Abstract
Accurately quantifying Legionella for regulatory purposes to protect public health is essential. Real-time PCR (qPCR) has been proposed as a better method for detecting and enumerating Legionella in samples than conventional culture method. However, since qPCR amplifies any target DNA in the sample, the technique’s inability to discriminate between live and dead cells means that counts are generally significantly overestimated. Propidium monoazide (PMA) has been used successfully in qPCR to aid live/dead discrimination. We tested PMA use as a method to count only live Legionella cells in samples collected from a modified chemostat that generates environmentally comparable samples. Counts from PMA-treated samples that were pretreated with either heat or three types of disinfectants (to kill the cells) were highly variable, with the only consistent trend being the relationship between biofilm mass and numbers of Legionella cells. Two possibilities explain this result: 1. PMA treatment worked and the subsequent muted response of Legionella to disinfection treatment is a factor of biofilm/microbiological effects; although this does not account for the relationship between the amount of biofilm sampled and the viable Legionella count as determined by PMA-qPCR; or 2. PMA treatment did not work, and any measured decrease or increase in detectable Legionella is because of other factors affecting the method. This is the most likely explanation for our results, suggesting that higher concentrations of PMA might be needed to compensate for the presence of other compounds in an environmental sample or that lower amounts of biofilm need to be sampled. As PMA becomes increasingly toxic at higher concentrations and is very expensive, augmenting the method to include higher PMA concentrations is both counterproductive and cost prohibitive. Conversely, if smaller volumes of biofilm are used, the reproducibility of the method is reduced. Our results suggest that using PMA is not an appropriate method for discriminating between live and dead cells to enumerate Legionella for regulatory purposes.
Keywords: Legionella, biofilm, qPCR, PMA, cooling tower
Introduction
Current and historical detection methods for Legionella from environmental sources have focused on culture as a standard method.1–6 Because of the complex nature of Legionella’s ecological niche, isolation and detection requires pretreatment of environmental samples before conventional culture methods can be used. These procedures tend to remove most competitive microorganisms, but often leave sporulating bacteria that multiply more rapidly than Legionella. This often renders identification and subculture difficult and enumeration of viable Legionella nearly impossible, particularly if motile organisms are present. Although successful isolation and detection by culture can be improved by passage through amoeba susceptible to Legionella parasitization, enumeration is not possible using this method.7 Detection of Legionella from industrial water samples is further confounded as the presence of disinfectants and other water treatment chemicals may render Legionella viable but not cultivable, leading to an unrealistically low number of visible colonies or false negatives, particularly in systems that are treated with monochloramine.8
More recently, PCR has begun to gain prominence as a method capable of detecting Legionella in complex samples. A sample of water is taken and filter concentrated, followed by DNA extraction and PCR amplification to detect the presence of short sequences of DNA originating from Legionella.9–11 Quantitative real-time PCR (qPCR) provides a further refinement of this method and can be used to estimate the number of copies of Legionella DNA present and hence the approximate number of bacteria in the original sample.12,13 Additionally, where it may take up to 7 days to obtain a result using culture-based detection of Legionella, PCR is comparatively rapid, providing results in under a day.
The major problem with PCR for enumeration is that it may result in false positives or unrealistically high estimation of Legionella numbers, as all DNA extracted from cells either live or dead will be amplified.14 The persistence of DNA from nonviable cells in environmental sources ranges from days to weeks depending on the microbial consortium present,15 particularly if a water system is treated with a nonoxidizing biocide. In order to counter the amplification of DNA originating from dead cells there is an increasing, albeit small, body of work demonstrating the use of either ethidium monoazide (EMA) or propidium monoazide (PMA) to selectively inhibit PCR amplification of DNA from dead cells allowing for viability-based discrimination.
EMA is a derivative of the commonly used DNA stain ethidium bromide, with the addition of an azide group allowing covalent bonding to DNA. Similarly, PMA is an azidified derivative of propidium iodide, a dye commonly used in microscopy for cell viability assays, which is also capable of covalently binding to DNA. EMA has been demonstrated to have a higher intrinsic toxicity to some bacteria,16 potentially causing an underestimation of the total viable DNA present in a sample. Because of its higher charge, PMA is less membrane permeable and less inherently toxic than EMA,17 allowing for greater efficiency of cell infiltration and for more accurate estimation of viable cell numbers with the generation of fewer false negatives.
Cells with intact membranes or cell walls are generally capable of excluding DNA-binding dyes, facilitating the use of dye exclusion as a method for microscopically assessing cell viability.18,19 Both EMA and PMA are DNA intercalating compounds, which covalently bind to DNA in the presence of bright visible light.17,20,21 When exposed to bright light, the azide group is converted to a highly reactive nitrene. This nitrene allows covalent bonding between base pairs, a process known as intercalation. Any supplementary unbound EMA or PMA is quenched with water to produce unreactive species, which do not continue to bind to DNA after extraction. This covalent DNA cross-linkage causes a structural change in nucleotide angle, rendering the DNA strands not only unavailable for elongation by polymerase but also insoluble in water causing them to be removed by DNA extraction procedures. By dissolving EMA and PMA in dimethyl sulfoxide (DMSO) to increase their bioavailability, complex cell samples can be efficiently exposed to these agents.16,20,22
To test the efficiency and accuracy of PMA-qPCR in detecting viable Legionella, three common industrial water treatment chemicals were chosen to disinfect samples of biofilms. These three chemicals (sodium hypochlorite, 2,2-dibromo-3-nitrilopropionamide (DBNPA), and isothiazolinone) were chosen because of their different modes of action, differences in their biofilm penetrative potential, and their common employment in reducing microbial load in cooling towers and industrial water systems. It was not the aim of this work to assess relative disinfectant effectiveness, but rather to use the disinfectants as a tool to assess the efficacy of novel molecular approaches for the quantification of Legionella in the built environment. As such, the disinfectants were used for the purpose of killing Legionella within biofilms in order to assess the use of PMA and qPCR to detect and enumerate viable Legionella contained in complex environmental sources.
As we move toward molecular methods as a new standard for detecting Legionella in the built environment, and begin to construct guidelines and risk assessments based on these results, it is imperative that we have an understanding of the limits of these techniques in their current form. We must establish the parameters within which these methods may operate and still return useful results, and refine them to better suit a wider range of circumstances. In order to assess the strength of PMA-qPCR in its current form, it was decided that extensive application to planktonic cells, or cells suspended in water, would be an inadequate measure of its accuracy and effectiveness. Previous work has demonstrated that under ideal conditions, PMA-qPCR was capable of discriminating between live and heat-killed Legionella suspended in water;23 however, to the best of the authors’ knowledge, its application directly on aggregations of biofilms has not been assessed.
