Abstract
The prostate epithelial lineage hierarchy remains inadequately defined. Recent lineage-tracing studies have implied the existence of prostate luminal epithelial progenitors with extensive regenerative capacity. However, this capacity has not been demonstrated in prostate stem cell activity assays, probably due to the strong susceptibility of luminal progenitors to anoikis. Here we show that constitutive expression of Notch1 intracellular domain impairs secretory function of mouse prostate luminal cells, suppresses anoikis of luminal epithelial cells by augmenting NF-κB activity independent of Hes-1, stimulates luminal cell proliferation by potentiating PI3K-AKT signaling, and rescues the capacities of the putative prostate luminal progenitors for unipotent differentiation in vivo and short-term self-renewal in vitro. Epithelial cell-autonomous AR signaling is dispensable for the Notch-mediated effects. As Notch activity is increased in prostate cancers and anoikis resistance is a hallmark for metastatic cancer cells, this study suggests a pro-metastatic function of Notch signaling during prostate cancer progression.
Introduction
Mammalian tissues consist of diverse cell types that form a defined lineage hierarchy through which tissue homeostasis is maintained and tissue repair and regeneration are executed with exquisite precision. Despite the extensive progress that has been made during the last decade, the prostate epithelial lineage hierarchy remains inadequately defined. Prostate epithelia consist of three types of cells: the columnar secretory luminal epithelial cells that form a continuous single layer surrounding the luminal space of prostate glands, the cuboidal basal epithelial cells that are aligned between the luminal cells and the basement membrane, and the rare neuroendocrine cells1. Early studies showed that prostate epithelia can regress and regenerate repeatedly in response to alternating androgen deprivation and replacement, suggesting the existence of cells that possess extensive regenerative potential2. Several lineage-tracing studies demonstrated that adult murine prostate basal and luminal cells are mainly self-sustained when residing in their native microenvironment under physiological conditions, suggesting the existence of stem cells or progenitors in both cell lineages3–6.
The stem cell activity within the basal cell lineage has been clearly demonstrated. A fraction of human and rodent basal epithelial cells can form serially passagable, clonogenic two-dimensional holoclones or three-dimensional spheroids in vitro, implying their capacity for self-renewal7. In addition, when human and rodent basal prostate epithelial cells are transplanted under the renal capsules of immunodeficient mice with embryonic urogenital sinus mesenchymal (UGSM) cells, they are capable of differentiating into all three prostate epithelial lineages8–13. Finally, in several recent lineage tracing studies basal cells are also shown to be capable of generating luminal cells, especially in the context of prostatic inflammation5,6,14.
In contrast, stem cells or progenitors within the luminal cell lineage remain poorly defined. Although recent lineage-tracing studies have clearly demonstrated that luminal cells residing in their native microenvironment are capable of undergoing extensive regeneration3–6, such capacity has not been recapitulated in various in vitro and in vivo assays. Unlike prostate basal cells, normal and cancerous luminal epithelial cells of both human and rodent origins rarely form colonies or spheres in 2-D or 3-D in vitro assays, or regenerate tissues in the prostate regeneration assay7,15. In addition, there are very few successful reports regarding the generation of immortalized normal prostate cell lines with a definitive luminal cell phenotype16,17.
The failure of luminal cells to expand or regenerate in these assays was considered as a feature associated with their terminal differentiation. Nevertheless, it may also reflect their strong susceptibility to anoikis. Anoikis is apoptosis induced in cells by insufficient or inappropriate cell-matrix interactions18. Compared to the luminal epithelial cells, dissociated basal epithelial cells are likely more resistant to anoikis due to several distinct intrinsic properties. First, basal cells express Bcl-2 at a higher level19. Second, basal cells express both adhesion-associated membrane receptors and their substrates in extracellular matrix20–23. Therefore, they are capable of establishing cell-matrix interactions cell-autonomously thereby antagonizing anoikis. Third, epithelial-mesenchymal transition has been shown to confer anoikis resistance 24. Compared to luminal cells, basal cells display a more mesenchymal phenotype and display a gene signature that promotes epithelial-mesenchymal transition. For example, basal cells express the miR-200 family members at a lower level compared to luminal cells25. Finally, many growth factor receptor tyrosine kinases are preferentially expressed in basal cells versus luminal cells in normal prostate tissues26,27. Therefore, basal cells possess higher levels of steady-state activities of MAPK and AKT, which also confer anoikis resistance.
The Notch signaling pathway plays an important role in specifying cell fate and regulating tissue homeostasis 28. Crosstalk between Notch and NF-κB has been extensively investigated and has been shown to play important roles in tissue development and disease progression29. Activation of NF-κB signaling following detachment of intestinal and mammary gland epithelial cells from their native environment upregulates various anti-apoptotic pathways and has been shown to delay anoikis23,30–32. In this study, we show that Notch delays anoikis of prostate luminal progenitors by augmenting NF-κB activity and rescues their capacities for short-term self-renewal and unipotent differentiation.
Results
Notch affects luminal cell growth and function in vivo
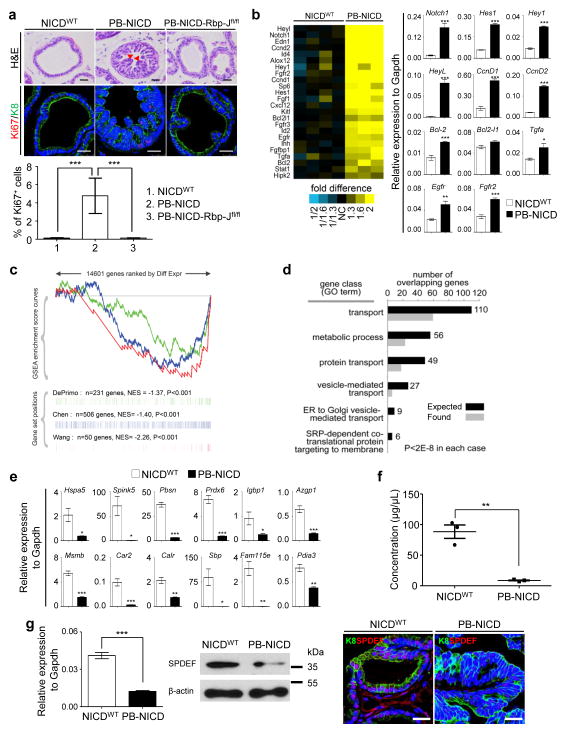
Previously, we showed that an ARR2PB-Cre;ROSA(N1IC) model (hereafter referred to as PB-NICD) that expressed the Notch1 intracellular domain (NICD or N1IC) in the prostate developed prostatic intraepithelial neoplasia (PIN lesions) 33. The prostates of PB-NICD mice were slightly heavier than those of the control ROSA(N1IC) mice (hereafter referred to as the NICDWT mice) (Supplementary Fig. 1a–b). PB-NICD mice developed PIN II lesions34 with numerous micro-papillary projections containing atypical cells with hyperchromasia and a high nuclear-cytoplasmic ratio (Fig. 1a). There was a significant increase in epithelial proliferation as shown by Ki67 staining in Fig. 1a and Supplementary Fig. 1c. Simultaneous knockout of the canonical Notch signaling transducer Rbp-J (PB-NICD-Rbp-Jfl/fl triple transgenic mice) abolished the phenotypes (Fig. 1a), demonstrating that NICD induces luminal cell proliferation via canonical Notch signaling. Notch activation is not sufficient to transform prostate epithelial cells since PB-NICD mice did not develop adenocarcinoma even by 1.5 years of age (Supplementary Fig. 1c). Despite a persistently elevated proliferation index during aging, epithelial tissues that shed into the luminal space of prostate glands were frequently observed and stained positive for the cleaved Caspase 3 (Supplementary Fig. 1c), suggesting that Notch does not confer long-term self-renewal capacity.
Fig. 1. Notch activation causes PIN lesion and impairs secretory function of prostate luminal cells.
(a) H&E staining and immunocostaining of Ki67 and K8 in dorsolateral lobes of NICDWT, PB-NICD, and PB-NICD-Rbp-jfl/fl mouse prostates. Bar graph shows means ± s.d. of percentage of Ki67+ cells from 3–5 mice per group. Arrowheads point to micro-papillary projections. (b) Heatmap from microarray data shows upregulation of cell cycle gene ontology group in PB-NICD mouse prostates. qRT-PCR validates expression changes of representative genes in microarray analysis. (c) GSEA shows that three independent androgen-induced gene sets are all repressed in PB-NICD mouse prostates. (d) GO analysis shows that gene ontology groups related to secretory function are downregulated in PB-NICD mouse prostates. (e) qRT-PCR validates expression changes of major secretory proteins in prostates. (f) Concentration of secretory proteins in anterior prostate lobes of 10-wk-old NICDWT and PB-NICD mice. Data represent means ± s.d. from 3 mice per group. (g) qRT-PCR, Western blot and IHC analysis of SPDEF expression in NICDWT and PB-NICD mouse prostates. All qRT-PCR data represent means ± s.d. from 3–4 mice per group. Scale bars, 25μm. Statistical significance was calculated using the t-test (*: p < 0.05, **: p < 0.01, ***: p < 0.001).
Gene expression profiling of prostate RNAs confirms that the expression of the cell cycle gene ontology group is significantly upregulated in PB-NICD prostates as compared to those of the controls, of which Notch1 and major Notch downstream targets (Hes1, Hey1, HeyL), anti-apoptotic protein Bcl-2, Cyclin Ds, and some growth factor receptors (Fgfr2 and Egfr) were verified by qRT-PCR (Fig 1b). Although the expression level of the androgen receptor (AR) was not significantly altered in PB-NICD mouse prostates (Supplementary Fig. 1d–e), gene set enrichment analysis (GSEA) revealed that the androgen receptor (AR)-induced genes from three independent datasets35 were all significantly down-regulated in PB-NICD prostates (Fig. 1c). This is consistent with the previous reports that Notch targets Hey1/2 and HeyL can act as AR corepressors36,37. Additionally, gene ontology groups related to secretory function, such as vesicle-mediated transport, were downregulated in PB-NICD prostates (Fig. 1d). We confirmed by qRT-PCR that transcripts for most of the major prostate-secreted proteins38 were drastically downregulated in PB-NICD prostates (Fig. 1e). In agreement with this, the concentration of secretory proteins within anterior prostate lobes of PB-NICD mice was 10 fold lower than that of NICDWT mice (Fig. 1f).
