI] [PubMed] [Google Scholar]
- 9.Luger K, Rechsteiner TJ, Richmond TJ. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 10.Gansen A, Toth K, Schwarz N, Langowski J. J Phys Chem B. 2009;113:2604–13. doi: 10.1021/jp7114737. [DOI] [PubMed] [Google Scholar]
- 11.Toth K, Brun N, Langowski J. Biochemistry. 2006;45:1591–8. doi: 10.1021/bi052110u. [DOI] [PubMed] [Google Scholar]
- 12.Lakowicz JR. Principles of Fluorescence Spectroscopy. Springer; 2006. [Google Scholar]
- 13.Pennings S, Allan J, Davey CS. Brief Funct Genomic Proteomic. 2005;3:351–61. doi: 10.1093/bfgp/3.4.351. [DOI] [PubMed] [Google Scholar]
- 14.Weber M, Schubeler D. Curr Opin Cell Biol. 2007;19:273–80. doi: 10.1016/j.ceb.2007.04.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.