Methods
Biofilm growth and harvesting
Biofilms were grown in modified chemostats, which were designed to mimic heat exchangers in their function and operation, and have been previously described in full.24 Each 10-L reservoir was filled with water collected from a cooling tower, and inoculated (20% final volume) with water sourced from a cooling tower associated with an outbreak to ensure that a representative microbial consortium was present. Reservoirs were maintained at 35°C and sparged to provide adequate aeration. Water was pumped at a rate of 1 L per minute through a series of opaque pipes (Reynold’s number <2,000), containing stainless steel coupons, which acted as sampling points for biofilm collection. Systems were continuously operated without further addition of nutrients or microbial inoculum for a period of one month before biofilms were sampled. Samples of biofilms were harvested by manual inversion of piping system 10 times. This was possible as the system was mounted with two rotating pivot points at either end. The total volume of water containing dislodged biofilms required for each set of experiments was then removed from the reservoir into a glass beaker containing a magnetic stirring bar. System water was transferred to 50-mL tubes, while stirring to keep the biofilm evenly suspended. Biofilm samples were vacuum filter concentrated onto preweighed 0.22 μm pore size polysulfonate filters (Millipore™, Kilsyth, VIC, Australia). Samples were filtered for 10 seconds, at which point biofilm was visibly free of water. Filters and biofilms were reweighed and total mass of biofilm determined (total mass—filter mass = mass of biofilm). This mass was then used to estimate the number of Legionella per mg of biofilm.
Legionella pneumophila culture conditions
Legionella cultures were grown on buffered charcoal yeast extract (BCYE) agar, supplemented with BCYE growth supplement, containing ACES buffer/potassium hydroxide, ferric pyrophosphate, L-cysteine HCl and α-ketoglutarate (Oxoid, Adelaide, SA, Australia). Plates were incubated at 35°C in a stationary incubator for 3–5 days until colonies appeared. Where required, cells were harvested by addition of 1 mL sterile water to plates and cells suspended by a cell scraper.
Heat inactivation of cells
Tubes containing samples were placed in a water bath heated to 80°C. Tubes were allowed 2 minutes to reach 80°C, followed by 10 minutes of treatment at 80°C. After treatment, tubes were immediately removed from the water bath and placed in ice to stop further degradation of samples.
Chemical disinfection of biofilms
Tubes containing suspended biofilms were placed in a water bath at 30°C. Samples of biofilms were treated with a series of disinfectants obtained from Hydrochem (Hydrochem, SA, Australia) and applied following the manufacturer’s specifications. Samples were removed from the water bath after 0, 30, 60, 120, and 440 minutes before further sample processing (PMA treatment, DNA extraction, and qPCR). 500 μL 30 mg/mL sodium thiosulfate was added to all samples to quench oxidizing biocides. Untreated biofilm samples were used as controls. Controls were harvested and placed in a water bath alongside treated samples and were treated with sodium thiosulfate in accordance with the described method.
PMA treatment
In order to determine whether PMA selectively inhibits amplification of DNA from unviable cells, water samples containing harvested biofilms or Legionella cultures were treated in two ways. DNA was extracted and amplified from biofilms or Legionella cultures using conventional qPCR and after treatment with PMA. Samples of biofilms were either heat killed or left untreated to assess the capability of PMA to amplify DNA exclusively from viable cells.
PMA (Biotium Australia, SA, Australia) solutions were prepared in 20% DMSO and stored in light shielded bottles at −20°C until required. Following PMA validation experiments (as described below), all further PMA-qPCR was conducted using 100 μM PMA solution.
Filter-bound biofilm samples were placed on aluminum foil covering a bed of ice in order to reduce detrimental impact of heat on cell viability during light treatment. Samples were treated with PMA loaded into an atomizer, which produced a fine mist containing 10–15 μL 100 μM PMA solution. In a fume hood, samples were sprayed with PMA and incubated in the dark for 5 minutes. Samples were then exposed to bright white light (500 W halogen lamp) for 5 minutes at a distance of 30 cm before proceeding with DNA extraction.
Legionella cultures and biofilm samples were heat killed as described or were left un-killed, before treatment (in triplicate) with PMA at four different concentrations (0, 30, 50, and 100 μM). Samples were PMA treated, and DNA extraction and qPCR analysis were performed. Copy numbers of Legionella DNA following qPCR were compared to assess the effect of the three different PMA concentrations compared with controls that received no PMA.
Quantitative real-time PCR enumeration of Legionella
qPCR detection of Legionella was carried out using the iQ-Check™ Quanti Legionella spp. PCR kit (Bio-Rad Laboratories Pty. Ltd., Gladesvilles, NSW, Australia), following methods supplied with the kit to produce a final reaction volume of 25 μL in each tube. PCR reactions were performed in a Corbett RotorGene 3000 real-time thermocycler (QIAGEN Pty Ltd, Victoria, Australia), using the following protocol: 1 cycle at 50°C for 2 minutes and 95°C for 15 minutes, followed by 50 cycles of 95°C for 15 seconds, 56°C for 30 seconds, 72°C for 30 seconds. A final melt was carried out at 72°C for 45 seconds, followed by a 1°C rise in temperature every 5 seconds to a final temperature of 95°C. Amplification data were acquired on the FAM channel (470 nm excitation, 510 nm detection, gain 4), with melt data acquired on the ROX channel (585 nm excitation, 610 nm detection, gain 9.33).
Statistical analyses
Statistical analysis was performed using Statistical Package for Social Sciences (SPSS) version 17.0 for Windows and GraphPad Prism version 5 for Windows. Graphs were prepared using GraphPad Prism. Normality and homogeneity of variance were tested prior to analysis of data. Effects of PMA concentration on detectable Legionella were compared using univariate analysis of variance (ANOVA) and Bonferroni post-hoc analysis to determine whether any significant differences occurred between treatments. Correlations between mass of biofilm sampled and enumerated Legionella were determined by Pearson correlation (rp) (two-tailed).
Results
PMA optimization and validation
Samples of Legionella harvested from pure plate cultures showed extremely high counts of viable cells (1.25 × 109–1.7 × 108), irrespective of heat killing or PMA treatment (Fig. 1). No statistically significant differences were found between results obtained with different PMA concentrations; however, a trend is visible whereby increasing the PMA concentration reduced the amount of DNA amplified from dead cells. The number of detectable Legionella in the sample was at most reduced by approximately 80% when using 100 μM PMA compared with non-PMA treated controls (Fig. 1).