Protein secretion is an important function of terminally differentiated prostate luminal cells. Notch impairs the terminal differentiation of goblet cells in the lung and small intestine by suppressing the SAM-pointed domain-containing Ets-like factor SPDEF39,40. SPDEF was shown recently to drive mammary gland luminal cell differentiation41. Interestingly, SPDEF positively regulates androgen receptor transcriptional activity42. Fig. 1g shows that Spdef was downregulated in PB-NICD mice at both the RNA and protein levels, suggesting that Notch may also impair luminal cell secretory function by negatively regulating AR activity via down-regulation of SPDEF. Collectively, these studies suggest that Notch promotes proliferation of luminal epithelial cells, impairs their secretory function, but is incapable of transforming naïve luminal epithelial cells.
Notch promotes luminal cell survival and growth in vitro
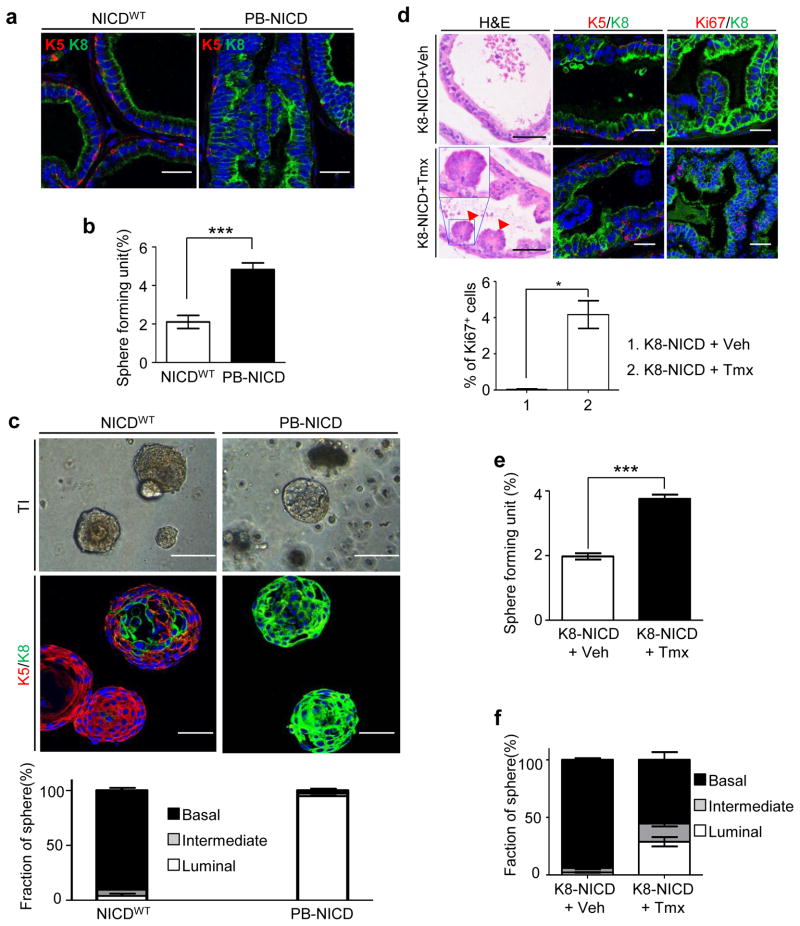
Rodent prostate epithelia are mainly composed of a single layer of cytokeratin 8 (K8)-expressing luminal cells surrounding the lumen and a discontinuous layer of cytokeratin 5 (K5)-expressing basal epithelial cells aligned between the luminal cells and the basement membrane (Fig. 2a). In comparison, the PIN lesions in PB-NICD mice consisted mainly of luminal cells while basal cells were significantly depleted (Fig. 2a). We showed previously that Notch suppressed prostate basal cell proliferation, and attributed the depletion of basal cells to the Cre activity in basal epithelial cells in the ARR2PB model33.
Fig. 2. Notch promotes formation of luminal prostate spheres.
(a) IHC analysis of K5 and K8 in NICDWT and PB-NICD mouse prostates. (b) Bar graph quantifies sphere-forming units in NICDWT and PB-NICD groups. (c) Transillumination (TI) images and IHC analysis of K5 and K8 in prostate spheres generated from NICDWT and PB-NICD mice. Bar graph quantifies types of spheres. (d) H&E staining and IHC analysis of K5, K8, and Ki67 in prostates of tamoxifen- (tmx) and control vehicle- (veh) treated 9-wk-old K8-NICD mice. Arrowheads point to PIN I lesions. Inset shows image at higher magnification. Bar graph shows means ± s.d. of percentage of Ki67+ cells from 3 mice per group. (e–f) Bar graphs quantify sphere-forming units and types of prostate spheres in tamoxifen- and vehicle-treated K8-NICD groups. Data represent means ± s.d. from 3–4 mice per group. Scale bars, 25μm (a, fluorescent images in d); 50μm (H&E images in d, fluorescent images in c); 100μm (TI images in c). Statistical significance was calculated using the t-test (*: p < 0.05, ***: p < 0.001).
When dissociated prostate epithelial cells from wild type mice are cultured in a prostate sphere assay, only prostate basal cells are capable of forming spheroids43. Surprisingly, although PB-NICD mouse prostates had fewer basal cells, the sphere-forming unit of dissociated prostate cells from PB-NICD mice was approximately 2.3 fold of that of NICDWT cells (Fig. 2b). NICDWT cells mainly generated spheres containing cells that only expressed the basal cell marker K5 (hereafter referred to as the basal prostate spheres) (Fig. 2c). A small fraction of basal spheres contained cells embedded in the center that expressed the luminal cell marker K8, which we termed intermediate spheres. Very few spheres in the NICDWT culture were comprised solely of K8-expressing luminal cells (hereafter referred to as the luminal prostate spheres). In contrast, approximately 5–8% of PB-NICD spheres were basal and intermediate prostate spheres, while the remaining spheres were all luminal spheres (Fig. 2c). The luminal spheres initially appeared hollow and bright but began to fill in noticeably starting at day 4 and became morphologically similar to basal prostate spheres by the end of the assay (Supplementary Fig. 2a). Formation of these luminal spheres was dependent on canonical Notch signaling because prostate cells of PB-NICD-Rbp-Jfl/fl triple transgenic mice did not form luminal prostate spheres (Supplementary Fig. 2b).
Since ectopic NICD expression in basal cells does not drive their differentiation into luminal cells33, we reasoned that luminal prostate spheres were derived from NICD-expressing luminal cells. To directly test this hypothesis, we generated K8-CreERT2; ROSA(N1IC) bigenic mice (hereafter referred to as K8-NICD), with which NICD can be specifically activated in the K8-expressing luminal cells upon tamoxifen induction44. As shown in Fig. 2d, three weeks after the induction of NICD expression, K8-NICD mice developed PIN I lesions and the proliferation index of epithelial cells was dramatically increased (0.03% in WT vs. 4.17% in PB-NICD). We performed a prostate sphere assay using prostate cells from K8-NICD mice that have been treated with tamoxifen or vehicle control. As shown in Fig. 2e, dissociated prostate cells from tamoxifen-treated K8-NICD mice displayed a 1.9 fold increase in sphere-forming capability. The control culture contained predominantly basal prostate spheres, while both basal and luminal spheres grew out in the tamoxifen-treated K8-NICD group (Fig. 2f). Since tamoxifen treatment did not induce the formation of luminal spheres from NICDWT prostate cells (Supplementary Fig. 2c), this directly demonstrates that luminal prostate spheres are derived from NICD-expressing luminal cells.
We also confirmed this conclusion using the PB-NICD model. In PB-NICD mice, prostate cells expressing NICD also express GFP simultaneously via an IRES (internal ribosomal entry site) expressing cassette. Since Sca-1 is mainly expressed in murine prostate basal cells and stromal cells10, we used Sca-1 and GFP to separate luminal cells that express NICD from other cell populations (Supplementary Fig. 2d). Immunocytochemical analyses (Supplementary Fig. 2e) corroborated that expression of the basal cell markers (K5 and K14) and the luminal cell markers (K8 and androgen receptor) were enriched in Sca-1+ and Sca-1− cells, respectively. Because stromal cells also express Sca-1, smooth muscle actin was detected in some Sca-1+ cells (Supplementary Fig. 2h). Individual FACS-isolated cell fraction was cultured in the sphere assay separately. If luminal prostate spheres are derived from Notch-expressing luminal cells, then the Sca-1−GFP+ cells should generate luminal spheres. As expected, Supplementary Fig. 2f shows that the spheres generated from the Sca-1− cells were luminal prostate spheres, while Sca-1+ cells formed mostly basal prostate spheres. We also noticed the formation of luminal spheres in the Sca-1−GFP− group, but the sphere-forming unit in this population is lower compared to that of the Sca-1−GFP+ group (Supplementary Fig. 2f). Because the fluorescence intensity of GFP is relatively low in this mouse model, the minor sphere-forming activity from the Sca-1−GFP− group may be attributed by the contaminated Sca-1−GFP+ cells during cell sorting. These results further corroborate that luminal prostate spheres were generated by NICD-expressing luminal cells. Interestingly, only 4.5% NICD-expressing luminal cells can survive and proliferate in vitro in response to Notch (Supplementary Fig. 2g). Similar observation was made when Lin−Sca-1−CD49flow luminal cells were used in the prostate sphere assay (Supplementary Fig. 2i). These sphere-forming luminal cells may be functionally distinct from the Notch-expressing luminal cells that did not form spheres and may represent the putative prostate luminal progenitors.
