Abstract
High-throughput RNA sequencing (RNA-seq) dramatically expands the potential for novel genomics discoveries, but the wide variety of platforms, protocols and performance has created the need for comprehensive reference data. Here we describe the Association of Biomolecular Resource Facilities next-generation sequencing (ABRF-NGS) study on RNA-seq. We tested replicate experiments across 15 laboratory sites using reference RNA standards to test four protocols (polyA-selected, ribo-depleted, size-selected and degraded) on five sequencing platforms (Illumina HiSeq, Life Technologies’ PGM and Proton, Pacific Biosciences RS and Roche’s 454). The results show high intra-platform and inter-platform concordance for expression measures across the deep-count platforms, but highly variable efficiency and cost for splice junction and variant detection between all platforms. These data also demonstrate that ribosomal RNA depletion can both enable effective analysis of degraded RNA samples and be readily compared to polyA-enriched fractions. This study provides a broad foundation for cross-platform standardization, evaluation and improvement of RNA-seq.
Introduction
RNA-seq is an important analytical technique that leverages the capacity of high-throughput sequencing instruments to quantitatively sample a population of RNA molecules with a large number of “reads” or parallel reactions on discrete templates1,2. Depending on experimental goals, sample types and read depths, results from RNA-seq data can be similar or superior to those from microarray data3-5. However, each sequencing platform has unique aspects of library synthesis, sequencing, alignment, and data processing6-9. Thus, many questions remain about RNA-seq in regards to inter-operability between platforms, cross-site reproducibility, bioinformatics methods and the sources of variance in results with both existing and emerging protocols, such as those for degraded RNA.
Notably, prior work comparing microarray platforms and methods showed high levels of inter-platform concordance for the ability to detect differentially expressed genes. The Microarray Quality Control (MAQC) Consortium landmark study10 examined the degree of variance within and across many different microarray platforms and found similar coefficients of variation between platforms. The MAQC data also provided an important benchmark for the application of microarray technologies to clinical assays. For high-throughput sequencing platforms, however, very little data exist about cross-site variation of expression measures. Only two inter-site variation studies are publicly available: the MAQC-III (a.k.a. the Sequencing Quality Control Consortium, SEQC)11 study and the GEUVARDIS Consortium12. These studies were either limited to one platform or did not assess some newer RNA-seq methods that are now widely used. Moreover, important RNA profiling parameters such as differential expression and splice variant detection have not been consistently evaluated. Thus, these studies do not answer key questions about the degree of concordance for RNA-seq across platforms and methods and also about the read depth, type, and length of sequence reads required to fully characterize a sample with current techniques13-16. Moreover, RNA-seq is an extremely useful method for exploring the expression of sequence variants, detecting novel RNAs and for discriminating between transcript splicing isoforms17-20, but there is no “gold standard” of reference data on the dynamic range of differential expression and splicing that includes different sample preparation protocols, instruments and data analysis strategies.
To address this challenge, members of the Association of Biomolecular Resource Facilities (ABRF)21 designed and conducted the first phase of a large-scale ABRF-NGS Study with a focus on RNA-seq. The goals of the ABRF-NGS Study are to evaluate the performance of NGS platforms and to identify optimal methods and best practices. A wide range of variables was evaluated, including library preparation methods (polyA-enriched and ribo-depleted), size-specific fractionation (1, 2 and 3 kb) and RNA integrity (using heat, RNase A and sonication to degrade the RNA). The latter variable was chosen to mimic some of the damaging effects of tissue fixation with formalin, which is a well-recognized issue for RNA profiling of formalin-fixed, paraffin-embedded (FFPE) clinical specimens22-24. Finally, we leveraged a data set of 18,124 PrimePCR reactions and used it with 802 previously published10 TaqMan RT-qPCR reactions as orthogonal measurements to gauge the linear response and dynamic range of the RNA-seq results from the different platforms and protocols. Both platform-agnostic and platform-specific aligners were also compared to support the validity of the conclusions. Taken together, these data represent a broad cross-platform characterization of widely used RNA standards and to our knowledge provide the largest comprehensive comparison of results from degraded, full-length and size-selected RNA across sequencing platforms and protocols.
Results
Platforms, RNA samples and sequencing protocols
Although comparisons of high-throughput sequencing platforms and sample preparation protocols have been reported in past studies6,5-27, no other study has been conducted using five platforms and two standardized RNA samples replicated at multiple sites (Fig. 1). Platforms evaluated included the Illumina HiSeq 2000/2500, Roche 454 GS FLX+, Life Technologies Ion Personal Genome Machine (PGM) and Proton, and the Pacific Biosciences RS (PacBio)6, 8, 28. Data were generated and analyzed by the members of five ABRF Research Groups, including 25 core facilities at 20 different institutions (Fig. 1 and Supplementary Table 1). Additional data from an Illumina MiSeq v2 instrument were used to compare metrics derived from different read lengths from the same Illumina library preparation and sequencing methods. Detection of differential RNA abundance was evaluated using two commercially available and very distinct RNA samples: A = RNA from cancer cell lines; B = RNA from pooled normal human brain tissues; and two pre-defined mixtures of these samples (C = [75% A + 25% B]; D = [25% A + 75% B]). All standardized RNA samples also contained synthetic RNA spike-ins from the External RNA Control Consortium (ERCC)10, 29, 30. Results from high-quality RNA on the Illumina HiSeq 2500 platform were compared to results on the same platform from RNAs degraded using three degradation conditions: heat, RNase and sonication. The RNA reference samples were degraded to a RIN (RNA integrity number) of 2 or less. In addition, results from ribosomal RNA-depleted and polyA-enriched libraries from intact RNA were compared using the Illumina HiSeq 2500 platform.
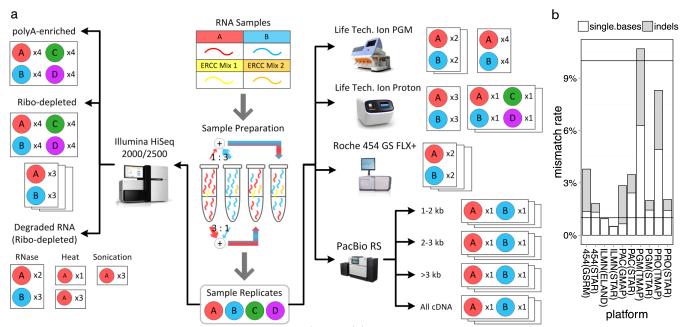
Figure 1. Experimental design and sequencing platforms.
(a) Two standard RNA samples (A = Universal Human Reference RNA and B = Human Brain Reference RNA) were combined with two sets of synthetic RNAs (ERCCs) to prepare a set of samples to be sequenced on five platforms: Illumina (ILMN) HiSeq 2000/2500, Life Technologies Personal Genome Machine (PGM), Life Technologies Proton (PRO), Pacific Biosciences (PacBio) RS (PAC), and the Roche 454 GS FLX+. Additional RNA samples were also generated: samples C and D were prepared as defined mixtures of A and B, while other aliquots of A and B were degraded by three methods. All these additional samples were ribo-depleted for RNA-seq on the HiSeq platform. The number of technical replicates (x2, x3 or x4) of each sample set is indicated for each platform and method. (b) Stacked bar plots of the sequencing platforms’ mismatch rates (y-axis) for single-base mismatches (white) and insertions/deletions (indels, grey) based on different aligners for each platform (x-axis). Q10 (90% accuracy) and Q20 (99% accuracy) are shown as the top and bottom line, respectively.