Figure 1.
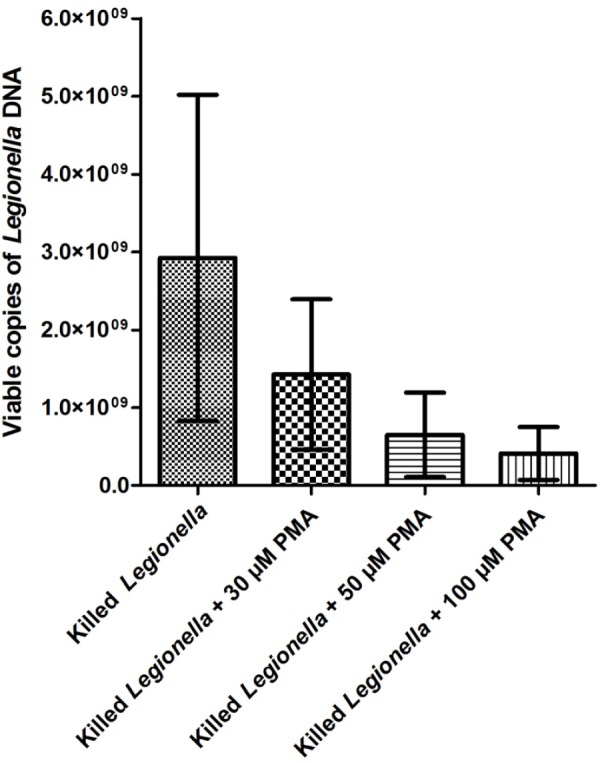
Number of viable Legionella (from pure plate cultures) detectable by PMA-qPCR after heat inactivation.
Results for heat-killed biofilm samples are shown in Figure 2. Samples of heat-killed biofilms all showed high variability in detected Legionella number, with no significant differences seen between treatments. Samples treated with PMA showed a decrease in the number of detected viable Legionella; however, because of the variability between samples, no clear trend is apparent between the different concentrations tested. Similar to heat-killed Legionella cells, biofilm samples showed roughly a maximum of 70% reduction in the number of detectable Legionella after PMA treatment.
Figure 2.
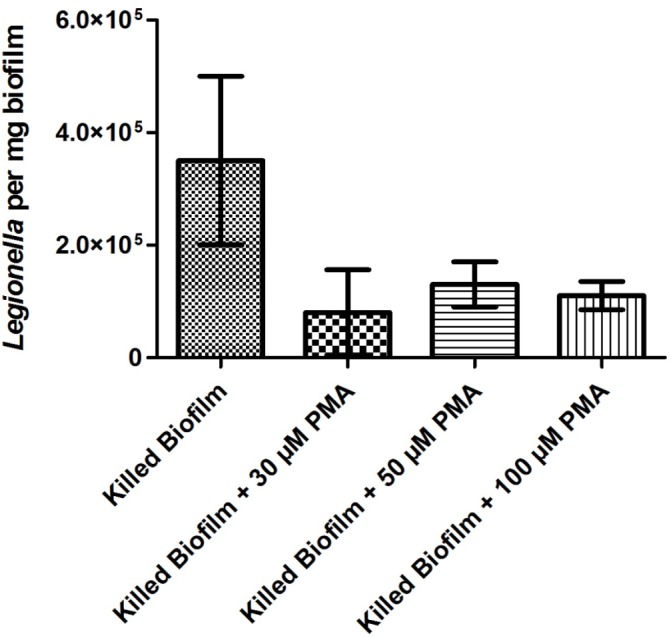
Number of viable Legionella in biofilms detectable by PMA-qPCR after heat inactivation.
Validation of PMA-qPCR for use on biocide-treated biofilm samples
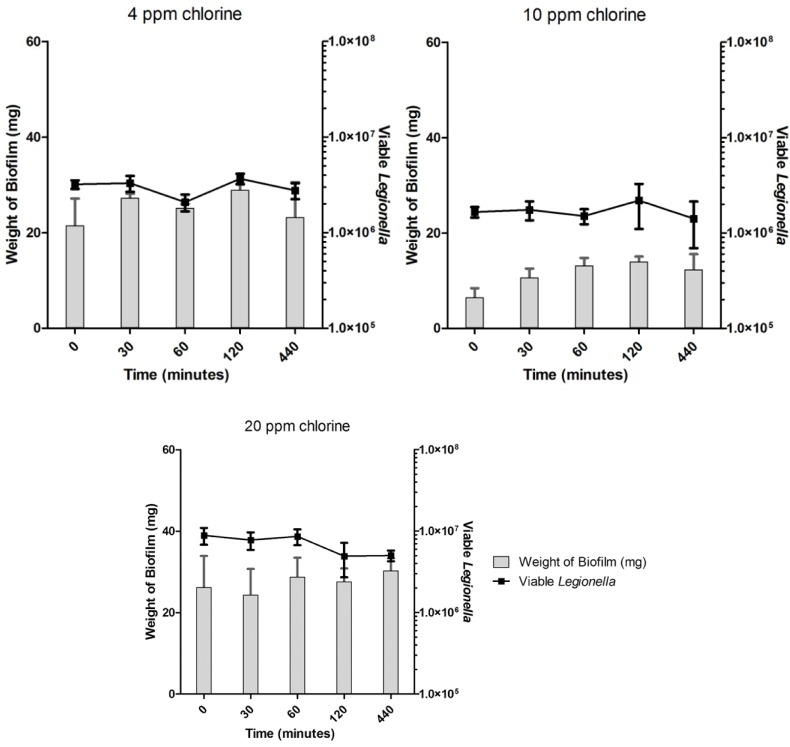
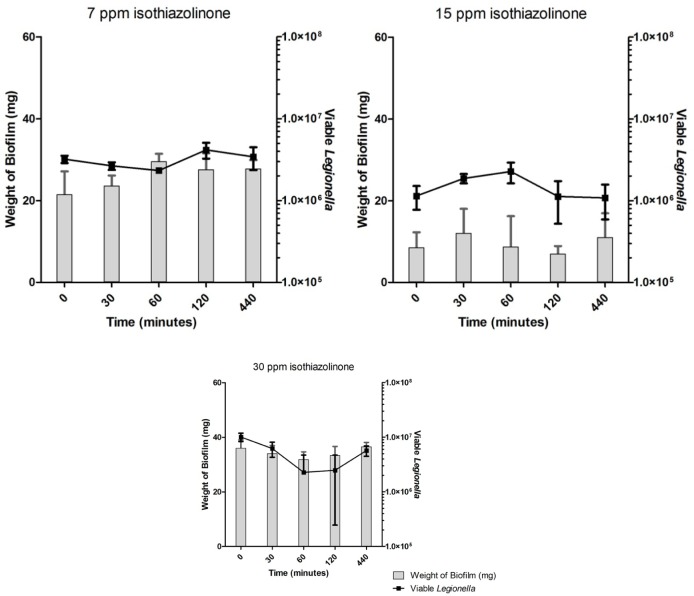
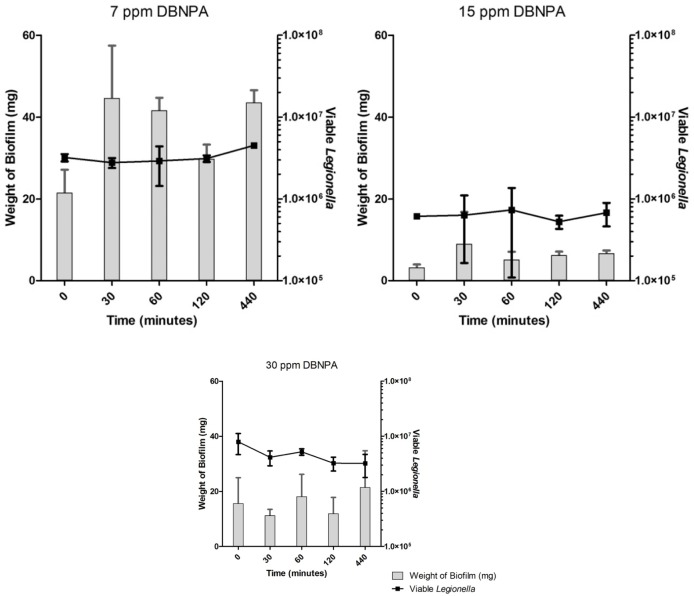
Untreated biofilms showed no decrease in the number of enumerated Legionella over the time-course of the experiments (Fig. 3). Disinfectant-treated biofilms showed no significant differences in the number of viable Legionella in comparison with untreated controls under any treatment regime (Figs. 4–6). Across all samples, the number of viable Legionella displayed a wide variability, with no clear trends apparent in relation to the concentration of disinfectant.
Figure 3.
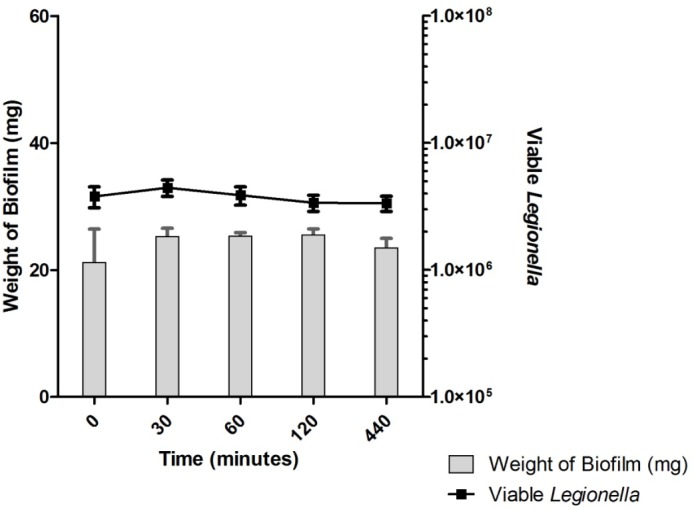
Representative example of an untreated biofilm control. The number of viable Legionella showed no significant change over a period of 440 minutes.
Figure 4.
Enumerated Legionella in biofilms after chlorine treatment.
Figure 6.
Enumerated Legionella in biofilms after isothiazolinone treatment.