Notch rescues prostate luminal progenitor activity
Previous studies showed that FACS-isolated luminal cells from intact mice were incapable of regenerating prostate tissues in a transplantation-based prostate regeneration assay8,9. Since Notch promotes survival and proliferation of luminal cells in the prostate sphere assay, we hypothesized that increased Notch activity might promote the in vivo survival of dissociated prostate luminal progenitors and rescue their regenerative capability in the prostate regeneration assay.
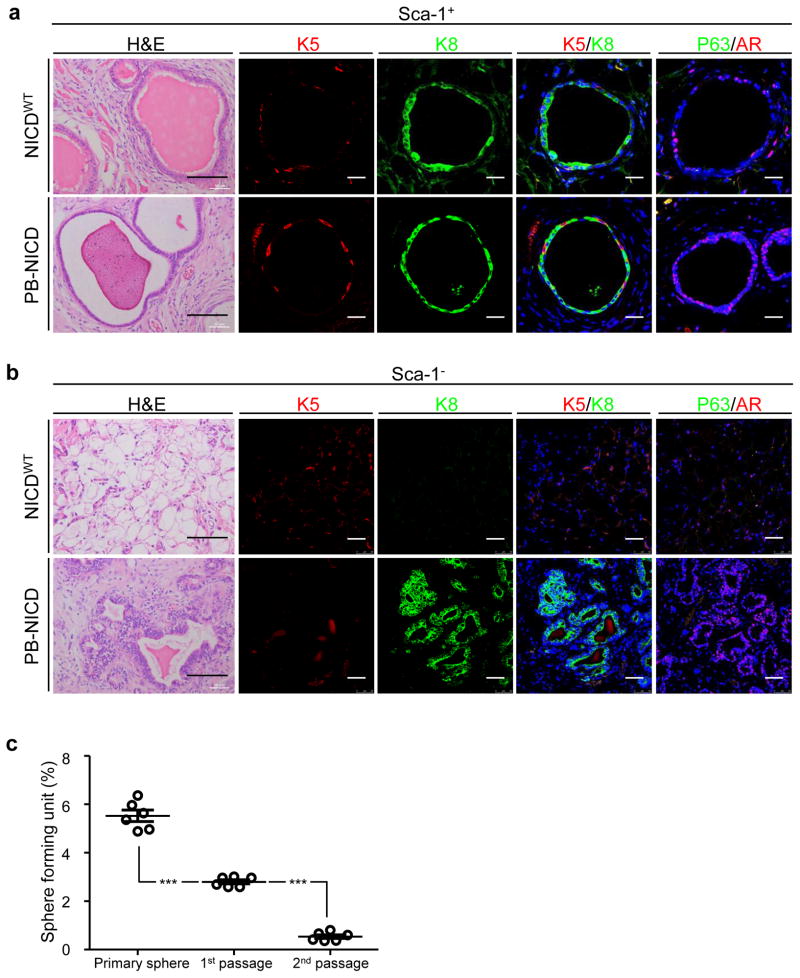
Sca-1+ (basal and stromal cells) and Sca-1−(luminal cells) cells were FACS isolated from wild type and PB-NICD mice. CD49f was not used during the sorting since Sca-1 is sufficient to separate luminal cells. Immunocytochemical analysis of specific lineage markers confirmed the identities of the FACS sorted cells (Supplementary Fig. 2e). Each cell fraction was mixed with urogenital sinus mesenchyme cells separately and incubated subcutaneously in immunodeficient male mice to allow regeneration. As shown in Fig. 3a, NICDWT Sca-1+ cells generated the typical prostatic glandular structures comprising K5+P63+ basal cells and K8+AR+ luminal cells10. PB-NICD Sca-1+ cells generated similar structures but the gland-forming unit was lower than that of NICDWT cells (0.03% in WT group vs. 0.015% in PB-NICD group), consistent with our previous study showing that Notch activity suppresses basal stem cell activity33. In addition, glands generated in the PB-NICD group often contained less eosinophilic secretions in luminal spaces (Fig. 3a) suggesting potential defects in secretory function of luminal epithelial cells. As expected, NICDWT Sca-1− luminal cells failed to regenerate discernible glandular structures (Fig. 3b). In contrast, Sca-1− luminal cells from PB-NICD mice regenerated tissues containing small glandular structures made solely of K8+ luminal cells that expressed AR (Fig. 3b). Many glands contained multiple layers of cells resembling the histopathology observed in the PB-NICD model. The average luminal gland-forming unit was approximately 0.3%. It should be noted that the estimated glandular-forming units represent the minimal values because we enumerated the numbers of the glandular structures from tissue sections that contained tissue specimens with the largest diameter instead of reconstructed three dimensional images. These results show that NICD can promote the in vivo survival and proliferation of a small fraction of prostate luminal cells in the regeneration assay. This is consistent with the fact that only a small fraction of NICD-expressing luminal cells can survive and proliferate in vitro in the sphere assay, and further implies a primitive lineage hierarchical status of these cells within the luminal cell lineage.
Fig. 3. Notch rescues the capacities of dissociated luminal progenitors for unipotent differentiation and short-term self-renewal.
(a–b) H&E staining and IHC analysis of K5, K8, AR and P63 in tissues regenerated from Sca-1+ and Sca-1− cells of NICDWT and PB-NICD mice. (c) Sphere-forming activity of serially-passaged luminal sphere cultures. Scale bars, 25μm (fluorescent images in a); 50μm (fluorescent images in b); 100μm (H&E images in a, b). Data represent means ± s.d. from 3 independent experiments. Statistical significance was calculated using the t-test (***: p < 0.001).
To determine whether NICD-expressing luminal sphere-forming cells possess self-renewal capacity, we examined whether luminal prostate spheres can be serially passaged. Prostate spheres were enzymatically dissociated into single cells and replated to form secondary and tertiary spheres. As shown in Figure 3c, primary luminal spheres can be passaged twice but the sphere-forming unit decreased dramatically. This result suggests that luminal sphere-forming cells only possess limited self-renewal capacity.
β-Catenin is not required for luminal sphere formation
Basal prostate sphere cells possess long-term self-renewal capability in vitro43. To gain insights into how and why NICD-expressing prostate luminal cells can survive and proliferate in vitro only for short term, we compared protein expression profiles of the luminal and the basal prostate spheres using a reverse phase protein array (RPPA) containing 170 different antibodies. The RPPA analysis shows that luminal prostate sphere cells expressed higher levels of several signaling components of the Wnt pathway including β-catenin, among which we confirmed by Western blot the expression of β-catenin (Supplementary Fig. 3a–b). We generated PB-Cre;Catnblox(ex3) bigenic mice that express a stabilized β-catenin specifically in the prostate epithelia45. Dissociated prostate cells from PB-Cre; Catnblox(ex3) mice did not form luminal prostate spheres in vitro (Supplementary Fig. 3c–d), suggesting that expression of β-catenin in prostate luminal cells is not sufficient to promote their in vitro survival. In addition, C57Bl/6 mouse prostate luminal cells infected with the lentivirus expressing a high level of constitutively active β-catenin driven by the human ubiquitin promoter also failed to form luminal spheres in vitro (Supplementary Fig. 3e–f). This excludes the possibility that the failure of PB-Cre; Catnblox(ex3) cells to generate luminal prostate spheres is due to insufficient expression of stabilized β-catenin driven by the endogenous promoter. To determine if β-catenin is essential for luminal sphere formation, we expressed a dominant negative β-catenin (dN-βCatenin)46 in the luminal prostate epithelial cells of PB-NICD mice. Expression of dN-βCatenin suppressed the Wnt target c-Myc (Supplementary Fig. 3g), but did not inhibit sphere formation or reduce sphere size (Supplementary Fig. 3h–j). Collectively, these results show that the transcriptional activity of β-Catenin is neither sufficient nor essential for the survival and proliferation of luminal sphere cells.
PI3K-AKT is necessary for luminal sphere cell proliferation
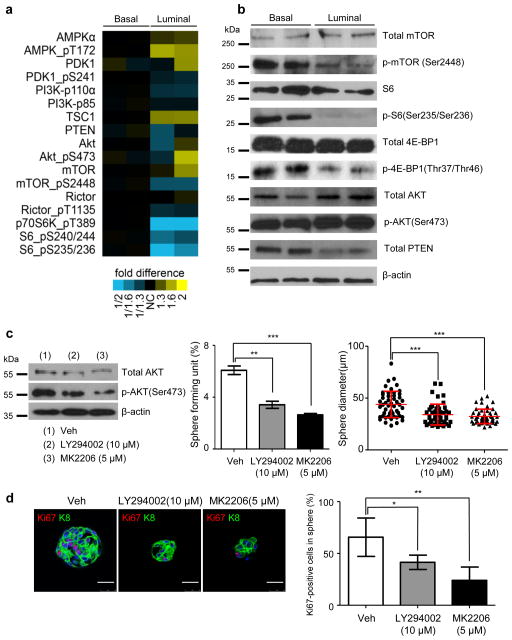
The RPPA assay also revealed that NICDWT basal prostate sphere cells possess a higher level of mTOR activity, although PB-NICD luminal sphere cells expressed a relatively higher level of pAKT probably due to Notch-mediated transcriptional repression of Pten47 (Fig. 4a),. This is probably because basal cells express various growth factor receptor kinases hence may have more signal entry and feedback inputs26,27. Western blot analysis confirmed the differential expression levels of several major signaling components (Fig. 4b). This difference in mTOR activity may partially explain why basal prostate sphere cells can proliferate extensively in vitro.