To map the sequencing reads to the human genome (hg19), we used both vendor-recommended alignment algorithms and ‘universal,’ platform-agnostic aligners. For gene expression quantification, the following aligners were evaluated: STAR31 (agnostic), ELAND (HiSeq), TMAP (PGM and Proton), GSRM (454) and GMAP (PacBio). With the exception of ELAND, each platform-specific algorithm produced better mapping rates, gene-body coverage evenness and Spearman correlations with PrimePCR quantification (Supplementary Tables 2–4) when compared to STAR applied uniformly across all platforms. However, the universal STAR alignments were used as input for shared junction detection (Supplementary Table 5), since these alignments always showed the lowest mapping error rate (Fig. 1). After mapping, additional processing for quantifying gene counts was performed using the open source r-make package (http://physiology.med.cornell.edu/faculty/mason/lab/data/r-make, and Online Methods) to calculate the reads and coverage for each gene feature based on GENCODE (v12) annotation. Quality control data were generated using the fastQC package (www.bioinformatics.babraham.ac.uk/projects/fastqc) to calculate a large set of performance metrics for sequence quality, gene coverage and transcriptome quantitation and characterization for all platforms (Fig. 1 and Supplementary Figs. 1–23).
Base qualities, data quality and duplicate rates
Quality Values (QV, a per base accuracy estimate) were calculated for all sample runs, for pre-alignment measures (Supplemental Figs. 1–6) and post-alignment measures (Fig. 1b). Results ranged from Q10 (90% accuracy) to Q60 (99.9999% accuracy) across platforms (Supplementary Figs. 1–6) and revealed three notable trends. First, most platforms show a biased QV distribution in the first 1–16 bases, a known effect from the reverse transcriptase (RT) priming step32. This RT bias can also affect the observed GC content (Supplementary Figs. 7–11) and base-frequency data11,33, 34 (Supplementary Figs. 12–17). Second, similar QV profiles were observed for samples A and B, and across different RNA size fractions. Third, although changes in library preparation techniques and sequencing chemistry for various platforms can affect the QVs, the largest increase in QVs came from the circular consensus sequencing (CCS) for the PacBio data (Supplementary Fig. 2), where median QVs near 40 were observed, though with a wide range of variation. Thus, for most platforms, the ends of the reads are where most “noise” was observed, but lower QVs also occurred at the beginnings of the reads. This results in a source of bias and noise for RNA-seq data that appears in all platforms and is usually addressed by appropriate sequence trimming.
The QVs for each base of a read, as well as the read length, alignment method and reference sequence quality, can all affect mapping accuracy. To estimate the platform-specific and aligner-specific impact of the sequencing error rate on alignment, we calculated the number of mismatches relative to the hg19 human reference genome, normalized by total mapped bases, for two aligners for each platform (Fig. 1b). These data showed that a tradeoff between higher mapping rate and accuracy can occur for RNA-seq, such as the increased mapping rate with TMAP/GSRM vs. STAR (Supplemental Table 2) that led to a higher empirically derived error rate (Fig. 1b). The most common type of mismatch for HiSeq was single-base substitutions, but the range between all platforms spanned 0.6–7.1%. Insertion/deletion (indel) type mismatch rates were also highly variable between platforms, spanning 0.017–4.4% of all mismatches observed. Moreover, for all platforms, the reported QVs were higher than the empirically derived QVs based on sequence mismatches, similar to the QV-inflation observed for DNA sequencing in the 1000 Genomes Project and GATK35, 36.
Previous work in RNA-seq has noted that duplicate reads may be a confounding factor in data analysis because reads with exactly the same start and end may arise from clonal copies produced during library amplification rather than from independently transcribed RNAs in the biological sample8, 33. However, unlike DNA sequencing of large diploid genomes, RNA-seq is expected to produce some reads from highly expressed transcripts that begin at the same nucleotide and are thus designated “duplicate.” An assessment of this question over a range of read lengths has not been previously reported, but is facilitated in this study by RNA-seq of the same samples over a range of varying read lengths (Supplementary Figs. 19–23). The read length distributions revealed distinct types for variable-read platforms, including Gaussian (454) and “ski-jump” (Proton and PGM), and the expected uniform lengths for Illumina platforms. Yet, all platforms showed no more than 51% of reads as putative duplicates (Supplementary Fig. 24), with the 454 and PacBio platforms showing the fewest duplicates (12–20%). PacBio library construction does not include any amplification step of the final cDNA library, while the reduced duplication with 454 is likely because the amplification step takes place after template attachment to single beads, so individual molecules in the library have less chance to spawn multiple reads. For the other platforms, this analysis cannot distinguish whether observed duplicates are due to independent transcripts or are a consequence of library amplification, but future datasets based on these same samples will support investigation of this question.
Coverage of genes
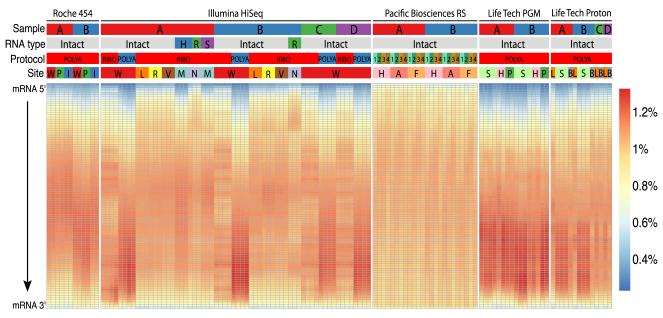
Next we examined the normalized coverage of all GENCODE gene transcripts from 5′ to 3′ termini for any bias in the number of mapped bases originating from different regions of the transcripts. Almost all samples showed a fairly similar distribution of coverage for genes (Fig. 2). Notably, the ribo-depleted RNA samples, whether degraded or not, consistently showed more-uniform gene coverage than did polyA-selected libraries. The data also showed “banding” or altered coverage distributions, likely caused by the use of a different library kit version at one of the test sites (C). This indicates that gene coverage can be affected by platform and preparation-dependent factors, but aligners can also play a role (Supplementary Table 3). Finally, the highest and most-uniform coverage of full-length transcripts came from preparing samples with enrichment for both the 3′ polyA tail and an antibody (Ab) for the 5′ methylguanylate cap (5′G cap), combined with long-read technology (see Online Methods for Pacific Biosciences).
Figure 2. Transcript coverage across all genes detected.
Each gene was examined as a set of 100 adjacent segments (percentiles of total transcript length). The relative number of reads that map to each segment was then plotted for each sample, platform, and technique (percent of all library reads per segment, see heatmap color key). Samples are categorized by five parameters (top): NGS platforms: Roche 454 GS FLX+, Illumina HiSeq 2000/2500, Pacific Biosciences RS, Life Technologies PGM and Proton; input RNA sample: samples A (red), B (blue), C (green), and D (purple); RNA type: intact or degraded by heat (H, blue), RNase (R, green) or sonication (S, purple); library protocol: polyA enrichment, ribosomal RNA depletion (ribo) or polyA plus 5′ cap enrichment with (1, 2, 3) or without (4) size fractionation; and site: 14 core facility sequencing laboratories. Most platforms showed less coverage at the 5′ and 3′ ends of the transcripts. Details on sequencing platforms, site abbreviations, sample type chemistries, and library preparations are listed in Table 2.
