Abstract
Plants are simultaneously subjected to a variety of stress conditions in the field and are known to combat the hostile conditions by up/down-regulating number of genes. There exists a significant level of cross-talk between different stress responses in plants. In this study, we predict the interacting pairs of transcription factors that regulate the multiple abiotic stress-responsive genes in the plant Arabidopsis thaliana. We identified the interacting pair(s) of transcription factors (TFs) based on the spatial proximity of their binding sites. We also examined the interactions between the predicted pairs of TFs using molecular docking. Subsequent to docking, the best interaction pose was selected using our scoring scheme DockScore, which ranks the docked solutions based on several interface parameters and aims to find optimal interactions between proteins. We analyzed the selected docked pose for the interface residues and their conservation.
Keywords: transcription factors, protein–protein interactions, Arabidopsis thaliana, docking, stress conditions
Introduction
Our understanding of the plant adaptation to various kinds of stress conditions at the molecular level has increased considerably over the years. Various kinds of abiotic stresses limit crop productivity in natural conditions. Abiotic stress negatively influences survival, biomass production and accumulation, and grain yield of most crops. Different crop ecosystems are affected by several abiotic stress factors, and to a differential extent. Insights into the responses elicited in plants by different types of stress have been obtained by studying the genes regulated (up/down) during these stress conditions.1–5 It has been documented that there exists a significant cross-talk between the signal transduction pathways activated during different abiotic stress conditions.6–12 There have been several attempts to explore and understand this cross-talk between signaling pathways using rice as a model organism.11
The current study aims at studying the transcription factors (TFs) that are responsive to multiple abiotic stress conditions in Arabidopsis thaliana. STIFDB13 is an in-house resource which compiles the genes known to be activated during eight different kinds of abiotic stress conditions in A. thaliana based on publicly available, genome-wide stress microarray data. Identification of the transcription factor binding sites (TFBS), upstream of these genes will provide important insights into the activated TFs, thereby regulating gene expression during multiple abiotic stress conditions. STIFDB also documents information on the TFBS, both 100 and 1000 base pairs (bp) upstream, for 2629 stress-responsive genes predicted using the algorithm named STIF.14 Large numbers of predicted TFBS are observed upstream of stress-upregulated genes.
The interaction between different TFs can also be exploited to generate diversity in controlling the gene expression.15,16 The diverse set of eukaryotic genes are regulated by small kinds of TFs and it is the different combination of these protein factors which regulate the expression of different genes.15,17,18 Physical interactions between the multiple TFs bound upstream of every gene may dictate the specific combination of conditions under which the gene will be expressed. Also, in 2011, the role of TFs in combating stress conditions by generating drought-resistant crops was stressed by manipulating the expression of drought-responsive genes with the help of TF DREB/CBF.16,19 There have been many other studies in the field, emphasizing the role of different TFs in regulating the stress responses in plants.16,20,21
Therefore, studying physical interactions between the pair of TFs, known to bind the multiple abiotic stress-upregulated genes, will further prove useful to understand how combinatorial regulation of gene expression is achieved in order to combat multiple stresses.
Protein–protein interactions constitute the “interactome” of the cell and dictate majority of cellular processes and are known to regulate responses of organisms to varied environments including stress conditions.20 There are excellent biochemical/experimental techniques like yeast two-hybrid, co-immunoprecipitation, which aim to identify the interacting pairs of proteins, but they are usually time-consuming. Further, not all pairs of proteins can be tested for their interactions using these methods. Therefore, it would be interesting to computationally predict the interacting pairs of proteins that might be regulating the multiple abiotic stress-responsive genes, followed by their detailed experimental validations.