Fig. 4. PI3K-AKT signaling is necessary for proliferation of luminal prostate sphere cells.
(a) Heatmap from RPPA analysis shows that basal sphere cells possess higher mTOR activity. (b) Western blot validates expression of several major PI3K-AKT-mTOR signaling components in basal and luminal prostate spheres. Individual lanes represent independent samples. (c) Western blot shows that LY294002 and MK2206 suppress AKT activity. Bar and dot graphs quantify sphere-forming activity and prostate sphere size from 3 independent experiments. (d) IHC analysis of Ki67 and K8 in luminal prostate spheres treated with vehicle, LY294002 and MK2206. Bar graph shows quantification. Data represent means ± s.d. from 3 independent experiments. Scale bars, 25μm. Statistical significance was calculated using the t-test (*: p < 0.05, **: p < 0.01, ***: p < 0.001).
We reasoned that PB-NICD luminal sphere cells are capable of proliferating in vitro partly because they possess a higher PI3K-AKT activity than NICDWT luminal cells. This hypothesis is supported by the fact that the gene signature up-regulated by AKT48 was enriched in prostate tissues of PB-NICD mice compared to WT mice (57 shared genes, one-sided Fisher’s exact test, P = 3.7e-10, Supplementary Fig. 4). Consistently, treating PB-NICD prostate sphere culture with the PI3K inhibitor LY294002 efficiently reduced AKT phosphorylation, suppressed luminal prostate sphere-forming activity by 45% and reduced sphere diameter by 33% (Fig. 4c). Loss of function of Pten is not sufficient to confer anoikis resistance on luminal cells, because Pten-null prostate luminal cells are incapable of forming luminal prostate spheres in vitro49. Therefore, LY294002 treatment may not directly impinge on the survival of prostate sphere-forming cells, but attenuates the sphere-forming activity by suppressing cell proliferation. Ki67 staining confirmed that LY294002 attenuated proliferation of luminal prostate sphere cells (Fig. 4d). Similarly, the AKT inhibitor MK2206 reduced AKT activity, suppressed luminal prostate sphere-forming activity by 57% and reduced sphere diameter by 37%, and inhibited cell proliferation as measured by Ki67 staining (Fig. 4c–d). These results show that the PI3K-AKT signaling is necessary for the in vitro proliferation of luminal prostate sphere cells.
Notch mediates anti-apoptotic signaling via NF-κB
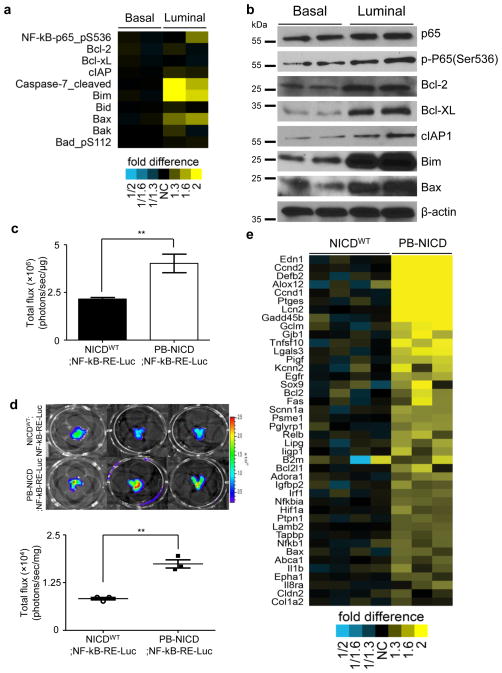
The RPPA assay also revealed that luminal sphere cells expressed phosphor-Ser536 P65 at a higher level (Fig. 5a). In addition, the expression levels of both pro-apoptotic and anti-apoptotic proteins were higher in the luminal sphere cells, suggesting an intense balance between the survival and apoptosis signals. Western blot analysis confirmed that luminal prostate sphere cells expressed higher levels of phosphorylated P65 as well as several direct NF-κB target genes such as Bcl-2, Bcl-xL and cIAP1 (Fig. 5b). The transcriptional activity of phosphorylated P65 can be affected by both nuclear localization and DNA binding. To determine definitively whether NICD expressing luminal prostate sphere cells possess a high level of NF-κB activity, we bred PB-NICD mice with an NF-κB-dependent in vivo luciferase reporter strain from Taconic (NF-κBRE-Luc). Fig. 5c showed that the bioluminescence signal of prostate sphere cells (95% of which were luminal spheres) derived from PB-NICD;NF-κB-RE-Luc mice was 1.9 fold higher than that of the basal spheres derived from age-matched control NICDWT;NF-κB-RE-Luc mice. Since NICD expression in basal spheres did not affect NF-κB activity (Supplementary Fig. 5), these results demonstrate that luminal sphere cells possess a higher NF-κB activity than basal sphere cells. The increased NF-κB activity in luminal cells is induced by NICD because the bioluminescence signal in the prostates of PB-NICD;NF-κB-RE-Luc mice is 2.1 fold that of the prostates in control NICDWT;NF-κB-RE-Luc mice (Fig. 5d). Finally, many known NF-κB-regulated genes (http://www.nf-kb.org) were enriched in the genes upregulated in the PB-NICD group as compared to the control (26 shared genes, one-sided Fisher’s exact test, P = 1.00e-6). Fig. 5e shows some representative NF-κB target genes that were upregulated in the PB-NICD group. Of note, both NF-κb1 and Relb were upregulated. This further corroborates that Notch promotes NF-κB activity in prostate luminal cells.
Fig. 5. Notch augments NF-kB activity in prostate luminal cells.
(a) Heatmap from RPPA analysis shows that luminal sphere cells express higher levels of phosphorylated P65 and proteins regulating apoptosis. (b) Western blot validates expression of representative proteins in RPPA assay. Individual lanes represent independent samples. (c) Bioluminescence assay of NF-κB activity in basal spheres generated from NICDWT mice and luminal spheres from PB-NICD mice. Bar graph shows means ± s.d. from 3 independent experiments. (d) Bioluminescence assay of NF-κB activity in 10-wk-old NICDWT;NF-κB-RELuc and PB-NICD;NF-κB-RE-Luc mice. Dot graph shows means ± s.d. from 3 pairs of mice. (e) Heatmap from microarray data shows upregulation of representative NF-κB target genes in PB-NICD mouse prostates. Statistical significance was calculated using the t-test (**: p < 0.01).
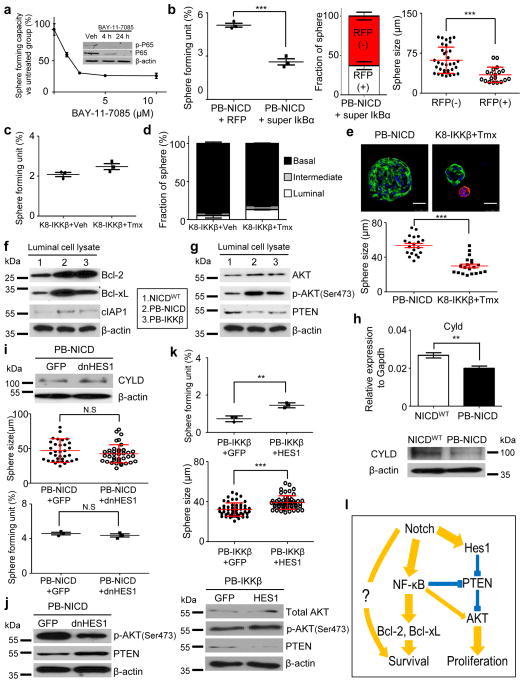
NF-κB signaling is essential for the in vitro survival of luminal prostate sphere-forming cells because the NF-κB inhibitor BAY-11-7085 suppressed P65 phosphorylation and luminal sphere forming activity in a dosage-dependent manner (Fig. 6a). WT basal prostate sphere cells also succumbed to NF-κB inhibition (Supplementary Fig. 6a). In addition, ectopic expression of a degradation-resistant super IκBα in PB-NICD luminal cells suppressed their sphere forming activity dramatically (Fig. 6b), corroborating that NF-κB is essential for the survival of luminal prostate sphere-forming cells in vitro.
Fig. 6. Notch delays anoikis and promotes proliferation by augmenting NF-κB activity.
(a) Inhibition of luminal prostate sphere formation by BAY-11-7085. Inset image: Western blot analysis of pho-P65. (b) Super IκBα suppresses luminal sphere formation. Data represent means ± s.d. from 3 independent experiments. (c–d) Quantitation of sphere-forming units (c) and prostate sphere types (d) from prostate cells of K8-IKKβ mice treated with tamoxifen (Tmx) and vehicle (Veh). Data represent means ± s.d. from 3 independent experiments. (e) Comparison of sizes of PB-NICD and K8-IKKβ luminal prostate spheres. Dot graph shows means ± s.d. from one of the two independent experiments. Scale bars, 25μm. (f) Western blot of expression of Bcl-2, Bcl-xL and cIAP1 in FACS-sorted prostate luminal cells from WT, PB-NICD and PB-IKKβ mice. (g) Western blot of Pten expression and AKT activity in FACS-sorted prostate luminal cells from WT, PB-IKKβ and PB-NICD mice. (h) qRT-PCR and Western blot of Cyld expression in NICDWT and PB-NICD mouse prostates. qRT-PCR data represent means ± s.d. from 3 independent experiments. (i) Expression of dominant negative Hes1 (dN-Hes1) in PB-NICD cells upregulates Cyld expression but does not significantly affect luminal sphere formation and size. Dot graphs show data representing means ± s.d. from 3 independent experiments. (j) Western blot analysis of Pten expression and AKT activity in luminal spheres derived from PB-NICD cells infected by GFP and dN-Hes1 lentivirus. (k) Ectopic expression of Hes1 in FACS-sorted PB-IKKβ prostate luminal cells enhances AKT activity and increases luminal sphere forming unit and sphere size. Dot graphs show data representing means ± s.d. from 3 independent experiments. (l) Schematic illustration of molecular mechanism through which Notch and NF-κB promote formation of luminal spheres. Boldness of arrow line reflects signaling contribution. Statistical significance was calculated using the t-test (**: p<0.01, ***: p < 0.001).