Transcriptome profiling and splice junction detection
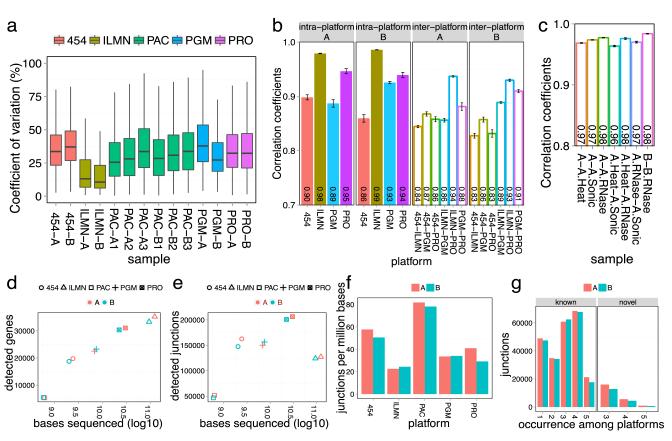
We investigated the ability of each platform to reproducibly detect and quantify genes and splice junctions across the transcriptome (Fig. 3). Data were restricted to genes that were observed at all test sites and in all technical replicates for each platform. The platforms showed a median range of 11–39% inter-site CV (Coefficient of Variation) in their quantification of detected genes using normalized gene expression values (Fig. 3a, Supplementary Methods), with HiSeq showing the lowest median CV. The Spearman correlations of normalized transcript levels were measured for samples A and B on different platforms (454, HiSeq, Proton and PGM) across multiple sites for Figure 3b; PacBio was not included because it displayed an (expected) low read count for many genes. The inter-platform correlation was high (R2 average of 0.83) for the same samples profiled on different platforms, and the intra-platform correlation was even higher (R2 average greater than 0.86). Each platform was also compared to normalized expression data from an orthogonal quantitation method (PrimePCR, Supplementary Fig. 25), and the Spearman correlations of the log2 fold differences were ranked as 454 < PGM < Proton, HiSeq, ranging from 0.83 to 0.89.
Figure 3. Intra- and inter-platform variation of RNA-seq transcript metrics.
The coefficients of variation (CV) of various metrics for transcripts detected across all sites were calculated for the Roche 454 GS FLX+, Illumina HiSeq 2000/2500 (ILMN), Pacific Biosciences RS (PAC), and Life Technologies PGM and Proton (PRO). (a) Inter-site CV of normalized gene expression for transcripts detected across all sites. The median CV for number of genes detected ranged from 10.70-38.68%, with many outlier genes present for each platform. (b) Inter-platform and intra-platform normalized gene expression Spearman correlation coefficients for samples A and B. (c) The degraded RNA profiles match the corresponding intact RNA profiles from HiSeq RNA-seq with very high correlation coefficients (0.975). Error bars are standard error of the mean. (d-e) Sequenced bases (log10) were plotted against the number of detected genes or the number of detected splice junctions for known GENCODE junctions. (f) More efficient splice junction detection (y-axis, number of junctions/Mb of sequence) was observed for long read platforms (PAC, 454). Detection efficiencies were calculated at comparable scales by constraining the total number of bases used from each platform to a range of 630-5451 × 106. (g) Most known junctions were detected by three or more platforms, indicating concordance among RNA-seq methods (left panel). The novel junctions (right) defined by independent observation on three or more platforms were less numerous than known junctions.
Next we examined the impact of read depth and length on transcript identification. A clear log-linear relationship was observed between sequence base depth and gene detection (Fig. 3d), showing that increasing the depth of sequencing for any platform is a quick means to find more genes. Characterizations of splice junction detection efficiency and inter-platform agreement have not been previously reported, so to account for each platform’s different read lengths, the effect of total sequenced base depth (rather than read count) was examined for previously annotated and new, unannotated splicing. Splice junction profiling showed an early plateau for detection of known junctions (Fig. 3e). The Proton, PGM and 454 platforms detected more known junctions despite fewer bases sequenced compared to Illumina HiSeq. However, a follow-up experiment with long-read Illumina MiSeq data (2×250 bp paired-end reads) showed a similar boost in junction identification (Supplementary Fig. 26), suggesting that splice junction detection is most affected by read length, rather than library preparation or sequencing chemistry. The ratio of the number of junctions detected as a function of total bases sequenced (junctions/Mb) revealed a wide range of values (Fig. 3f) but clearly demonstrated that longer reads are a more efficient way to capture junctions. This is reflected in the data from the long-read platforms and also in the comparison of the number of junctions detected in the Illumina HiSeq vs. MiSeq data from two aliquots of the same library (22.6 junctions/Mb for HiSeq vs. 33.9 junctions/Mb for MiSeq, Supplemental Fig. 26).
We also characterized the inter-platform agreement of known and novel junctions. The known GENCODE junctions (v12) showed higher inter-platform agreement, with most of these junctions detected by three or more platforms (Fig. 3g, left panel). However, unannotated junctions have lower concordance than known junctions across platforms (Fig. 3g). An examination of these rare isoforms revealed that the lower detection agreement is likely due to their lower expression levels (Supplementary Fig. 27), but they also may represent platform-specific artifacts. Therefore, only unannotated splice junctions observed on at least three platforms (which still includes >20,000 junctions per sample) are reported in this analysis.
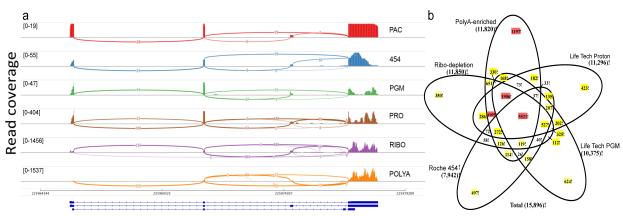
These cross-platform splicing data showed that the types of reads dramatically influenced each platform’s measure of low abundance transcripts. This effect was apparent for RNA splice isoforms such as SRP9 (Fig. 4a), suggesting that rare-isoform quantification benefits the most from greater read depths (such as from the Illumina HiSeq and Life Technologies Proton). However, uniformity of coverage across exons is improved with long-read technology such as PacBio (Fig. 4a and Supplementary Table 3), despite less read depth. An examination of the size-selected PacBio CCS libraries demonstrated that the polyA+5’G cap enrichment method captured the full lengths of expressed transcripts (Supplementary Fig. 28), with the majority (90%) showing complete transcript sequences in the 1–2 kb range or even longer. These results indicate that a combination of appropriate sample preparation and long reads can readily create cDNA profiles that approach the full-length sequences of mRNAs from complex samples, underlining the utility of long read platforms, despite the lower read depths they may produce37.
Figure 4. Inter-platform consistency of splicing and differential expression analysis.
(a) As a representative plot for RNA splicing, transcripts from the SRP9 gene are shown in a sashimi plot across five platforms and two Illumina library protocols. Pacific Biosciences (PAC), Roche 454 (454), and Life Technologies Ion PGM (PGM) detected the two most abundant isoforms. Life Technologies Proton (PRO) and Illumina ribo-depletion (RIBO) or polyA-enriched (POLYA) methods also detected a third isoform. PAC showed more uniform sequencing depth across the gene body. Read coverage as measured by the range of 19-1537 (coverage) is indicated in brackets. (b) Starting from the set of genes detected at any expression level on all platforms, the numbers of A vs. B differentially expressed genes uniquely or repeatedly detected at statistically significant thresholds (FDR <0.05 and fold change >2) are shown; sets of greater than 1000 genes are indicated in red, 100-999 in yellow.