There was a clear relationship between the amount of biofilms used for the sample and the number of detected viable Legionella, irrespective of disinfection or concentration used. All data obtained from qPCR and accompanying biofilm mass were pooled (N = 133). A significant positive correlation was found between the mass of biofilm and the number of copies of Legionella DNA in the sample (rp = 0.469; p < 0.001). This result is represented graphically in Figure 7. Data show some heteroscedasticity as the amount of biofilm in the sample increases.
Figure 7.
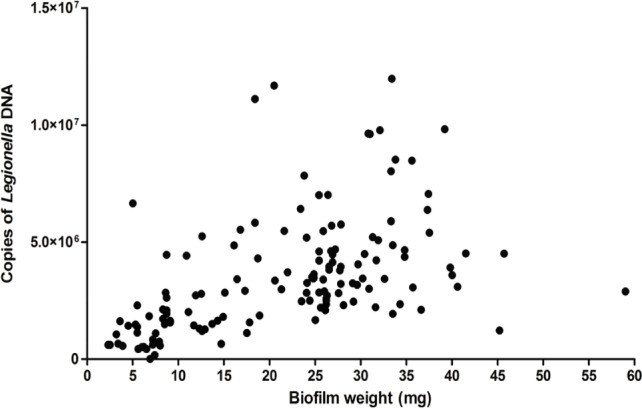
Pooled number of Legionella detected by qPCR vs. biofilm mass across all treatment combinations (N = 133).
Discussion
PMA-qPCR as a method for enumerating viable Legionella in complex biofilm samples
PMA treatment of biofilm samples coupled with qPCR (PMA-qPCR) proved to be an ineffective method for selectively enumerating viable Legionella. It was expected that increasing doses of biocides would generate a characteristic dose–response effect; however, in some cases (30 ppm isothiazolinone—Fig. 6), the greatest concentration of disinfectants showed increases in viability.
If disinfectant treatments significantly reduced cell viability, or if PMA was capable of discriminately removing DNA originating from dead cells, then the number of enumerated Legionella should have decreased over time in response to chemical disinfection. Furthermore, this relationship should have occurred independent of fluctuations in the amount of biofilm used for each sample.
Across all disinfection treatments, the number of overall detectable, viable Legionella was reduced by approximately 40–60% (Figs. 4–6) compared to untreated controls (Fig. 3), although fluctuations in the biofilm content of each sample and the number of detected Legionella make this trend more visible in heat-killed biofilm samples, where a maximum of 70% kill was achieved (Figs. 1 and 2). A similar relative decrease in viability across all treatments suggests either a systemic effect or a common factor affecting the number of enumerated Legionella.