To investigate whether enhanced NF-κB signaling is sufficient to promote in vitro survival and proliferation of prostate luminal cells, we took advantage of a K8-CreERT2;Rosa26-LSL-IKKβCa bigenic model (hereafter referred to as K8-IKKβ). Upon tamoxifen treatment, the constitutively activated IKKβ was specifically expressed in the K8-expressing prostate luminal cells. As shown in Fig. 6c, IKKβ activation did not substantially increase sphere forming activity, but did promote the formation of luminal sphere cells (5.3% vs. 13.1%) (Fig. 6d). Of note, the size of the K8-IKKβ luminal spheres was smaller than that of the PB-NICD luminal spheres (Fig. 6e). Similar observations were made when the ARR2PB-Cre; Rosa26-LSL-IKKβCa bigenic mice were examined (Supplementary Fig. 6b–c). No obvious phenotype was observed in PB-IKKβ and K8-IKKβ mice, consistent with a previous study showing that IKKβ alone is insufficient for transformation50. Collectively, these results demonstrate that increased NF-κB activity in prostate luminal cells is sufficient to delay anoikis of luminal sphere-forming cells. However, NF-κB is less potent than Notch, suggesting that Notch signaling mediates additional NF-κB-independent signaling to antagonize anoikis and promote proliferation of luminal sphere-forming cells.
NF-κB promotes the in vitro survival of luminal cells by upregulating anti-apoptotic proteins. The expression levels of the NF-κB target genes Bcl-2, Bcl-xL and cIAP1 in prostate lysates were all upregulated in the FACS-sorted luminal cells from the PB-IKKβ and PB-NICD models as compared to WT mice (Fig. 6f). NF-κB has also been shown to suppress Pten expression51 and increase AKT expression or activity52. However, down-regulation of Pten and increased AKT activity in PB-IKKβ mice compared to WT mice was not as significant as that seen in PB-NICD mice (Fig. 6g). This may partly explain why the size of luminal prostate spheres in PB-IKKβ group was smaller than that of the PB-NICD group.
Notch is capable of enhancing NF-κB activity by a Hes1-dependent mechanism (Hes1-mediated suppression of Cyld, a deubiquitinase that negatively regulates the IKK complex)53 and Hes1-independent mechanisms (NICD-induced expression of NF-κB, NICD-enhanced IKKα activity, or NICD-mediated NF-κB nuclear retention)54–56. Cyld was indeed down-regulated at both the RNA and protein levels in prostate tissues of PB-NICD mice (Fig. 6h). While ectopic expression of a dominant negative Hes1 (dN-Hes1) upregulated Cyld expression, luminal sphere-forming activity was not affected (Fig. 6i), suggesting that Hes1-mediated Cyld suppression does not play a role in NF-κB-mediated survival of luminal prostate sphere forming cells. The size of luminal spheres was slightly reduced though the change did not reach statistical significance (P=0.06) (Fig. 6i), which implies that Hes1 may regulate the proliferation of luminal sphere cells. This implication is supported by the observation that ectopic expression of dN-Hes1 slightly increased Pten expression and reduced AKT activity in PB-NICD luminal sphere cells (Fig. 6j). In addition, ectopic expression of Hes1 increases the sphere-forming unit (SFU) of FACS-sorted luminal cells of PB-IKKβ mice as well as the size of the generated luminal prostate spheres (Fig. 6k). The increased sphere size indicates that Hes-1-mediated signaling cooperates with NF-κB to promote proliferation, as corroborated by increased AKT activity (Fig. 6k). On the other hand, we believe that increased SFU is not a direct effect of Hes-1 on the survival of luminal sphere forming cells since ectopic Hes1 expression in WT prostate luminal cells was not sufficient to induce the formation of luminal prostate spheres in vitro (Supplementary Fig. 6d–e). Instead, the increased SFU reflects enhanced proliferation of some PB-IKKβ luminal progenitors that are otherwise incapable of doing so in the absence of Hes-1 mediated growth signaling. Collectively, as schematically illustrated in Fig. 6l our study demonstrates that Notch confers anoikis resistance to luminal prostate sphere-forming cells through Hes-1-independent NF-κB activation and probably also through some other unidentified signaling, while optimal proliferation of luminal sphere cells relies on an elevated activity of AKT regulated by both NF-κB and Hes-1.
Epithelial AR is dispensable for luminal sphere formation
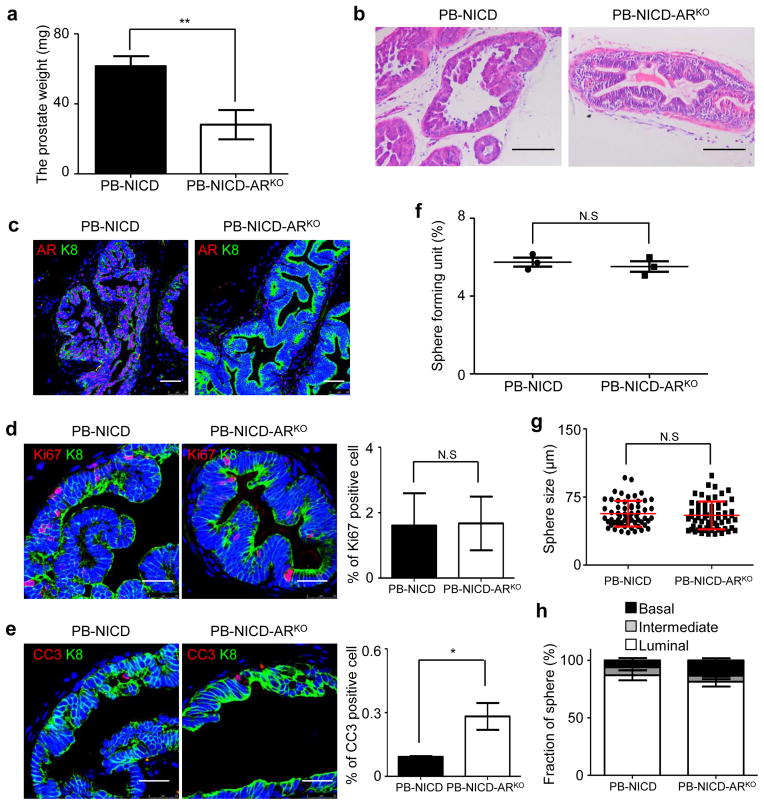
Luminal sphere-forming cells propagate in vitro without testosterone, and their proliferation is not affected by addition of di-hydro-testosterone (Supplementary Fig. 7). It was difficult to conclude via IHC analysis whether these cells completely lacked expression of the androgen receptor (AR) and its ligand-independent truncated isoforms or expressed them at very low levels. To determine definitively whether AR is essential for the survival and proliferation of luminal-sphere-forming cells, we eliminated epithelial AR in the PB-NICD model by generating ARR2PB-Cre;ROSA(N1IC);ARfl/Y triple transgenic mice (PB-NICD-ARKO). The prostates of PB-NICD-ARKO mice were half the weight of PB-NICD prostates, but H&E staining showed that both models developed PIN lesions with similar dynamics (Fig. 7a–b). AR was efficiently knocked out in the majority of prostate epithelial cells (Fig. 7c). The epithelial cells in the PIN lesions of the two models proliferated at a similar rate, but the apoptosis index in the PB-NICD-ARKO group was 3.1 fold higher (Fig. 7d–e). These results indicate that AR is not essential for the NICD-mediated mitogenic signaling, but positively regulates the survival of luminal epithelial cells. Dissociated prostate cells from PB-NICD-ARKO and PB-NICD mice were cultured in the sphere assay. Fig. 7f–h showed that sphere-forming units, sphere size, and the percentage of luminal spheres were all similar between the cultures of the two groups, demonstrating that epithelial AR signaling is dispensable for the in vitro survival and proliferation of luminal-sphere forming cells.
Fig. 7. Epithelial AR is dispensable for NICD-induced survival and proliferation of luminal-sphere-forming cells.
(a) Bar graph quantifies weight of 9-wk-old PB-NICD and PB-NICD-ARKO mouse prostates. Data represent means ± s.d. from 4 pairs of mice. (b). H&E staining of 9-wk-old PB-NICD and PB-NICD-ARKO mouse prostates. (c) IHC analysis of AR in PB-NICD and PB-NICD-ARKO mouse prostates. (d–e) IHC analyses of Ki67 and cleaved caspase 3 in PB-NICD and PB-NICD-ARKO mouse prostates. Bar graphs show means ± s.d. from 3 mice per group. (f–h) Bar and dot graphs quantify sphere-forming units, sphere size and types in PB-NICD and PB-NICD-ARKO groups. Scale bars, 100μm (a); 50μm (c); 25μm (d, e). Dot and bar graphs show data representing means ± s.d. from 3 independent experiments. Statistical significance was calculated using the t-test (*: p < 0.05, **: p < 0.01).
Discussion
Previous studies have suggested that Notch signaling may play an important role in prostate cancer progression. Notch activity has been shown to be increased in prostate cancer especially in advanced metastatic disease, although some cellular biological studies suggested otherwise57. More interestingly, Notch activity is higher in metastatic prostate tumor specimens than in primary tumor specimens58,59. Notch activity is enriched in the putative prostate cancer stem cells60 and docetaxol-resistant prostate tumor cells61. Our study in mouse models showed that increased Notch activity drives luminal epithelial proliferation and causes the formation of PIN lesions. More importantly, NICD confers anoikis resistance to dissociated luminal cells. Anoikis resistance is a hallmark for metastatic cancer cells. Therefore, our study supports a potential role of Notch in prostate cancer metastasis. In addition, we showed that Notch confers anoikis resistance to the putative luminal progenitor cells through activation of NF-κB signaling, which is also consistent with a previous study showing that prostate tumor-initiating stem-like cells exhibited increased NF-κB signaling62.