To examine the ability of each platform to detect differentially expressed genes (DEGs) (Fig. 4b, Supplementary Figs. 29–31), we used limma-voom38 to perform DEG analysis on the normalized counts for each platform. Although a majority of DEGs were observed by two or more methods, each produced unique DEGs at all statistical significance and fold-change cutoffs (Supplementary Figs. 30, 31). Thus, although high read–depth platforms showed greater DEG overlap, each platform produced unique subsets (from unique systematic effects) of statistically significant DEGs (FDR < 0.05, fold change > 2, Online Methods), ranging from 6–11% of all called DEGs detected uniquely by a platform or preparation method (Fig. 4b, peripheral sets). These instruments span different chemistries, measurement techniques (optical vs. electrical) and base-calling methods, all of which likely play roles in the system-specific noise profiles, as noted in Figures 1–3 and Supplementary Figures 1–24.
Influence of library preparation on transcriptome profiles
To examine other factors that affect DEG measurements, we prepared libraries using both polyA enrichment or ribosomal RNA depletion of the standard samples, and then performed sequencing on the same Illumina HiSeq 2500 instrument. Identical aliquots of the standards (A, B, C and D) were separated into quadruplicate sets for library preparation. All replicate libraries were then sequenced in a multiplexed assay on a full Illumina flow cell. The ribo-depletion library method produced a read source distribution very different from the polyA preparation method (Fig. 5a). The ribo-depleted libraries showed 40–47% of the bases mapping to introns vs. 7–12% for polyA RNA from the same sample (lower intronic reads were similarly observed for polyA RNA on the other platforms, Supplementary Fig. 32). Both methods produced fairly consistent measures of RNA abundance (FPKM, Online Methods), with a median FPKM difference of only 0.055 between all genes. However, more genes with lower levels of expression were observed with the ribo-depletion method, whereas the polyA libraries contained more highly expressed genes and 3′ untranslated regions (3′ UTR) Supplementary Fig. 33). As expected, the ribo-depleted libraries were enriched for non-coding RNAs, such as lncRNAs and snoRNAs (Supplementary Table 6), whereas the polyA libraries were enriched for protein-coding genes and mitochondrial genes (Supplementary Tables 6–8)39. Sequence annotations in GENCODE currently labeled as “intron” and other categories are likely to change as new non-coding RNAs (or new transcript classes) are identified.
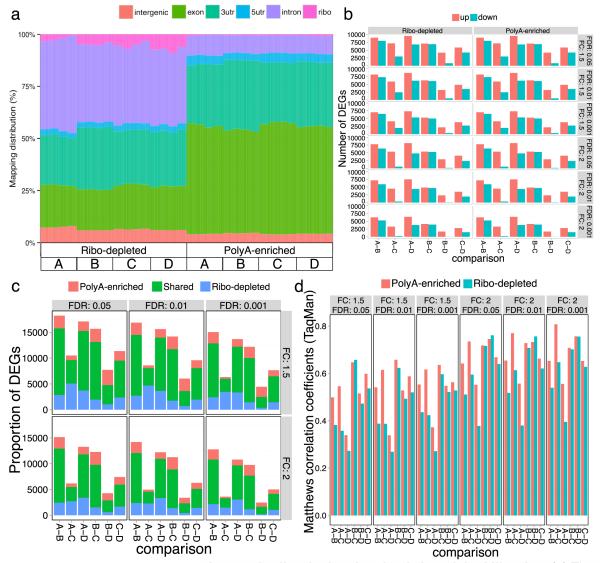
Figure 5. Differentially expressed genes in ribo-depleted and polyA-enriched libraries.
(a) The percentage of reads that map to various gene sequence categories was plotted. A greater number of intronic reads from ribo-depleted libraries was observed. The sequence type and read distribution of gene features detected in polyA-enriched and ribo-depleted libraries from the same sample were examined using GENCODE (v12) annotations. Mitochondrial RNA reads are present at trace levels (<0.1%, data not shown). (b) Differentially expressed genes (DEGs) were detected in all pairwise comparisons of the original (A, B) and mixed samples (C, D); (c) results were similar for both library types from the common set of detected genes at all fold-change (FC) and false discovery rate (FDR) thresholds tested. (d) Both library types show similar accuracy as evidenced by Matthews Correlation Coefficients (MCC) with RT-qPCR assays (see Suppl. Fig. 29b for expanded data). A subset of GENCODE mapped reads was used from each library (mean = 37.6 million reads, S.D. = 2.07 million per replicates) to ensure the same number of exon-mapped reads per sample was compared between all replicates.
Yet, few overall differences were observed between the polyA and ribo-depleted library preparations in gene quantification and detection of differentially expressed genes. Both data sets were evaluated using alignments from STAR and DEG calculations from limma-voom38, and surrogate variable analysis (SVA) was applied for the detection of latent variables (Online Methods)39. A pairwise comparison of the average normalized gene expression across replicates of the two library types for the four standard samples showed high Spearman correlation coefficients (sample A: 0.91, B: 0.93, C: 0.92, D: 0.93). The overall numbers of DEGs detected between the biologically distinct samples (A vs. B, A vs. D, etc.) were also consistent between library preparation methods (Fig. 5b, 5c). These DEG data were then compared to results from 802 TaqMan assays for these same RNA samples (GEO dataset GSE5350)10. Both library types had similar accuracy as measured by Matthews correlation coefficient (MCC, Fig. 5d)40, 41, which is a joint measure of the assay’s sensitivity and specificity. The corresponding DEGs without SVA analysis show similar but slightly lower overlap percentage and MCC (Supplementary Fig. 33). The median MCC is 0.659 before SVA and 0.678 after SVA, with an average increase of 0.015. Also, the percentage of shared DEGs ranges from 67– 81% at FDR < 0.01 and fold-change > 2, and similarly ranges from 68–81% after SVA. However, the synthetic RNAs spiked into these samples (ERCC controls) performed slightly better in the ribo-depletion protocol than the polyA-enrichment protocol (mean R2 = 0.91 and 0.82, respectively), although these ranges of correlation to TaqMan were similar to that observed for ERCCs sequenced on the PGM, where the mean R2 = 0.78 (Supplementary Figs. 34, 35).
Impact of RNA degradation on transcriptome profiling
As polyA and ribo-depleted gene quantifications were similar, we sought to test the effect of ribo-depletion on “low quality” or degraded RNAs. The reference samples A and B were degraded using heat, sonication or RNase-A until all samples showed a high level of degradation when evaluated on the Agilent Bioanlyzer 2100 (RIN≤2.0, Online Methods). Samples were ribo-depleted before library preparation and sequenced on the HiSeq platform at multiple sites. Multiple metrics indicated that the degraded RNA performed as well as the polyA-enriched or ribo-depleted libraries from intact RNA. First, sequencing of the degraded RNA, after ribo-depletion, fully covered the gene bodies (Fig. 2) and, similar to ribo-depleted libraries from intact RNA, more reads mapped to intronic areas of the genome (Supplementary Fig. 32). Second, the degraded RNA showed minor differences in gene detection or DEG accuracy, with high Spearman correlation (R2 >0.96, Fig. 3c) in expression comparisons to intact RNA samples. In addition, a comparison to the orthogonal PrimePCR dataset showed that the degraded RNA analysis was highly correlated (Pearson R2 >0.83) to the corresponding intact samples (Supplementary Table 4). However, the degraded RNA did have a lower Spearman rank-order correlation with quantitative PCR for the expression differences detected between samples A and B. The Spearman correlation was highest for heat degradation (R2 = 0.83, AH), followed by RNase A (R2= 0.79, AR), and then sonication (R2=0.74, AS) (Supplementary Figs. 36a–c). Comparison of the results from one degraded sample to the results from one intact sample, repeated at multiple laboratories (sites L, V and R), also produced an overall high average Spearman correlation coefficient (0.80, Supplementary Fig. 36d). These data indicate that although appropriate library preparation of degraded RNA can produce accurate expression measurements (Supplementary Fig. 36), but mixing intact and degraded samples (or samples degraded during different types of tissue processing) should be avoided.