To accomplish this, we selected A. thaliana as a model plant as its genome is fully sequenced and there is a wide variety of publicly available databases like Nottingham Arabidopsis Stock Centre,1 Database Resource for Analysis of Signal Transduction in Cells,4 Microarray Expression Data Search of the Riken Arabidopsis Genome Encyclopaedia,3 etc., which document the microarray data for gene expression under different stress conditions. The genes, which are authenticated to be upregulated during multiple stress conditions, were identified from a database STIFDB.13 TFBS were identified, which was followed by identification of pairs of TFs with spatially proximal binding sites. The selected pairs of TFs were further subjected to molecular docking that resulted in pool of docked poses. The near-native structure was selected from the pool using our scoring scheme DockScore.22 DockScore is an objective scoring scheme that is based on several interface features, and identifies optimal interactions between the interacting proteins in order to find near-native complexes from the pool of docked poses.22
Methods
Master genes identification
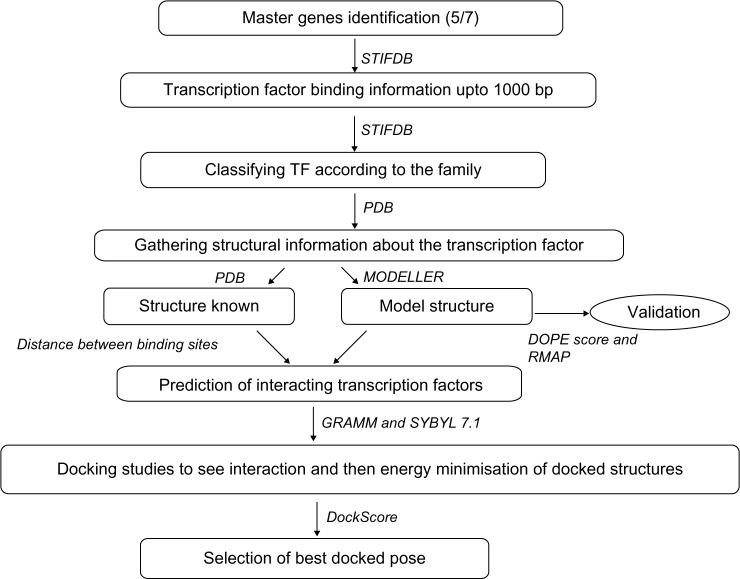
An in-house database STIFDB13 was employed to collect the genes that are regulated in abiotic stress conditions in A. thaliana. The multiple stress-responsive genes were identified from the set of 2629 genes documented in this database, which are known to be upregulated during any of the eight stress conditions considered in this database (drought, salinity, rehydration, abscisic acid (ABA), cold, high light, oxidative stress, and combination of cold–drought–salt stress). Figure 1 describes the work-flow adopted to perform the analysis. The genes overexpressed in at least five of the stress conditions were named as “master genes.”
Figure 1.
Workflow describing the method and tools/techniques adopted.
Notes: Multiple stress-responsive genes (master genes) were identified and their TFBS information was also obtained. Based on the spatial proximity between TFBS, we identified putative interacting pairs of transcription factors. The interactions between these pairs of TF were studied using docking.
Information on TFBS, 1000 bp upstream of the master genes, was also obtained using STIFDB (TFBS with Z-score >1.5 were used for further analysis).
TFBS on master genes
A pair of TF was expected to exhibit a combinatorial control of the master gene through physical interaction, if their TFBS were placed less than 50 bp apart. The 50 bp cut-off is used in some of the earlier studies to examine the formation of cis-regulatory modules.23–25 Following the cut-off of 50 bp, frequency matrix was prepared for every pair of TF. The pairs of TF observed with highest frequency from our dataset were selected as putative interacting pairs.
Molecular modeling and validation
The structural information for all the putative interacting pairs was gathered using PDB.26 In the case of TF with unknown structures, comparative modeling technique was used employing MODELLER 9v7.27 The template used to perform modeling was selected based on sequence homology and atomic resolution of the structure.
The five low-energy modeled structures obtained based on the Modeller’s DOPE score28 were further validated by performing Ramachandran map analysis (model structure with maximum number of amino acids in allowed regions) using PROCHECK server.29
Molecular docking
Docking was performed using GRAMM30 and 10 docked poses were obtained for all the putative interacting pairs. The docked model was further subjected to energy minimization using the SYBYL software package (Version 7.1) (Tripos Associates Inc., St. Louis, MO). Tripos force field was used for minimization (100 iterations with electrostatics off) to obtain final negative energy (kcal/mol) of the complex structure.
Scoring docked poses
The poses obtained subsequent to docking were scored using our scoring scheme DockScore in order to identify the optimal interactions between the putative interacting TFs. The top-ranking pose was selected as the near-native complex.