Several lines of evidence support the existence of a hierarchy within the luminal cell lineage. First, luminal cells are phenotypically heterogeneous. For example, they express varying levels of important transcription factors such as NKX3.163. Secondly, luminal cells behave differentially in response to androgen deprivation. Although the majority of luminal cells succumb to androgen deprivation, some luminal cells can survive. Third, only some luminal epithelial cells possess the capacity to retain a bromodeoxyuridine (BrdU) label64. Finally, recent in vivo lineage tracing studies have demonstrated the capability of the luminal lineage to self-sustain3–6. However, phenotypic and molecular features of these cells remain largely unknown. This is mainly due to a lack of in vitro or in vivo experimental systems through which the progenitor activity can be assayed quantitatively.
We showed that only a consistently small fraction of NICD-expressing luminal epithelial cells are capable of forming luminal spheres in vitro and generate glandular structures in vivo. These observations suggest that these cells possess the capacities for short-term self-renewal and unipotent differentiation, and imply that they are intrinsically distinct from those that are incapable of forming spheres or glands. Ectopic Notch activity promotes survival signaling partly by augmenting NF-κB, hence enabling their short-term in vitro propagation. We reason that these NICD-expressing luminal sphere-forming cells may represent the putative luminal progenitors or transit-amplifying/intermediate cells65, which probably constitute at least 4.5% of luminal cells based on our study. It is likely that the sphere-forming cells in vitro are the same cells that regenerate luminal glandular structures in vivo, which should be able to be directly interrogated once these cells are phenotypically characterized.
A more compelling and direct approach to characterize prostate luminal progenitors would be to physically isolate these cells and show that they possess progenitor activity in the presence of anti-anoikis signaling. As an attempt of this approach, we have prepared dissociated wild type prostate luminal cells and infected them with lentivirus expressing NICD. However, they failed to generate luminal prostate spheres efficiently. This observation implies that anoikis is initiated immediately after cell dissociation. Therefore the anti-anoikis and pro-proliferative signaling may have to be present prior to cell dissociation in order for these cells to survive and expand in vitro. Transgenic mouse models that harbor CreER transgene driven by specific promoters in conjunction with the ROSA-LSL-NICD model may be employed in the future to further determine the phenotypic identity of prostate luminal progenitors. It should be noted that our observation that Notch signaling confers anoikis resistance to some luminal epithelial cells does not imply that Notch signaling is essential for the maintenance of the luminal progenitors in vivo. Nor does it imply that luminal prostate epithelial cells possessing active Notch signaling in wild type mice are the luminal progenitors.
An interesting feature of luminal sphere cells is that they do not express AR or express AR at a very low level. In addition, AR is not essential for the survival and proliferation of luminal sphere-forming cells. These features resemble those of the putative prostate cancer stem cells that do not express AR or PSA62,66–68. These similarities imply that these luminal progenitors may serve as one of the origins for prostate cancer stem cells. Therefore, future phenotypic and molecular characterization of these cells will also provide novel insights into the properties and identities of prostate cancer stem cells.
Methods
Mice
All animals used in this study received humane care in compliance with the regulations relating to animals and experiments involving animals and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, NIH Publication, 1996 edition, and the protocol was approved by the Institutional Animal Care Committee of Baylor College of Medicine. The C57BL/6 and NOD/SCID mice were purchased from Charles River (Wilmington, MA). The Gt(ROSA)26Sortm1(Notch1)Dam/J (ICNf/f), and B6(Cg)-Gt(ROSA)26Sortm4(Ikbkb)Rsky/J mice were purchased from the Jackson Laboratory (Bar Harbor, ME). The NF-kB-RE-Luc mice were purchased from Taconic (Hudson, NY). The ARR2PB-Cre transgenic mice were from Dr. Fen Wang at the Institute of Bioscience and Technology, Texas A&M Health Science Center. The K8-CreERT2 mice were generated by our lab44. The RBP-Jf/f mice were form Dr. Tasuku Honjo at Kyoto University. The ARf/Y mice were generated by Drs. Guido Verhoeven and Karel De Gendt and were internally transferred from the laboratory of Dr. Alexander Agoulnik at Baylor College of Medicine69. The Catnblox(ex3) mice were internally transferred from the laboratory of Dr. David Spencer at Baylor College of Medicine. Mice were genotyped by polymerase chain reaction using mouse genomic DNA from tail biopsy specimens. The sequences of genotyping primers and the expected band sizes for PCR are listed in Supplementary Table 1. PCR products were separated electophoretically on 1% agarose gels and visualized via ethidium bromide under UV light.
Tamoxifen treatment
Tamoxifen (Sigma-Aldrich, St. Louis, MO) was dissolved into corn oil and was administrated i.p. into experimental mice at the age of 8 weeks (2–5 mg/40g/day for four consecutive days).
Lentivirus generation and infection
cDNA encoding Hes1 and dN-Hes1 were purchased from Addgene (Cambridge, MA) (#17624 and #24982). cDNAs encoding constitutively activated β-Catenin and dN-β-Catenin46 were from Drs. Roger Lo and Pierre McCrea, respectively. These cDNAs were PCR amplified, verified by sequencing, and cloned into the FU-CGW or FU-CRW lentiviral vectors70 using the EcoRI or XbaI sites. Dissociated prostate cells were infected by lentivirus at 1,500 g for 90 minutes at 25°C 70.
Prostate sphere and prostate regeneration assays
Prostate tissues were digested in DMEM/F12/Collagenase/Hyaluronidase/FBS (StemCell technologies, Vancouver, Canada) for 3 hours at 37°C, followed by an additional digestion in 0.25% Trypsin-EDTA (Invitrogen, Carlsbad, CA) on ice for 1 hour. Subsequently, digested cells were suspended in Dispase (Invitrogen, Carlsbad, CA, 5 mg/mL) and DNase I (Roche Applied Science, Indianapolis, IN, 1 mg/mL), and pipetted vigorously to dissociate cell clumps. Dissociated cells were then passed through 70 μm cell strainers (BD Biosciences, San Jose, CA) to get single cells.
Prostate sphere assays and prostate regeneration assays were performed using cells from 7–12 week old male experimental mice10,43. In the prostate sphere assay, 1–2 ×104 dissociated prostate cells were cultured in 1:1 Matrigel/PrEGM [Matrigel (BD Biosciences, Two Oak Park, Bedford, MA) / PrEGM (Lonza, Walkersville, MD)]. Prostate spheres were defined as spheroids with a diameter larger than 25 μm after a 6-day-culture. In the prostate regeneration assay, 1×104 to 1×105 dissociated cells or FACS-sorted cells were mixed with 1×105 mouse UGSM cells and resuspended in transplantation solution [PrEGM media (Lonza, Walkersville, MD):Matrigel (BD Biosciences, San Jose, CA), 1:1 (v/v)]. Cell mixtures were injected subcutaneously into 10-week old male NOD/SCID mice and incubated for 8 weeks. To semi-quantitate the number of glandular structures generated in the prostate regeneration assay, we enumerated the numbers of the glandular structures from tissue sections that contained tissue specimens with the largest diameter. Since we were not able to reconstruct the regenerated tissue in three dimensions, the estimated glandular-forming units should represent the minimal values.
Collection of prostatic secretions
Dissected anterior prostate lobes were cut into halves using syringe needles and prostate secretions were squeezed out by centrifugation at 6,000g for 1 minute. Protein concentrations were determined by a Bradford Assay kit (BioRad, Hercules, CA).
Pharmacological manipulation of signaling pathways
LY294002, MK2206, and BAY-11-7085 were purchased from Cell Signaling (Boston, MA), Selleck (Houston, TX), and Tocris Bioscience (Bristol, United Kingdom), respectively. Compounds were dissolved in DMSO and used in vitro at a concentration indicated in the Figures. Cell culture media was replaced every 24 hours.
FACS
For the separation of prostate cell lineages, dissociated murine prostate cells were stained with FITC- or Pacific blue-anti CD31, CD45 and Ter119 antibodies (eBioscience, San Diego, CA)], PE- or PE-Cy7-anti Sca-1 antibody (eBioscience, San Diego, CA) and Alexa 647-anti CD49f antibody (Biolegend, San Diego, CA). FACS analyses and sorting were performed by using the BD LSR II or BD LSR Fortessa and Aria I, respectively (BD Biosciences, San Jose, CA).
RNA isolation and quantitative RT-PCR
Total RNA was isolated from cells using the RNeasy Plus mini kit (Qiagen, Valencia, CA). Reverse transcription was performed using the iScript cDNA synthesis kit (BioRad, Hercules, CA). Quantitative RT-PCR was performed using the SYBR Premix Ex Taq (Perfect Real Time) (Takara Bio Inc., Otsu, Shiga, Japan) on a StepOne plus Real-Time PCR system (Applied Biosystems, Foster City, CA). Primer sequences for qPCR are listed in Supplementary Table 2.
Luciferase assay
Three mg of D-Luciferin (Promega, Madison, WI) in 100 μl PBS was intraperitoneally injected to 8–12 weeks old NICDWT:NF-kB-RE-Luc and PB- NICD;NF-kB-RE-Luc. After 10 min, prostates were harvested and bioluminescence imaging was performed with IVIS 200 (Caliper Lifer Sciences, Mountain View, CA). Imaged signals were quantitatively analyzed with IGOR-PRO Living Image software (Caliper Lifer Sciences). Luciferase assay was performed using the Luciferase Reporter Assay System (Promega). Luciferase activity was normalized by total cell numbers or tissue weight.