Discussion
This ABRF-NGS Study represents, to our knowledge, the largest reported cross-platform, cross-protocol, and cross-site examination of RNA-seq data performed to date. The results provide a unique opportunity to examine various aspects of the transcriptome, including the intra- and inter-site coefficients of variance of gene detection, gene expression quantification and RNA splicing between sequencing platforms, as well as the ability of long read lengths to enable complete isoform characterization. Comparisons of platform-specific aligners with STAR showed that mapping rates, error rates and transcript coverage are larger concerns when considering inter-platform data than is gene quantification. As such, the use of different alignment algorithms will have different influences on comparisons between experiments depending on the metric studied, and the importance of ‘bioinformatics noise’ can be placed alongside technical and biological noise as key factors in experimental design. Finally, the results expanded previous work26 by showing that gene detection and quantification with highly fragmented or degraded RNA samples (from three types of degradation) is strikingly similar to intact RNA, once ribosomal RNA is removed.
This study found similar RNA-seq results between the various NGS platforms and similar ranges in coefficients of variance across lab sites for each platform. These results indicate that both long- and short-read technologies measure gene expression with similar levels of statistical variation, although they show a ten-fold variation for error rates in indels. Using normalized gene expression as a comparison measure, we found high intra-platform consistency (R2>0.86) and high inter-platform concordance (R2> 0.83) measured by Spearman rank correlation (Fig. 3b). However, the results clearly show that deeper sequencing of the transcriptome is needed to reveal low abundance transcripts and splice junctions, indicating that read depth should be a key consideration when experimental goals include rarely expressed genes, coverage of introns and non-polyadenylated targets. Very deep sampling is not currently cost-effective with long-read platforms such as PacBio or 454 (Table 1), and thus the best discovery platforms for low-abundance targets are currently the shorter read platforms, as they can cover wider dynamic range of RNA molecules (i.e., generate more reads per sample).
Table 1.
Summary of RNA-seq platforms as deployed in the ABRF-NGS Study.
| Vendor | Instrument | Version | Run Time (hours) |
Read Length (mean) |
Reads per Run (million) |
Yield per Run |
Cost per Run ($)1 |
Cost per Mb ($) |
Paired- end |
Application | Strengths |
|---|---|---|---|---|---|---|---|---|---|---|---|
| lllumina | HiSeq 2000 | High Output | 132 | 50 | 6,000 | 300 Gb2 | 18,725.00 | 0.06 | Yes | Gene expression; splice junction detection; variant calling; fusions |
Deep read counts for transcript quantification |
| lllumina | HiSeq 2500 | High Output | 132 | 50 | 6,000 | 300 Gb2 | 18,725.00 | 0.06 | Yes | as above | as above |
| lllumina | MiSeq | v2 kit | 39 | 250 | 30 | 7.5 Gb | 982.75 | 0.13 | Yes | Splice junction detection; variant calling |
Rapid transcript quantification and variant detection |
|
| |||||||||||
| Life Technologies |
PGM | 318 chip | 7.3 | 176 | 6 | 1.056 Gb | 749.00 | 0.71 | No | Splice junction detection; variant calling |
Rapid transcript quantification and variant detection |
| Life Technologies |
Proton | Proton I chip | 2 to 4 | 81 | 70 | 5.67 Gb | 834.00 | 0.15 | No | Gene expression; variant calling |
Good read depth and length for transcript quantification |
|
| |||||||||||
| Pacific Biosciences |
RS | RS | 0.5 to 2 | 1,289 | 0.03 | 38.67 Mb | 136.38 | 3.53 | No | Splice junction detection; full length gene coverage |
Extended read lengths |
|
| |||||||||||
| Roche | 454 | Genome Sequencer FLX+ |
20 | 686 | 1 | 686 Mb | 5,985.00 | 8.72 | No | Splice junction detection | Read length |
sequencing reaction reagents, at academic list price U.S. dollars, and does not include library preparation reagents, labor, data storage or analysis, equipment or maintenance;
one sequencing run using 2 flowcells. Pacific Biosciences is calculated based on CCS reads; Gb: billion bases, Mb: million bases
Despite lower read depths and higher costs, the longer read NGS technologies have the best ability to efficiently catch the vast majority of known splice junctions (Fig. 3d–g), indicating that they can be an effective means to annotate splicing complexity. The ABRF-NGS Study’s results include a wealth of putative novel splice junctions, with more than one million such junctions observed in at least one platform (Gene Expression Omnibus GSE46876). These putative novel splice junctions displayed greater inter-platform disparity than the known splice junctions (Fig. 3e). This difference was likely due to the challenge of correctly predicting novel isoforms and also to the possibly high false-positive rate of such predictions, which is expected given their lower expression levels. However, a substantial number of the previously unannotated, predicted junctions are likely genuine, as they were observed using multiple platforms. The resulting data sets nearly double the catalog of splice junctions for these RNA standard samples. The junctions discovered on multiple platforms can be used alone or with previous data for future algorithm design and assay optimization, and as positive controls, to advance splicing isoform characterization by RNA-seq14, 42-44.
Perhaps most notably, the data demonstrated that results from polyA-enriched and ribo-depleted RNA libraries, and even libraries made from severely degraded RNAs, are comparable. Given sufficient depth of sequencing, results from ribo-depleted libraries can include almost all of the differentially detected genes identified by the polyA preparation method, without loss of sensitivity or specificity. This was evident not only in the overlap of DEGs, but also in comparisons to TaqMan and PrimePCR data. Furthermore, a near-complete reconstruction of the transcriptome profile was observed when using degraded RNA in the ribo-depletion protocol, with some variation between degradation treatments, as judged by correlations to the expression abundances measured in intact samples A and B by quantitative PCR and by the uniform coverage of full transcript lengths. Similar degraded RNA results were recently reported26, suggesting that low quality samples can now be considered for reliable RNA-seq expression profiling. This should support studies using old, degraded or fragmented RNAs, such as those from formalin-fixed, paraffin-embedded (FFPE) tissues in clinical archives. Although the degraded RNA samples were run only on the HiSeq platform, the clear utility of such an approach should spur the development of similar degraded RNA resources for analyses on all sequencing platforms.