Two possibilities could explain the results seen for the use of PMA-qPCR to enumerate viable Legionella in complex biofilms, either:
PMA treatment has worked and the subsequent muted response of Legionella to disinfection treatment is a factor of biofilm/microbiological effects; or
PMA treatment has not worked, and any measured decrease or increase in detectable Legionella is because of other factors affecting the method.
Each of these possibilities will be discussed separately.
1. PMA treatment has worked and the subsequent muted response of Legionella to disinfection treatment is a factor of biofilm/microbiological effects.
Assuming that PMA has selectively allowed enumeration of only viable Legionella, biofilm samples treated with PMA and untreated biofilms show a large discrepancy in detectable DNA. Figure 2 demonstrates that conventional qPCR returned a Legionella count roughly 50–65% higher than that for PMA-treated samples, suggesting that the majority of detectable Legionella DNA targets in biofilm samples originate from nonviable cells.
The relatively uniform survival across disinfection treatments may represent the fraction of cells that exist in a dormant state. Legionella’s nature as an opportunistic parasite may serve to protect it from chemical disinfection. When suitable hosts or microclimates for multiplication are presented, dormant Legionella will become active and multiply. In the absence of these triggers, Legionella exists in a viable, but nonculturable state with greatly reduced metabolic activity, accounting for a substantial portion of the sessile Legionella population.1,25–27 These nonproliferative cells, which carry out few cellular processes, show a vastly increased resistance to biocide activity.28,29 Application of biocides would reduce the complement of cells with higher metabolic activities (those most susceptible to disinfection), while cells with lower activity would survive disinfection.
The proportion of viable to nonviable cells should be relatively even across the biofilms sampled. If the presence of these low activity cells accounts for the reduced disinfection seen in tests, then determining the proportion of low activity cells in each biofilm would represent the true number of potentially infective organisms present.
Extracellular polymeric substances (EPS) have been demonstrated to reduce the effectiveness of biocides by a combination of quenching effects30–32 and a reduction in the penetration of active compounds into the film.33–36 As all biofilms were sampled from the same system, the chemical and microbiological makeup of each sample was relatively uniform, and an equivalent EPS quenching of disinfectants may account for the roughly equivalent reduction in viable cells across treatments. In this case, viable cells present in more superficial layers of biofilms would be killed while cells situated in deeper layers of biofilm would not receive a significant dose of active compound.
Legionella content of biofilm
These microbiological effects may explain some overall trends present in the number of enumerated viable Legionella, but cannot account for the relationship between the amount of biofilm sampled and the viable Legionella count as determined by PMA-qPCR. This relationship demonstrates that the amount of Legionella detected by PMA-qPCR is more strongly affected by the quantity of biofilm in the sample than by the strength of the disinfection treatment it is exposed to.
The mass of each biofilm was used to estimate the number of viable Legionella per mg of biofilm, and therefore the amount of biofilm sampled will determine the corresponding number of Legionella. This relationship should break down as disinfectants are applied, and even more markedly so after heat treatment, which is unaffected by the amount of biofilm present in the sample. As Legionella are inactivated by disinfectants or heat, the viable Legionella count per mg of biofilm should decrease. This expected result is not reflected in the data, as heating or increasing doses of biocides over increasing exposure times did not significantly affect the number of detected viable Legionella. However, the expected relationship between disinfectant treatments and die off became more noticeable for amounts of biofilm below 10 mg, which only make-up a small proportion of the samples taken. This strongly suggests that variations in enumerated Legionella and their relationship with the amount of biofilm sampled are the results of a systemic effect inherent in the sampling procedure or in the application and action of PMA on each sample.
Factors affecting PMA effectiveness
PMA does not selectively interact with Legionella and is not specifically applicable to prokaryotic DNA. PMA will interact with all biofilm components containing suitable binding sites. These include all bacterial DNA contained within nonviable cells, any damaged eukaryotic cells with exposed nuclei, any naked DNA accumulated in pockets of cellular debris or entrained within EPS as well as EPS itself, and any DNA structures excreted by bacteria that may be used to adhere to surfaces.37
As Legionella only comprised a small percentage of total biofilm mass, the majority of available PMA could be quenched in non-Legionella targets upon exposure to light. This process would result in a relatively uniform decrease in detectable Legionella numbers to a lower limit determined by the amount of PMA added, at which point all available PMA will have bonded to the vast majority of non-Legionella DNA. This would produce a result where the apparent viability of Legionella was increased with increasing amounts of biofilm, regardless of the disinfection treatment used, as seen in the current study. This relationship became apparent when the total available dataset was pooled to compare the viable Legionella count of each sample with the amount of biofilm in the sample (Fig. 7). As increasing amounts of biofilm were used, the viable Legionella count increased, regardless of the treatment used. In samples with decreased biofilm mass, the apparent viable Legionella count sharply dropped as the number of non-Legionella targets was reduced, thus reducing the PMA quenching effect. This effect was most notable below 10 mg of biofilm, where the viable Legionella count sharply declined with each mg of biofilm used.
In order to compensate for this effect, either higher concentrations of PMA or lower amounts of biofilm would need to be sampled. As PMA becomes increasingly toxic at higher concentrations and because of the expense of the reagent ($115 per mg [Biotium Australia, South Australia]), augmenting the method to include higher PMA concentrations is both counterproductive and cost prohibitive. However, as smaller volumes of biofilm are used, the reproducibility of the method is reduced. Because of the sparse distribution of Legionella microcolonies in biofilms, water samples containing larger volumes of biofilm are more likely to be representative of the system’s microbial population.
The physical properties of biofilms may further reduce the applicability of PMA treatment, as film infiltration may be substantially reduced in thicker or more complex films. The dark incubation period may need to be lengthened to allow for full penetration of PMA into deeper layers of film. Alternatively, increasing DMSO concentrations may increase biofilm infiltration by PMA, although DMSO has been shown to decrease PCR efficiency at higher concentrations, and may potentiate the toxicity of PMA leading to the generation of false negatives.38
Additionally, the thickness of the sample as well as the content of pigmented microorganisms and algae will alter the penetration of light, which is required for PMA to covalently intercalate DNA. In thicker, more complex biofilms, brighter light may be required to sufficiently treat each sample. Care should be taken to ensure that samples do not dry out or become damaged by the increased temperature generated by prolonged close proximity to a light source.
Further possibilities
If we assume that PMA treatment did not provide any discrimination between viable and dead cells, any other appreciable decreases in quantifiable Legionella not explained by fluctuations in the volume of biofilm sampled might have been caused by processes that make the DNA in the sample unavailable for PCR amplification.