Adenovirus infection
Dissociated primary prostate cells were infected with Ad5-CMV or Ad5-CMV-Cre (Viral Vector Production Core Laboratory, Baylor College of Medicine, Houston, TX) at an MOI of 50 at 37°C for 1 hour in DMEM. Cells were washed twice with DMEM and resuspended in PrEGM media to be used in the prostate sphere assay.
Western blot
Prostate tissues were lysed in RIPA buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4) with protease inhibitors and phosphatase inhibitors (Roche Applied Science, Indianapolis, IN) using Tissuelyser LT (Qiagen, Valencia, CA). Protein concentrations were determined by a Bradford Assay kit (BioRad, Hercules, CA). Protein was separated by 8% or 12% SDS/PAGE and transferred onto a PVDF membrane (Amersham Biosciences, Arlington Heights, IL). The membrane was blocked in 5% skim milk, and subsequently incubated with primary antibodies listed in Supplementary Table 3 at 4°C overnight followed by incubation with peroxidase-conjugated goat anti-mouse IgG or goat anti-rabbit IgG (Jackson ImmunoResearch, Inc., West Grove, PA), and developed with Pierce ECL reagent (Thermal Scientific, Rockford, IL).
Reverse phase protein array
Reverse-phase protein arrays (RPPA) were performed and analyzed by the RPPA core of the MD Anderson Cancer Center. Briefly, protein lysates obtained from primary samples were printed onto slides, together with normalization and expression controls, in 5 serial dilutions. The slides were probed with strictly validated primary antibodies against total or phosphorylated protein and a secondary antibody was used to amplify the signal. The stained slides were scanned using a scanner and the images were analyzed using the MicroVigene Version 3.4 software program (VigeneTech) to produce quantified data. The “supercurve” algorithm was used to estimate the sample-specific protein expression levels. The RPPA data were normalized by median centering the results for each sample across all antibodies.
Histology and immunostaining
H&E staining and immunofluorescence staining were performed using standard protocols on 5–μm paraffin sections. Primary antibodies and dilutions used are listed in the Table below. Slides were incubated with 5% normal goat serum (Vector Labs) and with primary antibodies diluted in 2.5% normal goat serum overnight at 4°C. Slides then were incubated with secondary antibodies (diluted 1:500 in PBST) labeled with Alexa Fluor 488 or 594 (Invitrogen/Molecular Probes). Sections were counterstained with 4,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, St. Louis, MO). Immuno-fluorescence staining was imaged using an Olympus BX60 fluorescence microscope or a Leica EL6000 confocal microscope. Images of IHC were analyzed by Image-Pro Plus version 6.3 by Media Cybernetics. Cell number was determined by using the count feature in the software which asks for the user to indicate the color that would be used to indicate a positive cell (For example: blue would be indicated to count nuclei and thus indicate total numbers of cells). Borders were created such that only epithelial cells would be analyzed.
Expression microarray
Expression microarray assays were performed using 4×44K Whole Mouse Genome Oligo Microarray chip (Agilent Technologies, Santa Clara, CA) and the Feature Extraction Software v9.1.3.1 (Agilent Technologies) was used to extract and analyze the signals.
Statistics
All experiments were performed using 2–8 mice in independent experiments. Data are presented as mean ± SD. Student’s t test was used to determine significance between groups. For all statistical tests, the 0.05 level of confidence was accepted for statistical significance. One-sided Fishers exact tests also assessed enrichment between genes high in our PB-NICD group (544 unique genes with P<0.01, fold difference>1.5) and genes high in Akt or NF-κB signatures (710 and 270 genes, respectively).
Supplementary Material
Acknowledgments
Source of support: NIDDK, NCI
We thank Dr. Tasuku Honjo for the Rbp-J conditional mice, Drs. Julienne Carsten and David Spencer for the Catnblox(ex3) mice, Drs. Guido Verhoeven and Karel De Gendt for the AR conditional mice, Dr. Pierre McCrea for the dN-βCatenin cDNA, Dr. Jeffrey Whitsett for the antibody against SPDEF, Dr. Olga Dakhova and Ms. Yiqun Zhang for performing microarray analyses. We thank the technical support from the Cytometry and Cell Sorting Core at Baylor College of Medicine (NIH AI036211, CA125123, and RR024574) and the expert assistance of Joel M. Sederstrom. This work is supported by CPRIT RP110005, NIH R00 CA125937, R01 DK092202 (L.X.), NIH U01 CA141497 (M.M.I.), and NIH P30 CA125123 (the Cancer Center Shared Resources Grant).
Footnotes
Author Contributions
L.X., O.K. and J.M.V. designed the experiments.
O.K., J.M.V., L.Z., B.Z., X.W. and Q.S. performed experiments.
O.K., J.M.V., M.M.I and C.J.C. analyzed the data.
L.X. wrote the manuscript.
O.K. and J.M.V. contributed equally to the work.
Accession codes
Array data have been deposited in Gene Expression Omnibus under accession number GSE53036.
Conflict of Interest
The authors have no conflict of interest to declare.
References
- 1.Abate-Shen C, Shen MM. Molecular genetics of prostate cancer. Genes & development. 2000;14:2410–2434. doi: 10.1101/gad.819500. [DOI] [PubMed] [Google Scholar]
- 2.Isaacs JT. Control of cell proliferation and death in normal and neoplastic prostate: A stem cell model. In: Rodgers CH, CDS, Cunha GR, editors. Benigh prostatic hyperplasia. Bethesda: 1985. pp. 85–94. [Google Scholar]
- 3.Choi N, Zhang B, Zhang L, Ittmann M, Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21:253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, et al. Regenerated luminal epithelial cells are derived from preexisting luminal epithelial cells in adult mouse prostate. Mol Endocrinol. 2011;25:1849–1857. doi: 10.1210/me.2011-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang ZA, et al. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nature cell biology. 2013;15:274–283. doi: 10.1038/ncb2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu TL, et al. Conditionally ablated Pten in prostate basal cells promotes basal-to-luminal differentiation and causes invasive prostate cancer in mice. Am J Pathol. 2013;182:975–991. doi: 10.1016/j.ajpath.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Lawson DA, Witte ON. Stem cells in prostate cancer initiation and progression. The Journal of clinical investigation. 2007;117:2044–2050. doi: 10.1172/JCI32810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein AS, et al. Purification and direct transformation of epithelial progenitor cells from primary human prostate. Nature protocols. 2011;6:656–667. doi: 10.1038/nprot.2011.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci U S A. 2007;104:181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xin L, Lawson DA, Witte ON. The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proc Natl Acad Sci U S A. 2005;102:6942–6947. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leong KG, Wang BE, Johnson L, Gao WQ. Generation of a prostate from a single adult stem cell. Nature. 2008;456:804–808. doi: 10.1038/nature07427. [DOI] [PubMed] [Google Scholar]
- 12.Burger PE, et al. Sca-1 expression identifies stem cells in the proximal region of prostatic ducts with high capacity to reconstitute prostatic tissue. Proc Natl Acad Sci U S A. 2005;102:7180–7185. doi: 10.1073/pnas.0502761102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richardson GD, et al. CD133, a novel marker for human prostatic epithelial stem cells. Journal of cell science. 2004;117:3539–3545. doi: 10.1242/jcs.01222. [DOI] [PubMed] [Google Scholar]
- 14.Kwon OJ, Zhang L, Ittmann MM, Xin L. Prostatic inflammation enhances basal-to-luminal differentiation and accelerates initiation of prostate cancer with a basal cell origin. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1318157111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xin L. Cells of origin for cancer: an updated view from prostate cancer. Oncogene. 2013;32:3655–3663. doi: 10.1038/onc.2012.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salm SN, et al. Generation of active TGF-beta by prostatic cell cocultures using novel basal and luminal prostatic epithelial cell lines. J Cell Physiol. 2000;184:70–79. doi: 10.1002/(SICI)1097-4652(200007)184:1<70::AID-JCP7>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 17.Tokar EJ, Ancrile BB, Cunha GR, Webber MM. Stem/progenitor and intermediate cell types and the origin of human prostate cancer. Differentiation; research in biological diversity. 2005;73:463–473. doi: 10.1111/j.1432-0436.2005.00047.x. [DOI] [PubMed] [Google Scholar]
- 18.Frisch SM, Screaton RA. Anoikis mechanisms. Current opinion in cell biology. 2001;13:555–562. doi: 10.1016/s0955-0674(00)00251-9. [DOI] [PubMed] [Google Scholar]
- 19.McDonnell TJ, et al. Expression of the protooncogene bcl-2 in the prostate and its association with emergence of androgen-independent prostate cancer. Cancer Res. 1992;52:6940–6944. [PubMed] [Google Scholar]
- 20.Carroll DK, et al. p63 regulates an adhesion programme and cell survival in epithelial cells. Nature cell biology. 2006;8:551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- 21.Mazzone M, et al. Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells. Proc Natl Acad Sci U S A. 2010;107:5012–5017. doi: 10.1073/pnas.1000896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfohler C, et al. In situ hybridization analysis of genes coding collagen IV alpha1 chain, laminin beta1 chain, and S-laminin in prostate tissue and prostate cancer: increased basement membrane gene expression in high-grade and metastatic lesions. Prostate. 1998;36:143–150. doi: 10.1002/(sici)1097-0045(19980801)36:3<143::aid-pros1>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 23.Weaver VM, et al. beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2002;2:205–216. doi: 10.1016/s1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frisch SM, Schaller M, Cieply B. Mechanisms that link the oncogenic epithelial-mesenchymal transition to suppression of anoikis. Journal of cell science. 2013;126:21–29. doi: 10.1242/jcs.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Zhao W, Valdez JM, Creighton CJ, Xin L. Low-density Taqman miRNA array reveals miRNAs differentially expressed in prostatic stem cells and luminal cells. Prostate. 2009;70:297–304. doi: 10.1002/pros.21064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harper ME, Glynne-Jones E, Goddard L, Mathews P, Nicholson RI. Expression of androgen receptor and growth factors in premalignant lesions of the prostate. The Journal of pathology. 1998;186:169–177. doi: 10.1002/(SICI)1096-9896(1998100)186:2<169::AID-PATH164>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 27.Sroka IC, McDaniel K, Nagle RB, Bowden GT. Differential localization of MT1-MMP in human prostate cancer tissue: role of IGF-1R in MT1-MMP expression. Prostate. 2008;68:463–476. doi: 10.1002/pros.20718. [DOI] [PubMed] [Google Scholar]