However, despite their overall similarities, distinct transcriptomes are represented in libraries prepared by polyA enrichment, ribo-depletion or combined polyA and 5′G cap enrichment. The dual enrichment method for PacBio libraries provided superior 5′ to 3′ coverage of the sequenced transcripts, as illustrated by comparisons across platforms for genes consistently detected by PacBio (Supplementary Fig. 37). The revised version of the Illumina library kit (v2 vs. v3) includes built-in ribo-depletion and tags cDNA strand orientation, and the two protocols produced differences in gene-body coverage. A comparison of polyA and ribo-depleted libraries showed different detection of nonpolyadenylated transcripts, 3′UTRs and introns. The former is an intentional consequence of the enrichment protocol, but it is not clear if the 3′UTR coverage bias is due to different efficiencies of priming during reverse transcription or to skewed sampling caused by a higher concentration of structural and non-coding RNAs in ribo-depleted libraries. Owing to the higher rate of intron-mapped reads, RNA-seq of total RNA will require greater read depths for ribo-depleted libraries (~2.5X) than for polyA libraries to achieve equal coverage of exons. Transcriptome measurement variations demonstrated between the reference datasets are easily avoided by consistently using the same protocols, platform, and analysis pipeline for all samples in an experiment. Nonetheless, if this is not possible, surrogate variable analysis enabled removal of latent variables from the data for ribo-depleted and polyA-enriched libraries, producing nearly indistinguishable lists of DEGs and illustrating the utility of surrogate variable analysis as a powerful and strongly recommended method for ameliorating the effects of inter-batch and cross-protocol noise.
The results presented here also highlight additional variables that should be considered when aligning library protocols and platforms with research goals. The reported QV values of all platforms are all higher than empirically derived error rates, indicating that a splicing-aware, base quality score recalibration may be needed for RNA-seq, as is already done for DNA-seq with GATK. Long-read sequencing effectively cataloged splicing isoforms, but had less dynamic range for transcript quantitation and discovery due to lower read depths. The use of the ERCCs is generally recommended as a good QC metric check, but these standards performed better in ribo-depleted libraries than in polyA libraries, and this should also be considered during experimental design. In summary, the priorities for biological interpretation are essential when considering the protocols and methods that will be used in an RNA-seq experiment. Some of these priorities are summarized in Table 1, which provides a cross platform summary of the strengths and relative costs of the sequencing technologies included in this study.
The ABRF-NGS Study is not intended to be a “bake-off” between NGS platforms, but rather is an effort to establish a useful reference data set for each platform which will assist laboratories in improving their methods and in evaluating new chemistries, protocols and instruments. It is encouraging that comparison of gene expression quantification, including results from intra-platform, inter-platform, inter-protocol and even inter-aligner comparisons, demonstrated high correlations overall. This result suggests broader inter-study analyses and data mining can be successfully carried out across multiple platforms despite intrinsic differences between technologies, methods and aligners.
Reference data resources, such as the results from this ABRF-NGS Study, are key to understanding the effects of variable sample quality, changes to platform protocols and the adoption of new technologies. Given the rapid pace of advancement in sequencing technologies, techniques and bioinformatics tools, the methods and data described here can facilitate the development of best practices for gene quantification, isoform characterization, dynamic range comparisons, managing inter-site and intra-site variation, analysis pipeline refinement, and cross-platform testing of transcriptome hypotheses. These data can also be used to address other aspects of RNA-seq, including polymorphism detection, allele-specific expression, intron retention, RNA editing and gene fusions, and provide an immediately useful resource that can complement current databases, such as the RNA-Seq Atlas45. These and other applications, especially clinical molecular diagnostics that rely on nucleic acid biomarkers, will require a level of technical stability across time and both within and between studies, which this study helps to establish. Reference data resources are key to monitoring platform stability, and widespread adoption of standard samples and routine reference benchmarking are challenges that must be addressed to advance genomics technologies further.
Supplementary Material
Table 2.
Sequencing platforms, chemistries and library preparations.
| Illumina Hiseq2000/2500 and MiSeq | ||||||
|---|---|---|---|---|---|---|
| Labs | Samples | Libraries per Sample |
Preparation | Read Length | Number of Reads per Library |
Output (Mb) |
| 1(L) | MAQC A 1 | 3 | Ribo-depleted | 100 bp (2 x 50) | 386,726,967 | 38,673 |
| MAQC B 1 | 3 | Ribo-depleted | 100 bp (2 x 50) | 251,724,566 | 25,172 | |
|
| ||||||
| 2(R) | MAQC A 1 | 3 | Ribo-depleted | 100 bp (2 x 50) | 229,131,233 | 22,913 |
| MAQC B 1 | 3 | Ribo-depleted | 100 bp (2 x 50) | 229,591,730 | 22,959 | |
| MAQC B 1 | 1 | Ribo-depleted | 500 bp (2 x 250) | 7,848,217 | 3,924 | |
|
| ||||||
| 3(V) | MAQC A 1 | 3 | Ribo-depleted | 100 bp (2 x 50) | 207,603,620 | 20,760 |
| MAQC B 1 | 3 | Ribo-depleted | 100 bp (2 x 50) | 239,930,780 | 23,993 | |
|
| ||||||
| 4(N) | MAQC A 1,3 | 2 | Ribo-depleted | 100 bp (2 x 50) | 215,903,801 | 21,590 |
| MAQC B 1,3 | 3 | Ribo-depleted | 100 bp (2 x 50) | 219,257,190 | 21,926 | |
| MAQC A 1,4 | 1 | Ribo-depleted | 100 bp (2 x 50) | 183,811,383 | 18,381 | |
|
| ||||||
| 5(M) | MAQC A 1,4 | 3 | Ribo-depleted | 100 bp (2 x 50) | 386,726,967 | 38,673 |
| MAQC A 1,5 | 3 | Ribo-depleted | 100 bp (2 x 50) | 181,740,643 | 18,174 | |
|
| ||||||
| 6(W) | MAQC A 2 | 4 | Ribo-depleted | 100 bp (2 x 50) | 128,133,887 | 12,813 |
| MAQC B 2 | 4 | Ribo-depleted | 100 bp (2 x 50) | 137,096,343 | 13,710 | |
| MAQC C 2 | 4 | Ribo-depleted | 100 bp (2 x 50) | 142,135,538 | 14,214 | |