Application of oxidizing biocides can degrade DNA, rendering it unable to be amplified by PCR.39 However, this possibility is unlikely to have occurred as any destruction of DNA caused by exposure to biocides should be dose dependent, a response not seen in the current data. Furthermore, samples treated with nonoxidizing biocides should not show a decrease in available DNA by this mechanism.
2. PMA treatment has not worked, and any measured decrease or increase in detectable Legionella is because of other factors affecting the method.
Legionella detection and enumeration protocols are beginning to shift away from microbiological isolation and culture-based techniques to predominantly more molecular methods such as PCR and fluorescence microscopy.3,9,40,41 These newer methods are generally more sensitive than culture-based methods and are more high-throughput, requiring less time before a result is obtained.41
Rather than developing a protocol using novel or previously published primer sets to enumerate Legionella after a disinfection event, it was decided that an industry-recognized method would provide a more realistic result with regard to current Legionella testing protocols. The Bio-Rad iQ-Check™ PCR kit was specifically chosen for use in this study, as it has been independently validated42 for detection of Legionella in water sources, and is designed to be used for risk assessment and cooling tower investigation. After application of this kit to the samples tested in this study, it appears that at best this kit is able to detect the presence of Legionella DNA originating from living or dead cells, or aggregations of cellular debris in EPS. No discriminatory capability between viable and nonviable cells is possible, even with the inclusion of PMA treatment using current methodology.
If PMA-qPCR is not capable of showing a definitive decrease in viable Legionella residing in biofilms, as the results of the current study indicate, then conventional PCR-based enumeration methods are unsuitable for reliable assessment of water sources because of the inherent likelihood of generating false positives. This method must be refined further to ensure that the gap between false negatives resulting from conventional culture-based enumeration and false positives generated from PCR is narrowed. The discrepancy between false positives and true viable counts is also of legislative and regulatory concern. Under Australian law, a cooling tower that returns a Legionella count of 100–1000 CFU/mL is deemed to be potentially hazardous, while a count of 10–99 indicates that maintenance protocols may not be sufficient.43 A 60% margin of error in enumeration may potentially span this boundary and result in incorrect remedial maintenance measures being undertaken.
Currently, the literature demonstrates that detection of Legionella in water sources by PCR is inherently more likely to generate false positives and overestimate Legionella numbers. This is because of the presence of DNA originating from dead cells, and from naked DNA entrained in EPS and cellular debris, particularly in areas of biofilm showing low microbial activity. DNA in this form has been demonstrated to persist in environmental sources for up to two weeks.15 However, in terms of public health, the generation of false positives and an overestimation of risk is a comparatively more desirable outcome than an underestimation of risk.
Results of PMA-qPCR for disinfectant-treated biofilm samples displayed large variability between replicates. Legionella sampled and enumerated from environmental water sources (both constructed and natural) will vary greatly depending upon the sampling location and procedure. A total comparison of all enumerated Legionella with their respective biofilm mass (Fig. 7) demonstrated considerable heteroscedasticity. Increasing biofilm mass generated higher Legionella counts and greatly magnified the spread of the data. The complexity inherent in biofilm communities demonstrates a similar inherent variability in samples. Numerous factors alter biofilm thickness and microbial density (aeration, flow rate, nutrient availability, etc.). These factors are difficult to adequately standardize at all points in a biofilm and will generate variations in the placement and proliferation of organisms within the biofilm. Even given perfectly uniform conditions, it is unlikely that Legionella or any other organism would uniformly inhabit biofilms at the same density in all locations.
In addition to variation in the biofilm itself, sampling procedure and location also exert an influence on the results obtained. Water samples taken from the bulk recirculating water will contain Legionella that has been shed from biofilms, is contained within protozoa, or has become flagellated after lysis of a host. This subpopulation will be significantly different from Legionella sampled from surfaces and contained within biofilms. Sessile Legionella are afforded significant protection by biofilms in comparison to planktonic suspensions of Legionella, requiring an exposure time of at least four times as long for an equivalent decrease in viability to occur.44 This result is corroborated in other studies demonstrating that planktonic Legionella cells were more than seven times more sensitive to chlorine disinfection than biofilm-bound Legionella (15 minutes exposure to 0.4 mg/L chlorine for planktonic cells compared with 3 mg/L for sessile cells).45,46
Legionella shed from biofilms and recirculated within cooling water should show a reduced variability in number as microbes would be capable of even distribution in a rapidly moving body of water. This would further differentiate the populations and relative counts of Legionella sampled from either bulk recirculating water or from biofilms within industrial water systems.
It is conceivable that Legionella may benefit from the formation of large temperature gradients within water systems. Previous work has shown that Legionella is capable of parasitizing organisms that would not be present in the warmest parts of a system, and only under temperature conditions that are favorable to Legionella and unfavorable to the host.47–49 Because of this adaptation to parasitization, systems that contain both cooler areas capable of supporting temperature-sensitive hosts and warmer areas that may harbor Legionella and stress these hosts will naturally be advantageous to Legionella growth.
Conclusions
The employment of molecular methods (conventional PCR, qPCR, etc.) for detection and quantification of Legionella, and their use as risk assessment tools, is an important step toward more rapid and accurate assessment of industrial water systems. Accurately quantifying Legionella cells for regulatory purposes to protect public health is essential. We tested PMA use as a method to count only live Legionella cells in samples collected from a modified chemostat that generates environmentally comparable samples. Counts from PMA-treated samples that were pretreated with either heat or three types of disinfectants (to kill the cells) were highly variable, with the only consistent trend being the relationship between biofilm mass and numbers of Legionella cells. Two possibilities explain this result: 1. PMA treatment worked and the subsequent muted response of Legionella to disinfection treatment is a factor of biofilm/microbiological effects; although this does not account for the relationship between the amount of biofilm sampled and the viable Legionella count as determined by PMA-qPCR; or 2. PMA treatment did not work, and any measured decrease or increase in detectable Legionella is because of other factors affecting the method. This is the most likely explanation for our results, suggesting that higher concentrations of PMA might be needed to compensate for the presence of other compounds in an environmental sample or that lower amounts of biofilm need to be sampled. As PMA becomes increasingly toxic at higher concentrations and is very expensive, augmenting the method to include higher PMA concentrations is both counterproductive and cost prohibitive. Conversely, if smaller volumes of biofilm are used, the reproducibility of the method is reduced. Our results suggest that using PMA is not an appropriate method for discriminating between live and dead cells to enumerate Legionella for regulatory purposes.