- 28.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ang HL, Tergaonkar V. Notch and NFkappaB signaling pathways: Do they collaborate in normal vertebrate brain development and function? BioEssays : news and reviews in molecular, cellular and developmental biology. 2007;29:1039–1047. doi: 10.1002/bies.20647. [DOI] [PubMed] [Google Scholar]
- 30.Mehrotra S, et al. IAP regulation of metastasis. Cancer Cell. 2010;17:53–64. doi: 10.1016/j.ccr.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan SR, et al. Activation of NF-kappaB following detachment delays apoptosis in intestinal epithelial cells. Oncogene. 2005;24:6482–6491. doi: 10.1038/sj.onc.1208810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leong KG, et al. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. The Journal of experimental medicine. 2007;204:2935–2948. doi: 10.1084/jem.20071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valdez JM, et al. Notch and TGFbeta form a reciprocal positive regulatory loop that suppresses murine prostate basal stem/progenitor cell activity. Cell stem cell. 2012;11:676–688. doi: 10.1016/j.stem.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park JH, et al. Prostatic intraepithelial neoplasia in genetically engineered mice. Am J Pathol. 2002;161:727–735. doi: 10.1016/S0002-9440(10)64228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creighton CJ. Multiple oncogenic pathway signatures show coordinate expression patterns in human prostate tumors. PLoS One. 2008;3:e1816. doi: 10.1371/journal.pone.0001816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavery DN, et al. Repression of androgen receptor activity by HEYL, a third member of the Hairy/Enhancer-of-split-related family of Notch effectors. The Journal of biological chemistry. 2011;286:17796–17808. doi: 10.1074/jbc.M110.198655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belandia B, et al. Hey1, a mediator of notch signaling, is an androgen receptor corepressor. Molecular and cellular biology. 2005;25:1425–1436. doi: 10.1128/MCB.25.4.1425-1436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujimoto N, Suzuki T, Ohta S, Kitamura S. Identification of rat prostatic secreted proteins using mass spectrometric analysis and androgen-dependent mRNA expression. Journal of andrology. 2009;30:669–678. doi: 10.2164/jandrol.108.005553. [DOI] [PubMed] [Google Scholar]
- 39.Chen G, et al. SPDEF is required for mouse pulmonary goblet cell differentiation and regulates a network of genes associated with mucus production. The Journal of clinical investigation. 2009;119:2914–2924. doi: 10.1172/JCI39731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noah TK, Kazanjian A, Whitsett J, Shroyer NF. SAM pointed domain ETS factor (SPDEF) regulates terminal differentiation and maturation of intestinal goblet cells. Experimental cell research. 2010;316:452–465. doi: 10.1016/j.yexcr.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buchwalter G, et al. PDEF Promotes Luminal Differentiation and Acts as a Survival Factor for ER-Positive Breast Cancer Cells. Cancer Cell. 2013;23:753–767. doi: 10.1016/j.ccr.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oettgen P, et al. PDEF, a novel prostate epithelium-specific ets transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. The Journal of biological chemistry. 2000;275:1216–1225. doi: 10.1074/jbc.275.2.1216. [DOI] [PubMed] [Google Scholar]
- 43.Xin L, Lukacs RU, Lawson DA, Cheng D, Witte ON. Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cells. 2007;25:2760–2769. doi: 10.1634/stemcells.2007-0355. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, et al. Targeting CreER(T2) expression to keratin 8-expressing murine simple epithelia using bacterial artificial chromosome transgenesis. Transgenic research. 2012 doi: 10.1007/s11248-012-9598-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harada N, et al. Lack of tumorigenesis in the mouse liver after adenovirus-mediated expression of a dominant stable mutant of beta-catenin. Cancer Res. 2002;62:1971–1977. [PubMed] [Google Scholar]
- 46.Montross WT, Ji H, McCrea PD. A beta-catenin/engrailed chimera selectively suppresses Wnt signaling. Journal of cell science. 2000;113 ( Pt 10):1759–1770. doi: 10.1242/jcs.113.10.1759. [DOI] [PubMed] [Google Scholar]
- 47.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Majumder PK, et al. Prostate intraepithelial neoplasia induced by prostate restricted Akt activation: the MPAKT model. Proc Natl Acad Sci U S A. 2003;100:7841–7846. doi: 10.1073/pnas.1232229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mulholland DJ, et al. Lin-Sca-1+CD49fhigh stem/progenitors are tumor-initiating cells in the Pten-null prostate cancer model. Cancer Res. 2009;69:8555–8562. doi: 10.1158/0008-5472.CAN-08-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Birbach A, et al. Persistent inflammation leads to proliferative neoplasia and loss of smooth muscle cells in a prostate tumor model. Neoplasia. 2011;13:692–703. doi: 10.1593/neo.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghosh-Choudhury N, Mandal CC, Ghosh-Choudhury N, Ghosh Choudhury G. Simvastatin induces derepression of PTEN expression via NFkappaB to inhibit breast cancer cell growth. Cellular signalling. 2010;22:749–758. doi: 10.1016/j.cellsig.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meng F, Liu L, Chin PC, D’Mello SR. Akt is a downstream target of NF-kappa B. The Journal of biological chemistry. 2002;277:29674–29680. doi: 10.1074/jbc.M112464200. [DOI] [PubMed] [Google Scholar]
- 53.Espinosa L, et al. The Notch/Hes1 pathway sustains NF-kappaB activation through CYLD repression in T cell leukemia. Cancer Cell. 2010;18:268–281. doi: 10.1016/j.ccr.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shin HM, et al. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. The EMBO journal. 2006;25:129–138. doi: 10.1038/sj.emboj.7600902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng P, et al. Notch-1 regulates NF-kappaB activity in hemopoietic progenitor cells. J Immunol. 2001;167:4458–4467. doi: 10.4049/jimmunol.167.8.4458. [DOI] [PubMed] [Google Scholar]
- 56.Vacca A, et al. Notch3 and pre-TCR interaction unveils distinct NF-kappaB pathways in T-cell development and leukemia. The EMBO journal. 2006;25:1000–1008. doi: 10.1038/sj.emboj.7600996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shou J, Ross S, Koeppen H, de Sauvage FJ, Gao WQ. Dynamics of notch expression during murine prostate development and tumorigenesis. Cancer Res. 2001;61:7291–7297. [PubMed] [Google Scholar]
- 58.Danza G, et al. Notch3 is activated by chronic hypoxia and contributes to the progression of human prostate cancer. International journal of cancer. Journal international du cancer. 2013 doi: 10.1002/ijc.28293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu H, Zhou X, Redfield S, Lewin J, Miele L. Elevated Jagged-1 and Notch-1 expression in high grade and metastatic prostate cancers. American journal of translational research. 2013;5:368–378. [PMC free article] [PubMed] [Google Scholar]
- 60.Patrawala L, et al. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2− cancer cells are similarly tumorigenic. Cancer Res. 2005;65:6207–6219. doi: 10.1158/0008-5472.CAN-05-0592. [DOI] [PubMed] [Google Scholar]
- 61.Domingo-Domenech J, et al. Suppression of Acquired Docetaxel Resistance in Prostate Cancer through Depletion of Notch- and Hedgehog-Dependent Tumor-Initiating Cells. Cancer Cell. 2012;22:373–388. doi: 10.1016/j.ccr.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rajasekhar VK, Studer L, Gerald W, Socci ND, Scher HI. Tumour-initiating stem-like cells in human prostate cancer exhibit increased NF-kappaB signalling. Nature communications. 2011;2:162. doi: 10.1038/ncomms1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsujimura A, et al. Proximal location of mouse prostate epithelial stem cells: a model of prostatic homeostasis. J Cell Biol. 2002;157:1257–1265. doi: 10.1083/jcb.200202067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uzgare AR, Xu Y, Isaacs JT. In vitro culturing and characteristics of transit amplifying epithelial cells from human prostate tissue. Journal of cellular biochemistry. 2004;91:196–205. doi: 10.1002/jcb.10764. [DOI] [PubMed] [Google Scholar]
- 66.Gu G, Yuan J, Wills M, Kasper S. Prostate cancer cells with stem cell characteristics reconstitute the original human tumor in vivo. Cancer Res. 2007;67:4807–4815. doi: 10.1158/0008-5472.CAN-06-4608. [DOI] [PubMed] [Google Scholar]
- 67.Qin J, et al. The PSA(-/lo) prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell stem cell. 2012;10:556–569. doi: 10.1016/j.stem.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stoyanova T, et al. Prostate cancer originating in basal cells progresses to adenocarcinoma propagated by luminal-like cells. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1320565110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Gendt K, et al. A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. Proc Natl Acad Sci U S A. 2004;101:1327–1332. doi: 10.1073/pnas.0308114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xin L, Ide H, Kim Y, Dubey P, Witte ON. In vivo regeneration of murine prostate from dissociated cell populations of postnatal epithelia and urogenital sinus mesenchyme. Proc Natl Acad Sci U S A. 2003;100 (Suppl 1):11896–11903. doi: 10.1073/pnas.1734139100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.