Results
Identification of master genes and their TFBS
Fifteen master genes, expressed in five or more stress conditions, were identified from STIFDB (Table 1). TFBS data, 1000 bp upstream of these genes, were also collected from STIFDB to identify the regulators of master genes (Supplementary Table 1 and Supplementary Table 2). The schematic exemplifies the nature and position of TFBS for one of the master gene (AT4G27410, Supplementary Fig. 1). The distances between TFBS were analyzed and the frequency matrix was constructed for the number of pairs of TFBS on master genes located <50 nucleotides apart (Table 2). Four pairs of TFs MYB-bHLH, MYB-ARF, HSF-WRKY, and bHLH-bZIP, having TFs from six different families (the highest frequency in the matrix), were selected (Table 2) and designated as putative interacting pairs. These four putative interacting pairs were further tested for the presence and the nature of interactions.
Table 1.
Master genes with stress conditions they are upregulated in Identification of 15 master genes selected out of 2629 abiotic stress responsive genes in Arabidopsis thaliana documented in STIFDB. The gene IDS are mentioned in first column and the different stress conditions are marked in first row. (“+” indicates upregulation of the gene and ABA: Abscisic acid).
| ABA | COLD | DROUGHT | LIGHT | SALT | REHYDRATION | |
|---|---|---|---|---|---|---|
| AT4G27410 | + | + | + | + | + | |
| AT1G20100 | + | + | + | + | + | |
| AT5G15850 | + | + | + | + | + | |
| AT3G12740 | + | + | + | + | + | |
| AT1G51760 | + | + | + | + | + | |
| AT3G05880 | + | + | + | + | + | |
| AT1G16850 | + | + | + | + | + | |
| AT5G52310 | + | + | + | + | + | |
| AT4G26080 | + | + | + | + | + | |
| AT5G39590 | + | + | + | + | + | |
| AT2G21620 | + | + | + | + | + | |
| AT1G19180 | + | + | + | + | + | |
| AT1G73390 | + | + | + | + | + | |
| AT1G78070 | + | + | + | + | + | |
| AT4G37980 | + | + | + | + | + |
Table 2.
Frequency matrix for interactions between transcription factors. The transcription factors predicted to bind 15 master genes belong to 9 classes. For each of the master genes if the distance between the two successive binding sites is ≤50, then it is given the score of 1. This matrix records the score for every 45 possible pairs of transcription factors (frequency matrix). The score marked with asterisk corresponds to the pair having maximum frequency and were named as “putative interacting.”
| MYB | BHLH | NAC | BZIP | ARF | HSF | WRKY | EREBP | HB | |
|---|---|---|---|---|---|---|---|---|---|
| MYB | – | 6* | 1 | 1 | 3* | 1 | 2 | 1 | 0 |
| bHLH | – | 1 | 4* | 2 | 1 | 1 | 1 | 0 | |
| NAC | – | 1 | 1 | 0 | 1 | 1 | 0 | ||
| bZIP | – | 0 | 2 | 1 | 1 | 0 | |||
| ARF | – | 1 | 1 | 0 | 0 | ||||
| HSF | – | 3* | 0 | 0 | |||||
| WRKY | – | 1 | 0 | ||||||
| EREBP | – | 0 | |||||||
| HB | – |
Interactions among TFs
TFs regulating master genes, as documented in STIFDB, belong to nine different families. Out of the above-mentioned six families of TFs, structures are available for two TFs (MYB and WRKY) and for rest of the four families, structures were modeled by comparative modeling techniques (Table 3). Out of five best models obtained for each of the TF (please see Methods for details), one model with least DOPE score and highest percentage of allowed regions in Ramachandran map was selected as the best (Fig. 2). The TFs, bHLH and bZIP, are known to exist as either homo or heterodimers in the cell. However, we modeled bHLH both as a homodimer using multi-chain modeling and single chain so as to study its interactions with different TFs, MYB and bZIP. As bZIP and bHLH are known to heterodimerize,31 bZIP was modeled as single chain to study its interaction with bHLH TF. Among these four pairs of TFs studied, we also validated their interaction using the BioGrid database. bHLH is reported to form homodimer as well as it can heterodimerize with the bZIP TF.32,33 In the BioGrid database, the interaction data for WRKY is still not curated; however, there is an earlier report that stated WRKY and HSF co-express during oxidative stress conditions.34
Table 3.