Figure 5.
Enumerated Legionella in biofilms after 2,2-dibromo-3-nitrilopropionamide (DBNPA) treatment.
Footnotes
ACADEMIC EDITOR: Raul Rivas, Editor in Chief
FUNDING: This work was supported by funding from the Victorian Department of Human Services. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
This paper was subject to independent, expert peer review by a minimum of two blind peer reviewers. All editorial decisions were made by the independent academic editor. All authors have provided signed confirmation of their compliance with ethical and legal obligations including (but not limited to) use of any copyrighted material, compliance with ICMJE authorship and competing interests disclosure guidelines and, where applicable, compliance with legal and ethical guidelines on human and animal research participants.
Author Contributions
Conceived and designed the experiments: MT, RB, KR. Analyzed the data: MT. Wrote the first draft of the manuscript: MT. Contributed to the writing of the manuscript: MT, KR. Agree with manuscript results and conclusions: MT, RB, KR. Jointly developed the structure and arguments for the paper: MT, KR. Made critical revisions and approved final version: MT, KR. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Hussong D, Colwell RR, O’Brien M, et al. Viable Legionella pneumophila not detectable by culture on agar media. Nat Biotechnol. 1987;5(9):947–950. [Google Scholar]
- 2.Steele TW, Moore CV, Sangster N. Distribution of Legionella longbeachae sero-group 1 and other legionellae in potting soils in Australia. Appl Environ Microbiol. 1990;56(10):2984–2988. doi: 10.1128/aem.56.10.2984-2988.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartie C, Venter SN, Nel LH. Identification methods for Legionella from environmental samples. Water Res. 2003;37(6):1362–1370. doi: 10.1016/S0043-1354(02)00220-8. [DOI] [PubMed] [Google Scholar]
- 4.Bopp CA, Sumner JW, Morris GK, Wells JG. Isolation of Legionella spp. from environmental water samples by low-pH treatment and use of a selective medium. J Clin Microbiol. 1981;13(4):714–719. doi: 10.1128/jcm.13.4.714-719.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Touron-Bodilis A, Pougnard C, Frenkiel-Lebossé H, Hallier-Soulier S. Usefulness of real-time PCR as a complementary tool to the monitoring of Legionella spp. and Legionella pneumophila by culture in industrial cooling systems. J Appl Microbiol. 2011;111(2):499–510. doi: 10.1111/j.1365-2672.2011.05063.x. [DOI] [PubMed] [Google Scholar]
- 6.Bonetta S, Bonetta S, Ferretti E, Balocco F, Carraro E. Evaluation of Legionella pneumophila contamination in Italian hotel water systems by quantitative realtime PCR and culture methods. J Appl Microbiol. 2010;108(5):1576–1583. doi: 10.1111/j.1365-2672.2009.04553.x. [DOI] [PubMed] [Google Scholar]
- 7.Rowbotham TJ. Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J Clin Pathol. 1983;36(9):978–986. doi: 10.1136/jcp.36.9.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alleron L, Merlet N, Lacombe C, Frère J. Long-term survival of Legionella pneumophila in the viable but nonculturable state after monochloramine treatment. Curr Microbiol. 2008;57(5):497–502. doi: 10.1007/s00284-008-9275-9. [DOI] [PubMed] [Google Scholar]
- 9.Ballard AL, Fry NK, Chan L, et al. Detection of Legionella pneumophila using a real-time PCR hybridization assay. J Clin Microbiol. 2000;38(11):4215–4218. doi: 10.1128/jcm.38.11.4215-4218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishimatsu S, Miyamoto H, Hori H, Tanaka I, Yoshida SI. Sampling and detection of Legionella pneumophila aerosols generated from an industrial cooling tower. Ann Occup Hyg. 2001;45(6):421–427. [PubMed] [Google Scholar]
- 11.Declerck P, Verelst L, Duvivier L, Van Damme A, Ollevier F. PCR as a test for the presence or absence of Legionella in (cooling) water. Water Sci Technol. 2003;47(3):103–107. [PubMed] [Google Scholar]
- 12.Yanez MA, Carrasco-Serrano C, Barbera VM, Catalan V. Quantitative detection of Legionella pneumophila in water samples by immunomagnetic purification and real-time PCR amplification of the dotA gene. Appl Environ Microbiol. 2005;71(7):3433–3441. doi: 10.1128/AEM.71.7.3433-3441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joly P, Falconnet PA, André J, et al. Quantitative real-time Legionella PCR for environmental water samples: data interpretation. Appl Environ Microbiol. 2006;72(4):2801–2808. doi: 10.1128/AEM.72.4.2801-2808.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whiley H, Taylor M. Legionella detection by culture and qPCR: comparing apples and oranges. Crit Rev Microbiol. 2014 Mar 3; doi: 10.3109/1040841X.2014.885930. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 15.Josephson KL, Gerba CP, Pepper IL. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl Environ Microbiol. 1993;59(10):3513–3515. doi: 10.1128/aem.59.10.3513-3515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nocker A, Camper AK. Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Appl Environ Microbiol. 2006;72(3):1997–2004. doi: 10.1128/AEM.72.3.1997-2004.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nocker A, Cheung C-Y, Camper AK. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J Microbiol Methods. 2006;67(2):310–320. doi: 10.1016/j.mimet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Walker JT, Keevil CW. Study of microbial biofilms using light microscope techniques. Int Biodeterior Biodegradation. 1994;34:223–236. [Google Scholar]
- 19.Altman SA, Randers L, Rao G. Comparison of trypan blue dye exclusion and fluorometric assays for mammalian cell viability determinations. Biotechnol Prog. 1993;9(6):671–674. doi: 10.1021/bp00024a017. [DOI] [PubMed] [Google Scholar]