| MAQC D 2 | 4 | Ribo-depleted | 100 bp (2 x 50) | 128,040,437 | 12,804 | |
| MAQC A 2 | 4 | polyA-enriched | 100 bp (2 x 50) | 106,762,840 | 10,676 | |
| MAQC B 2 | 4 | polyA-enriched | 100 bp (2 x 50) | 111,430,017 | 11,143 | |
| MAQC C 2 | 4 | polyA-enriched | 100 bp (2 x 50) | 108,582,900 | 10,858 | |
| MAQC D 2 | 4 | polyA-enriched | 100 bp (2 x 50) | 105,978,082 | 10,598 | |
| Life Technologies Ion Torrent PGM and Proton | ||||||
|---|---|---|---|---|---|---|
| Labs | Samples6 | Libraries per Sample |
Preparation7 | Median Read Length |
Mean Number of Reads |
Output (Mb) |
| 1(P) Ion PGM |
MAQC A | 2 | polyA-enriched | 161 | 5,323,672 | 857 |
| MAQC B | 2 | polyA-enriched | 184 | 5,802,563 | 107 | |
| ERCC 1 | 1 | polyA-enriched | 189 | 4,188,385 | 792 | |
| ERCC 2 | 1 | polyA-enriched | 158 | 3,231,475 | 511 | |
| ERCC 1 | 1 | polyA-enriched | 180 | 4,442,093 | 800 | |
| ERCC 2 | 1 | polyA-enriched | 189 | 4,310,663 | 815 | |
|
| ||||||
| 2(H) Ion PGM |
MAQC A | 3 | polyA-enriched | 128 | 3,374,068 | 445 |
| MAQC B | 3 | polyA-enriched | 129 | 3,409,662 | 436 | |
| ERCC 1 | 2 | polyA-enriched | 152 | 2,538,594 | 810 | |
| ERCC 2 | 2 | polyA-enriched | 112 | 2,119,884 | 514 | |
| VL A | 1 | polyA-enriched | 187 | 3,965,022 | 770 | |
| VL B | 1 | polyA-enriched | 162 | 4,138,326 | 687 | |
|
| ||||||
| 3(S) Ion PGM |
MAQC A | 3 | polyA-enriched | 198 | 5,049,998 | 1,000 |
| MAQC B | 3 | polyA-enriched | 199 | 5,743,028 | 1,140 | |
| ERCC 1 | 1 | polyA-enriched | 206 | 6,835,287 | 1,410 | |
| ERCC 2 | 2 | polyA-enriched | 207 | 7,119,023 | 1,480 | |
| ERCC 1 | 1 | polyA-enriched | 182 | 6,525,478 | 1,190 | |
| ERCC 2 | 1 | polyA-enriched | 191 | 5,490,495 | 1,050 | |
|
| ||||||
| 4(S) Proton | MAQC A | 3 | polyA-enriched | 78 | 50,063,784 | 3,900 |
| MAQC B | 3 | polyA-enriched | 85 | 53,203,028 | 4,497 | |
|
| ||||||
| 5(B) Proton | MAQC A | 1 | polyA-enriched | 95 | 57,701,947 | 4,864 |
| MAQC B | 1 | polyA-enriched | 75 | 39,099,605 | 2,946 | |
| MAQC C | 1 | polyA-enriched | 64 | 41,308,206 | 6412,641 | |
| MAQC D | 1 | polyA-enriched | 53 | 46,665,851 | 3,160 | |
|
| ||||||
| 6(L) Proton | MAQC A | 1 | polyA-enriched | 99 | 60,106,614 | 5,978 |
| MAQC B | 1 | polyA-enriched | 100 | 60,769,231 | 6,085 | |
| MAQC C | 1 | polyA-enriched | 107 | 60,353,696 | 6,454 | |
| MAQC D | 1 | polyA-enriched | 106 | 69,977,984 | 7,413 | |
| Pacific Biosciences RS | ||||||
|---|---|---|---|---|---|---|
| Labs | Samples | Libraries per Sample: Size Fractionation |
Preparation8 | Avg. Read Length |
Reads/Mb | Output (Mb) |
| 1(A) | MAQC A | 1: >3 kb | polyA + 5'G cap | 3,983 | 251 | 663 |
| MAQC A | 1: 2-3 kb | polyA + 5'G cap | 3,513 | 284 | 520 | |
| MAQC A | 1: 1-2 kb | polyA + 5'G cap | 2,811 | 356 | 780 | |
| MAQC B | 1: >3 kb | polyA + 5'G cap | 3,467 | 288 | 536 | |
| MAQC B | 1: 2-3 kb | polyA + 5'G cap | 3,223 | 310 | 459 | |
| MAQC B | 1: 1-2 kb | polyA + 5'G cap | 3,112 | 321 | 638 | |
|
| ||||||
| 2(F) | MAQC A | 1: >3 kb | polyA + 5'G cap | 3,472 | 288 | 634 |
| MAQC A | 1: 2-3 kb | polyA + 5'G cap | 3,644 | 274 | 555 | |
| MAQC A | 1: 1-2 kb | polyA + 5'G cap | 2,792 | 358 | 927 | |
| MAQC A | 1: unfractionated | polyA + 5'G cap | 2,832 | 353 | 260 | |
| MAQC B | 1: >3 kb | polyA + 5'G cap | 3,578 | 280 | 667 | |
| MAQC B | 1: 2-3 kb | polyA + 5'G cap | 3,523 | 284 | 594 | |
| MAQC B | 1: 1-2 kb | polyA + 5'G cap | 2,844 | 351 | 991 | |
| MAQC B | 1: unfractionated | polyA + 5'G cap | 2,814 | 355 | 251 | |
|
| ||||||
| 3(H) | MAQC A | 1: >3 kb | polyA + 5'G cap | 3,201 | 312 | 528 |
| MAQC A | 1: 2-3 kb | polyA + 5'G cap | 3,135 | 319 | 344 | |
| MAQC A | 1: 1-2 kb | polyA + 5'G cap | 2,761 | 362 | 767 | |
| MAQC A | 1: unfractionated | polyA + 5'G cap | 2,998 | 334 | 477 | |
| MAQC B | 1: >3 kb | polyA + 5'G cap | 3,189 | 314 | 572 | |
| MAQC B | 1: 2-3 kb | polyA + 5'G cap | 2,952 | 339 | 383 | |
| MAQC B | 1: 1-2 kb | polyA + 5'G cap | 2,779 | 360 | 660 | |
| MAQC B | 1: unfractionated | polyA + 5'G cap | 3,069 | 326 | 395 | |
| Roche 454 FLX | ||||||
|---|---|---|---|---|---|---|
| Labs | Samples | Libraries per Sample |
Preparation9 | Median Read Length |
Total Reads per PicoTiterPlate |
Output (Mb) |
| 1(I) | MAQC A | 1 | polyA-enriched | 520 | 1,061,320 | 552 |
| MAQC B | 1 | polyA-enriched | 494 | 1,001,678 | 495 | |
| MAQC A | 1 | polyA-enriched | 497 | 805,399 | 400 | |
| MAQC B | 1 | polyA-enriched | 496 | 1,076,634 | 534 | |
|
| ||||||
| 2(P) | MAQC A | 1 | polyA-enriched | 455 | 832,580 | 379 |
| MAQC B | 1 | polyA-enriched | 470 | 1,181,610 | 555 | |
|
| ||||||
| 3(C) | MAQC A | 2 | polyA-enriched | 505 | 1,294,497 | 654 |
| MAQC B | 2 | polyA-enriched | 358 | 293,471 | 105 | |
Illumina RNA TruSeq v2 library kit,
Illumina RNA TruSeq v3 library kit,
RNaseA degraded,
heat degraded,
sonicated
ERCC: synthetic standards only (External RNA Control Consortium); VL: pilot data for sample A or B;
Ion Total RNA-Seq v2 library kit
PacBio Large Insert Template Prep Kit
Roche cDNA Rapid Library kit
Acknowledgements
We greatly appreciate the contribution and distribution of reference sample RNA from Leming Shi (FDA) and his valuable interactions to assist in the planning of this study. This work was supported with funding from the National Institutes of Health (NIH), including R01HG006798, R01NS076465, R24RR032341, as well as funds from the Irma T. Hirschl and Monique Weill-Caulier Charitable Trusts and the STARR Consortium (I7-A765).
We thank the following contributors for their technical wisdom, including laboratory expertise, data analysis and bioinformatics contributions, and technical design guidance and consultation. Without their help, this study would not have been possible: Diane Stopka (Memorial Sloan-Kettering Cancer Institute), Gregory Grove (Penn State Univ.), Daniel Hannon (Penn State Univ.), Kristine Jones (NIH/NCI/SAIC), Castle Raley (NIH/NCI/SAIC), Henriette O’Geen (UC Davis), Danman Zheng (Univ. Illinois-Urbana), Oanh Nguyen (UC Davis), Zhi-Wei Lu (UC Davis), Jezra Spisak (Cornell Univ.), Dawei Lin (NIH/NIAID), Jaroslaw Pillardy (Cornell Univ.), Po-Yen Wu (Georgia Institute of Technology), John Phan (Emory Univ.), Dayna Oschwald (New York Genome Center), Hugh Arnold (Perkin Elmer), Selene Tyndale (Univ. Southern California), Helen Truong (Univ. Southern California), Yanping Zhang (Univ. Florida), Nedka Panayotova (Univ. Florida), David Moraga (Univ. Florida), Savita Shanker (Univ. Florida), and Natalie Barker (U.S. Army Environmental Quality Research Program).