Putative interacting pairs of transcription factors and their details of their structural data. The table highlights the PDB ID of the template used for modeling the transcription factor along with its percentage identity with the query and resolution of the template. It is appropriately listed, if the structure of a transcription factor is already deposited in PDB. For transcription factors, MYB and WRKY, the structures were there in PDB, whereas for bHLH, bZIP, ARF and HSF, the structures were modeled using comparative modeling.
| PUTATIVE INTERACTING PAIRS | CRYSTAL STRUCTURE | PDB ID* | ORGANISM TO WHICH TEMPLATE BELONG | % IDENTITY WITH TEMPLATE | RESOLUTION (Å) |
|---|---|---|---|---|---|
| MYB | Yes | 2AJE | – | – | – |
| bHLH | No | 1R05 | Homo sapiens | 29.4 | NMR |
|
| |||||
| bZIP | No | 1I04 | Homo sapiens | 39.7 | 3.0 |
| bHLH | No | 1R05 | Homo sapiens | 29.4 | NMR |
|
| |||||
| MYB | Yes | 2AJE | – | – | – |
| ARF | No | 1WID | Arabidopsis thaliana | 34.8 | NMR |
|
| |||||
| WRKY | Yes | 2AYD | – | – | – |
| HSF | No | 1HKS | Drosophila melanogaster | 44.4 | NMR |
Note:
PDB ID of crystal structure if known else of the template used for modeling.
Figure 2.
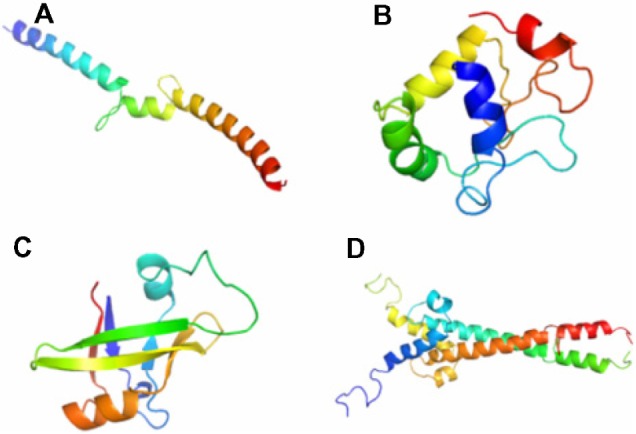
Modeled structures of transcription factors using Modeller (9v7) (A). bZIP (B). HSF (C). ARF (D). bHLH dimer.
Notes: Using comparative modeling, the structures for four transcription factors were modeled and were further validated by checking the percentage allowed regions in Ramachandran map. The best model was selected on the basis of highest percentage allowed regions and least DOPE score as given by Modeller.
Using these structures, interactions between the four pairs of TFs were studied with the help of molecular docking using GRAMM. Ten docked poses for each of the TF docking were generated which were further selected by implementing our scoring scheme DockScore.
TF pairs subjected to DockScore
After testing and assessing the performance of DockScore on the testing dataset comprising of 30 protein–protein complexes,22 the four pairs of TFs identified as putative interacting pairs were subjected to DockScore in order to identify the docked pose with optimal interactions. The docked pose obtaining the highest score was selected as the best-docked pose (Fig. 3).
Figure 3.
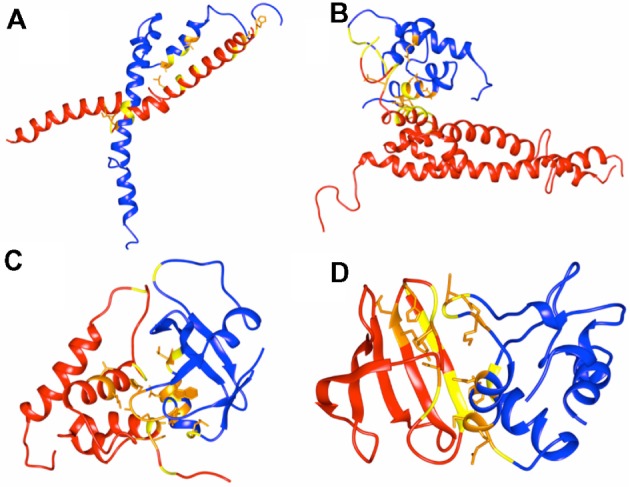
Docked posed pairs of transcription factors and their interface analysis.