- 20.Hein I, Schneeweiss W, Stanek C, Wagner M. Ethidium monoazide and propidium monoazide for elimination of unspecific DNA background in quantitative universal real-time PCR. J Microbiol Methods. 2007;71(3):336–339. doi: 10.1016/j.mimet.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Nocker A, Sossa-Fernandez P, Burr MD, Camper AK. Use of propidium monoazide for live/dead distinction in microbial ecology. Appl Environ Microbiol. 2007;73(16):5111–5117. doi: 10.1128/AEM.02987-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudi K, Moen B, Dromtorp SM, Holck AL. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl Environ Microbiol. 2005;71(2):1018–1024. doi: 10.1128/AEM.71.2.1018-1024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yáñez MA, Nocker A, Soria-Soria E, Múrtula R, Martínez L, Catalán V. Quantification of viable Legionella pneumophila cells using propidium monoazide combined with quantitative PCR. J Microbiol Methods. 2011;85(2):124–130. doi: 10.1016/j.mimet.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Taylor M, Ross K, Bentham R. Spatial arrangement of Legionella colonies in intact biofilms from a model cooling water system. Microbiol Insights. 2013;6:49–57. doi: 10.4137/MBI.S12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hambleton P, Broster MG, Dennis PJ, Henstridge R, Fitzgeorge R, Conlan JW. Survival of virulent Legionella pneumophila in aerosols. J Hyg. 1983;90(3):451–460. doi: 10.1017/s0022172400029090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohno A, Kato N, Yamada K, Yamaguchi K. Factors influencing survival of Legionella pneumophila serotype 1 in hot spring water and tap water. Appl Environ Microbiol. 2003;69(5):2540–2547. doi: 10.1128/AEM.69.5.2540-2547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abu Kwaik Y, Gao L-Y, Stone BJ, Venkataraman C, Harb OS. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl Environ Microbiol. 1998;64(9):3127–3133. doi: 10.1128/aem.64.9.3127-3133.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 29.Lewis K. Persister cells and the riddle of biofilm survival. Biochemistry (Moscow) 2005;70(2):267–274. doi: 10.1007/s10541-005-0111-6. [DOI] [PubMed] [Google Scholar]
- 30.Chen Z, Luo J, Sun Y. Biocidal efficacy, biofilm-controlling function, and controlled release effect of chloromelamine-based bioresponsive fibrous materials. Biomaterials. 2007;28(9):1597–1609. doi: 10.1016/j.biomaterials.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campanac C, Pineau L, Payard A, Baziard-Mouysset G, Roques C. Interactions between biocide cationic agents and bacterial biofilms. Antimicrob Agents Chemother. 2002;46(5):1469–1474. doi: 10.1128/AAC.46.5.1469-1474.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mah T-FC, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9(1):34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 33.De Beer D, Srinivasan R, Stewart PS. Direct measurement of chlorine penetration into biofilms during disinfection. Appl Environ Microbiol. 1994;60(12):4339–4344. doi: 10.1128/aem.60.12.4339-4344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart PS, Rayner J, Roe F, Rees WM. Biofilm penetration and disinfection efficacy of alkaline hypochlorite and chlorosulfamates. J Appl Microbiol. 2001;91(3):525–532. doi: 10.1046/j.1365-2672.2001.01413.x. [DOI] [PubMed] [Google Scholar]
- 35.Szomolay B, Klapper I, Dockery J, Stewart PS. Adaptive responses to antimicrobial agents in biofilms. Environ Microbiol. 2005;7(8):1186–1191. doi: 10.1111/j.1462-2920.2005.00797.x. [DOI] [PubMed] [Google Scholar]
- 36.Walters MC, III, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47(1):317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vilain S, Pretorius JM, Theron J, Brozel VS. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl Environ Microbiol. 2009;75(9):2861–2868. doi: 10.1128/AEM.01317-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gelfand DH. Taq DNA polymerase. In: Erlich H, editor. PCR Technology: Principles and Applications for DNA Amplification. New York: M. Stockton Press; 1989. pp. 17–22. [Google Scholar]
- 39.Epe B. Reviews of Physiology Biochemistry and Pharmacology. Vol. 127. Berlin: Springer; 1996. DNA damage profiles induced by oxidizing agents; pp. 223–249. [DOI] [PubMed] [Google Scholar]
- 40.Behets J, Declerck P, Delaedt Y, Creemers B, Ollevier F. Development and evaluation of a Taqman duplex real-time PCR quantification method for reliable enumeration of Legionella pneumophila in water samples. J Microbiol Methods. 2007;68(1):137–144. doi: 10.1016/j.mimet.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 41.Buchbinder S, Trebesius K, Heesemann J. Evaluation of detection of Legionella spp. in water samples by fluorescence in situ hybridization, PCR amplification and bacterial culture. Int J Med Microbiol. 2002;292(3–4):241–245. doi: 10.1078/1438-4221-00213. [DOI] [PubMed] [Google Scholar]
- 42.AFNOR . Water quality: detection and quantification of Legionella and/or L. pneumophila by concentration and gene amplification by polymerase chain reaction (PCR). XP T90-471. Association Française de Normalisation, Saint-Denis La Plaine; France: 2006. [Google Scholar]
- 43.Australian/New Zealand Standard 3666.3 (AS/NZS3666.3) Air-Handling and Water Systems of Buildings—Microbial Control. Part 3: Performance-Based Maintenance of Cooling Water Systems. Standards of Australia; 1995. [Google Scholar]
- 44.Wright JB, Ruseska I, Costerton JW. Decreased biocide susceptibility of adherent. Legionella pneumophila J Appl Microbiol. 1991;71(6):531–538. doi: 10.1111/j.1365-2672.1991.tb03828.x. [DOI] [PubMed] [Google Scholar]
- 45.Skaliy P, Thompson TA, Gorman GW, Morris GK, McEachern HV, Mackel DC. Laboratory studies of disinfectants against. Legionella pneumophila Appl Environ Microbiol. 1980;40(4):697–700. doi: 10.1128/aem.40.4.697-700.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muraca P, Stout JE, Yu VL. Comparative assessment of chlorine, heat, ozone, and UV light for killing Legionella pneumophila within a model plumbing system. Appl Environ Microbiol. 1987;53(2):447–453. doi: 10.1128/aem.53.2.447-453.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbaree JM, Fields BS, Feeley JC, Gorman GW, Martin WT. Isolation of protozoa from water associated with a legionellosis outbreak and demonstration of intracellular multiplication of Legionella pneumophila. Appl Environ Microbiol. 1986;51(2):422–424. doi: 10.1128/aem.51.2.422-424.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fields BS, Shotts EB, Jr, Feeley JC, Gorman GW, Martin WT. Proliferation of Legionella pneumophila as an intracellular parasite of the ciliated protozoan Tetrahymena pyriformis. Appl Environ Microbiol. 1984;47(3):467–471. doi: 10.1128/aem.47.3.467-471.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith-Somerville HE, Huryn VB, Walker C, Winters AL. Survival of Legionella pneumophila in the cold-water ciliate Tetrahymena vorax. Appl Environ Microbiol. 1991;57(9):2742–2749. doi: 10.1128/aem.57.9.2742-2749.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]