We would also like to thank the platform vendors, Illumina, Life Technologies, Pacific Biosciences and Roche Life Sciences, for their support of this study, and their distinguished scientists for providing technical expertise and assistance in study designs, protocols, new methods development and significant contributions of reagents and sequencing kits. In particular, alphabetically by vendor: Gary Schroth (Illumina); Michael Gallad, Jeff Smith, Tom Bittick and Robert Setterquist (Life Technologies); Jonas Korlach, Steve Turner and Elizabeth Tseng (Pacific Biosciences); and Karin Fredrickson and Clotilde Teiling (Roche Life Sciences).
We are sincerely appreciative of the Association of Biomolecular Resource Facilities (ABRF) for supporting this study and the contributing ABRF Research Groups. Special thanks to our ABRF Executive Board liaison, Anoja Perera (Stowers Institute for Medical Research), and to Michelle Carr (Cornell Univ.) for administrative support.
Footnotes
Competing Financial Interests The authors declare that they have no relevant competing financial interests.
References
- 1.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagalakshmi U, Waern K, Snyder M. RNA-Seq: a method for comprehensive transcriptome analysis. Curr Protoc Mol Biol. 2010;11:11–13. doi: 10.1002/0471142727.mb0411s89. Chapter 4, Unit 4. [DOI] [PubMed] [Google Scholar]
- 3.Liu S, Lin L, Jiang P, Wang D, Xing Y. A comparison of RNA-Seq and high-density exon array for detecting differential gene expression between closely related species. Nucleic Acids Res. 2011;39:578–588. doi: 10.1093/nar/gkq817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 6.Liu L, et al. Comparison of next-generation sequencing systems. J. Biomed. Biotechnol. 2012;2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ratan A, et al. Comparison of sequencing platforms for single nucleotide variant calls in a human sample. PloS one. 2013;8:e55089. doi: 10.1371/journal.pone.0055089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quail MA, et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loman NJ, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotechnol. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 10.Shi L, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(MAQC-3), S.Q.C.C. Power and limitations of RNA-Seq. Nature biotechnology. 2014 [Google Scholar]
- 12.t Hoen PA, et al. Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nature biotechnology. 2013 doi: 10.1038/nbt.2702. [DOI] [PubMed] [Google Scholar]
- 13.Tarazona S, Garcia-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNAseq: a matter of depth. Genome research. 2011;21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labaj PP, et al. Characterization and improvement of RNA-Seq precision in quantitative transcript expression profiling. Bioinformatics. 2011;27:i383–391. doi: 10.1093/bioinformatics/btr247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McIntyre LM, et al. RNA-seq: technical variability and sampling. BMC Genomics. 2011;12:293. doi: 10.1186/1471-2164-12-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang R, et al. An RNA-Seq strategy to detect the complete coding and non-coding transcriptome including full-length imprinted macro ncRNAs. PloS one. 2011;6:e27288. doi: 10.1371/journal.pone.0027288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toung JM, Morley M, Li M, Cheung VG. RNA-sequence analysis of human B-cells. Genome Res. 2011;21:991–998. doi: 10.1101/gr.116335.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 21.Angeletti RH, et al. Research technologies: fulfilling the promise. Faseb J. 1999;13:595–601. doi: 10.1096/fasebj.13.6.595. [DOI] [PubMed] [Google Scholar]
- 22.Moelans CB, Oostenrijk D, Moons MJ, van Diest PJ. Formaldehyde substitute fixatives: effects on nucleic acid preservation. J Clin Pathol. 2011;64:960–967. doi: 10.1136/jclinpath-2011-200152. [DOI] [PubMed] [Google Scholar]
- 23.Opitz L, et al. Impact of RNA degradation on gene expression profiling. BMC Med Genomics. 2010;3:36. doi: 10.1186/1755-8794-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morlan JD, Qu K, Sinicropi DV. Selective depletion of rRNA enables whole transcriptome profiling of archival fixed tissue. PloS one. 2012;7:e42882. doi: 10.1371/journal.pone.0042882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pareek CS, Smoczynski R, Tretyn A. Sequencing technologies and genome sequencing. J Appl Genet. 2011;52:413–435. doi: 10.1007/s13353-011-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adiconis X, et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat Methods. 2013;10:623–629. doi: 10.1038/nmeth.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boland JF, et al. The new sequencer on the block: comparison of Life Technology’s Proton sequencer to an Illumina HiSeq for whole-exome sequencing. Human genetics. 2013 doi: 10.1007/s00439-013-1321-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glenn TC. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 2011;11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 29.Jiang L, et al. Synthetic spike-in standards for RNA-seq experiments. Genome research. 2011;21:1543–1551. doi: 10.1101/gr.121095.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zook JM, Samarov D, McDaniel J, Sen SK, Salit M. Synthetic spike-in standards improve runspecific systematic error analysis for DNA and RNA sequencing. PloS one. 2012;7:e41356. doi: 10.1371/journal.pone.0041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 2010;38:e131. doi: 10.1093/nar/gkq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aird D, et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Risso D, Schwartz K, Sherlock G, Dudoit S. GC-content normalization for RNA-Seq data. BMC bioinformatics. 2011;12:480. doi: 10.1186/1471-2105-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genomes Project C, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing nextgeneration DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharon D, Tilgner H, Grubert F, Snyder M. A single-molecule long-read survey of the human transcriptome. Nat Biotechnol. 2013 Nov;31(11):1009–14. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smyth GK. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 39.Cui P, et al. A comparison between ribo-minus RNA-sequencing and polyA-selected RNAsequencing. Genomics. 2010;96:259–265. doi: 10.1016/j.ygeno.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 40.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baldi P, Brunak S, Chauvin Y, Andersen CA, Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- 42.Shi L, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nature biotechnology. 2010;28:827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haas BJ, Zody MC. Advancing RNA-Seq analysis. Nature biotechnology. 2010;28:421–423. doi: 10.1038/nbt0510-421. [DOI] [PubMed] [Google Scholar]
- 44.Wenger Y, Galliot B. RNAseq versus genome-predicted transcriptomes: a large population of novel transcripts identified in an Illumina-454 Hydra transcriptome. BMC Genomics. 2013;14:204. doi: 10.1186/1471-2164-14-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pipes L, et al. The non-human primate reference transcriptome resource (NHPRTR) for comparative functional genomics. Nucleic acids research. 2013;41:D906–914. doi: 10.1093/nar/gks1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krupp M, et al. RNA-Seq Atlas--a reference database for gene expression profiling in normal tissue by next-generation sequencing. Bioinformatics. 2012;28:1184–1185. doi: 10.1093/bioinformatics/bts084. [DOI] [PubMed] [Google Scholar]
- 47.Van Peer G, Mestdagh P, Vandesompele J. Accurate RT-qPCR gene expression analysis on cell culture lysates. Sci. Rep. 2012;2:222. doi: 10.1038/srep00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative realtime PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 50.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23:2881–2887. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- 52.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9:321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 53.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 55.Canales RD, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat. Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 56.Dvinge H, Bertone P. HTqPCR: high-throughput analysis and visualization of quantitative realtime PCR data in R. Bioinformatics. 2009;25:3325–3326. doi: 10.1093/bioinformatics/btp578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.