Notes: All four-transcription factor pairs were subjected to docking and the docked pose was selected using the scoring scheme DockScore. The figure shows the selected pose with highest score. (A) bHLH and bZIP, (B) MYB and bHLH, (C) MYB and ARF, and (D) WRKY and HSF. Chain A of each docked pose is in red and second chain in blue. The interface residues from both the chains are colored in yellow. In orange are the atoms of conserved interface residues from both the chains.
The best-docked poses selected for the four pairs were further analyzed for the interface residues and their conservation using ConSurf.35 The interface formed upon interaction is rich in conserved residues (marked in orange in Fig. 3).
For the TF pair MYB-bHLH, it is previously reported that the N-terminus region of bHLH is involved in interaction with MYB TF.36 The docked pose we selected (for bHLH and MYB) also bears the interface residues at the N-terminus of the bHLH. The literature evidence also supports interaction between the TFs MYB-ARF and MYB is known to interact with C-terminus of ARF.37 The docked pose we obtained for this pair of TFs possess interface residues at the C-terminus of ARF. bZIP and bHLH are reported to form high molecular weight complexes, suggesting functional relationship between the two.31
Discussion
Plants respond to different stress conditions by either upregulating or downregulating the expression of some genes. The regulation of gene expression is a very vexed mechanism in eukaryotes and it is accomplished with the help of different TFs. The function of genes is highly orchestrated by the action of these protein factors. Therefore, to understand the details of gene regulation, studying interactions between different TFs will be of great value.
Also, as a response to different stress conditions, plants are known to upregulate some general as well as stress-specific genes. The present study deals with the genes elicited in response to multiple stress conditions. We observed that some genes were upregulated in multiple abiotic stress conditions and we named them as “master genes.” There were 15 master genes, identified in A. thaliana from STIFDB database, which were upregulated in five different abiotic stress conditions – ABA, cold, drought, light, and salt stress. For these genes, the presence of TFBS was searched 1000 bp upstream region using the algorithm STIFAL and the predicted positions of various TFBS were obtained. These TFs were observed to belong to nine different families. Where possible, the structures for the TFs predicted to bind master genes were obtained from PDB. In the absence of the known structure, the TF was modeled based on comparative modeling using Modeler (9v7). The physical interactions, between the pairs of factors having predicted binding sites less than 50 bp apart, were studied using molecular docking.
Since accurate structure determination of macromolecular complexes are highly challenging, prediction of protein–protein interactions through molecular docking is highly appropriate. However, implementing molecular docking, for studying the interactions between a pair of proteins poses a challenge to identify the best-docked pose out of the pool of various poses suggested by the docking program GRAMM. For identifying the best-docked pose, we devised an objective scoring scheme, named DockScore, which takes into account several interface parameters and hence ranks the docked poses. The four pairs of TFs were subjected to DockScore and the best-docked pose was selected as the one with the highest DockScore.
In future, more TF pairs will be analyzed in detail to validate the interactions between them, even as recorded in STIFDB2 database.38 Also, employing the similar approach, interactions between the TFs upregulating stress-specific genes and other multiple stress-responsive genes can be studied.
These kinds of studies will aim to provide detailed insights into the regulation of stress-responsive genes at the level of transcription. Also, the existence of interaction among protein factors regulating the responses of plant under stress conditions will provide an additional level of regulation on one hand, and will also lead to combinatorial diversity of regulatory complexes. With different combinations of these factors, regulation of diverse numbers of genes can be achieved. Therefore, studying physical interactions among TFs will provide useful insights into unraveling the basis of this combinatorial diversity in eukaryotes.
Conclusions
Plants are continuously exposed to a number of stress conditions in the fields and they have developed stress resistance or tolerance mechanisms since they are sessile in nature. They achieve this by up/downregulating some genes, which are termed as stress-responsive genes. We have studied transcriptional regulation of multiple stress-responsive genes in A. thaliana. This regulation is accomplished by various TFs and in order to do so they are known to interact with each other. The structural analysis of the TF complex provides details into the functional mechanism of transcriptional regulation. In future, these predictions can be verified using biochemical experiments.
Supplementary Data
Supplementary Table 1. Transcription factor binding site data (from STIFDB) for one of the master gene.
Supplementary Table 2. The TFBS information for rest 14 master genes. The URL provides 1000 bp upstream TFBS information for the respective master genes.
Supplementary Figure 1. Transcription factor binding site data (from STIFDB) for one of the master gene.
Acknowledgments
The authors thank NCBS (National Centre for Biological Sciences) for infrastructure and other facilities.
Footnotes
Author Contributions
Conceived and designed the experiments: RS. Analyzed the data: SM. Wrote the first draft of the manuscript: SM. Contributed to the writing of the manuscript: RS. Agree with manuscript results and conclusions: SM, RS. Jointly developed the structure and arguments for the paper: SM, RS. Made critical revisions and approved final version: RS. Both authors reviewed and approved of the final manuscript.
ACADEMIC EDITOR: JT Efird, Associate Editor
FUNDING: SM is supported by a fellowship from the Department of Biotechnology, Ministry of Science and Technology. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
This paper was subject to independent, expert peer review by a minimum of two blind peer reviewers. All editorial decisions were made by the independent academic editor. All authors have provided signed confirmation of their compliance with ethical and legal obligations including (but not limited to) use of any copyrighted material, compliance with ICMJE authorship and competing interests disclosure guidelines and, where applicable, compliance with legal and ethical guidelines on human and animal research participants.
REFERENCES
- 1.Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S. NASCArrays: a repository for microarray data generated by NASC’s transcriptomics service. Nucleic Acids Res. 2004;32(Database issue):D575–7. doi: 10.1093/nar/gkh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W, GENEVESTIGATOR Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136(1):2621–32. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakurai T, Satou M, Akiyama K, et al. RARGE: a large-scale database of RIKEN Arabidopsis resources ranging from transcriptome to phenome. Nucleic Acids Res. 2005;33(Database issue):D647–50. doi: 10.1093/nar/gki014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Button DK, Gartland KMA, Ball LD, Natanson L, Gartland JS, Lyon GD. DRASTIC – INSIGHTS: querying information in a plant gene expression database. Nucleic Acids Res. 2006;34(suppl 1):D712–6. doi: 10.1093/nar/gkj136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dash S, Van Hemert J, Hong L, Wise RP, Dickerson JA. PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Res. 2011;40(D1):D1194–201. doi: 10.1093/nar/gkr938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol. 2000;3(3):217–23. [PubMed] [Google Scholar]
- 7.Chinnusamy V, Schumaker K, Zhu JK. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot. 2004;55(395):225–36. doi: 10.1093/jxb/erh005. [DOI] [PubMed] [Google Scholar]
- 8.Ludwig AA, Saitoh H, Felix G, et al. Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proc Natl Acad Sci U S A. 2005;102(30):10736–41. doi: 10.1073/pnas.0502954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smékalová V, Doskočilová A, Komis G, Samaj J. Crosstalk between secondary messengers, hormones and MAPK modules during abiotic stress signalling in plants. Biotechnol Adv. 2014;32(1):2–11. doi: 10.1016/j.biotechadv.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Xiao J, Cheng H, Li X, Xiao J, Xu C, Wang S. Rice WRKY13 regulates cross talk between abiotic and biotic stress signaling pathways by selective binding to different cis-elements. Plant Physiol. 2013;163(4):1868–82. doi: 10.1104/pp.113.226019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma R, De Vleesschauwer D, Sharma MK, Ronald PC. Recent advances in dissecting stress-regulatory crosstalk in rice. Mol Plant. 2013;6(2):250–60. doi: 10.1093/mp/sss147. [DOI] [PubMed] [Google Scholar]
- 12.O’Brien JA, Benková E. Cytokinin cross-talking during biotic and abiotic stress responses. Front Plant Sci. 2013;4:451. doi: 10.3389/fpls.2013.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shameer K, Ambika S, Varghese SM, Karaba N, Udayakumar M, Sowdhamini R. STIFDB-Arabidopsis stress responsive transcription factor database. Int J Plant Genomics. 2009;2009:583429. doi: 10.1155/2009/583429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sundar AS, Varghese SM, Shameer K, Karaba N, Udayakumar M, Sowdhamini R. STIF: identification of stress-upregulated transcription factor binding sites in. Arabidopsis thaliana Bioinformation. 2008;2(10):431–7. doi: 10.6026/97320630002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reményi A, Schöler HR, Wilmanns M. Combinatorial control of gene expression. Nat Struct Mol Biol. 2004;11(9):812–5. doi: 10.1038/nsmb820. [DOI] [PubMed] [Google Scholar]
- 16.Hussain SS, Kayani MA, Amjad M. Transcription factors as tools to engineer enhanced drought stress tolerance in plants. Biotechnol Prog. 2011;27(2):297–306. doi: 10.1002/btpr.514. [DOI] [PubMed] [Google Scholar]
- 17.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424(6945):147–51. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 18.Hu Z, Gallo SM. Identification of interacting transcription factors regulating tissue gene expression in human. BMC Genomics. 2010;11(1):49. doi: 10.1186/1471-2164-11-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwal PK, Agarwal P, Reddy MK, Sopory SK. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006;25(12):1263–74. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- 20.Singh K, Foley RC, Oñate-Sánchez L. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol. 2002;5(5):430–6. doi: 10.1016/s1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 21.Vinocur B, Altman A. Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol. 2005;16(2):123–32. doi: 10.1016/j.copbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Malhotra S, Sankar K, Sowdhamini R. Structural interface parameters are discriminatory in recognising near-native poses of protein-protein interactions. PLoS One. 2014;9(2):e80255. doi: 10.1371/journal.pone.0080255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marinescu VD, Kohane IS, Riva A. The MAPPER database: a multi-genome catalog of putative transcription factor binding sites. Nucleic Acids Res. 2005;33(suppl 1):D91–7. doi: 10.1093/nar/gki103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riva A. The MAPPER2 database: a multi-genome catalog of putative transcription factor binding sites. Nucleic Acids Res. 2012;40(Database issue):D155–61. doi: 10.1093/nar/gkr1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannenhalli S, Levy S. Predicting transcription factor synergism. Nucleic Acids Res. 2002;30(19):4278–84. doi: 10.1093/nar/gkf535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 28.Shen M, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15(11):2507–24. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26(2):283–91. [Google Scholar]
- 30.Tovchigrechko A, Vakser IA. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006;34(Web Server issue):W310–4. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuras L, Barbey R, Thomas D. Assembly of a bZIP—bHLH transcription activation complex: formation of the yeast Cbf1–Met4–Met28 complex is regulated through Met28 stimulation of Cbf1 DNA binding. EMBO J. 1997;16(9):2441–51. doi: 10.1093/emboj/16.9.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(suppl 1):D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arabidopsis Interactome Mapping Consortium Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333(6042):601–7. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller G, Mittler R. Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann Bot. 2006;98(2):279–88. doi: 10.1093/aob/mcl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38(suppl 2):W529–33. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pattanaik S, Xie CH, Yuan L. The interaction domains of the plant Myc-like bHLH transcription factors can regulate the transactivation strength. Planta. 2008;227(3):707–15. doi: 10.1007/s00425-007-0676-y. [DOI] [PubMed] [Google Scholar]
- 37.Shin R, Burch AY, Huppert KA, et al. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell. 2007;19(8):2440–53. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naika M, Shameer K, Mathew OK, Gowda R, Sowdhamini R. STIFDB2: an updated version of plant stress-responsive transcription factor database with additional stress signals, stress-responsive transcription factor binding sites and stress-responsive genes in Arabidopsis and rice. Plant Cell Physiol. 2013;54(2):e8. doi: 10.1093/pcp/pcs185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Transcription factor binding site data (from STIFDB) for one of the master gene.
Supplementary Table 2. The TFBS information for rest 14 master genes. The URL provides 1000 bp upstream TFBS information for the respective master genes.
Supplementary Figure 1. Transcription factor binding site data (from STIFDB) for one of the master gene.