Abstract
Purpose
To compare the frequency of ERG rearrangement, PTEN deletion, SPINK1 overexpression, and SPOP mutation in prostate cancer in African American and Caucasian men.
Experimental design
Dominant tumor nodules from radical prostatectomy specimens of 105 African American men (AAM) were compared to 113 dominant nodules from Caucasian men (CaM). Clinical and pathologic characteristics of the two groups were similar. SPINK1 overexpression was evaluated by immunohistochemistry, ERG rearrangement and PTEN deletion by FISH, and SPOP mutation by Sanger sequencing.
Results
ERG rearrangement was identified in 48/113 tumors (42.5%) in CaM and 29/105 tumors (27.6%) in AAM (p=0.024). PTEN deletion was seen in 19/96 tumors (19.8%) in CaM and 7/101 tumors (6.9%) in AAM (p=0.011). SPINK1 overexpression was present in 9/110 tumors (8.2%) in CaM and 25/105 tumors (23.4%) in AAM (p=0.002). SPOP mutation was identified in 8/78 (10.3%) tumors in CaM and 4/88 (4.5%) tumors in AAM (p=0.230). When adjusted for age, BMI, Gleason score, and pathologic stage, ERG rearrangement and SPINK1 overexpression remain significantly different (p=0.018 and p=0.008, respectively), and differences in PTEN deletion and SPOP mutation approach significance (p=0.061 and p=0.087, respectively).
Conclusions
Significant molecular differences exist between prostate cancers in AAM and CaM. SPINK1 overexpression, an alteration associated with more aggressive prostate cancers, was more frequent in AAM, while ERG rearrangement and PTEN deletion were less frequent in this cohort. Further investigation is warranted to determine if these molecular differences explain some of the disparity in incidence and mortality between these two ethnic groups.
Keywords: African American, prostate cancer, race, pathology, molecular
Introduction
Prostate cancer is known to exhibit differences among racial/ethnic groups
African American men (AAM) have a higher incidence and mortality from prostate cancer than that observed in Caucasian men (CaM) as well as other ethnicities (1). Many factors have been postulated to contribute to incidence and/or mortality differences, such as access to care, attitudes toward care, socioeconomic and educational disparities, differences in type and aggressiveness of treatment, and dietary fat intake (2). Some studies have shown that when these factors are controlled for, there is no difference in mortality, but the incidence of prostate cancer in AAM has consistently been shown to be higher (3).
Biochemical recurrence has also been demonstrated to be higher in locally advanced disease in AAM, although a difference in biochemical recurrence was not detected between AAM and CaM with organ-confined prostate cancer after radical prostatectomy (4). PSA levels have also been demonstrated to be higher in AAM than CaM with locally advanced prostate cancer (4). Genetic differences in prostate cancer between AAM and CaM are postulated to contribute to these disparities.
Genetic differences in prostate cancer
Differences in genes involved in the androgen signaling pathways have been observed between AAM and CaM, favoring increased androgen activity in AAM (5–7). Also, increased testosterone levels in AAM as compared to CaM have been shown in some studies (8,9). Several more recent studies have demonstrated differences in gene methylation and aggressive biomarker expression in prostate cancers between CaM and AAM (10–13), strongly suggesting that genetic differences do exist and at least partially contribute to differences observed in clinical outcomes between these two populations.
More is now known about specific molecular aberrations in prostate cancer, with several new discoveries over the past decade. These include recurrent gene fusions involving androgen regulated genes (i.e. TMPRSS2) and ETS family genes(14), PTEN genomic deletion (15–18), overexpression of SPINK1 (a low molecular weight trypsin inhibitor)(19,20), and, more recently, non-synonymous somatic mutations of SPOP (21). Several previous studies have examined the prevalence of ERG rearrangement in AAM (22–24), all of which found a lower frequency of ERG rearrangement and/or ERG overexpression in AAM. Similarly, another study found increased ERG gene expression in prostate cancers among CaM relative to AAM when gene expression profiling was performed (24). To our knowledge, our study is the first to compare the prevalence of PTEN deletion, SPINK1 overexpression, and SPOP mutation between AAM and CaM. Furthermore, our study reports on all four of these molecular aberrations in AAM and CaM who were treated at a single academic medical center and demonstrated similar pre- and post-operative clinicopathologic features.
Materials and Methods
Case selection
All parts of this retrospective study were carried out following Institutional Review Board approval. Archival formalin-fixed, paraffin-embedded (FFPE) radical prostatectomy (RP) specimens from 105 consecutive self-identified AAM who underwent RP between 2001 and 2011 were retrieved. Archival FFPE specimens from an existing tissue microarray cohort of 113 representative self-identified CaM who underwent RP from 2007 to 2009 were included as controls. Although year of surgery was more variable in the AAM cohort, the remaining clinical and pathologic characteristics of the two groups were similar (Table 1). All patients were treated at our institution, a tertiary care academic medical center, and all patients had pre-existing health insurance, , suggesting equal access to care. Furthermore, there was no significant difference in the type of primary insurance between the two groups (private versus government-sponsored) with 81/105 AAM (77%) and 81/113 CaM (72%) having only private insurance (p=0.36). No patients received hormonal or radiation therapy prior to surgery.
Table 1. Clinical and Pathologic Characteristics of 218 Men with Prostate Cancer Treated by Radical Prostatectomy.
| Caucasian | African American | p-value | |
|---|---|---|---|
| Number of men | 113 | 105 | |
| Age at Surgery | 0.148 | ||
| Mean ± SD | 61.2 ± 7.1 | 59.6 ± 7.6 | |
| Range | 45.6-75.5 | 37.0-73.1 | |
| Body Mass Index (BMI) | 0.924 | ||
| Mean ± SD | 26.9 ± 3.2 | 27.0 ± 5.4 | |
| Range | 22.0-38.0 | 19-68.0 | |
| Gleason Score [no.(%)] | 0.601 | ||
| 6 | 18 (16) | 19 (18) | |
| 7(3+4) | 63 (56) | 57 (54) | |
| 7(4+3) | 18 (16) | 21 (20) | |
| 8 and 9 | 14 (12) | 8 (8.0) | |
| Pathological Stage [no. (%)] | 0.160 | ||
| T2 | 80 (71) | 85 (81) | |
| T3a | 25 (22) | 13 (12) | |
| T3b | 8 (7.1) | 7 (6.6) | |
| Margin Positivity [no./total (%)] | 18/113 (16) | 13/105 (12) | 0.561 |
| Biochemical Recurrence [no./total (%)] | 14/110 (12) | 12/81 (15) | 0.665 |
Biochemical recurrence information was available for the majority of men; however, these rates were not adjusted for post-RP treatment, as post-RP treatment was administered at the discretion of the treating physicians. Biochemical recurrence was defined as a post-operative PSA value of >0.2 ng/mL on two separate occasions. The median follow-up time in the CaM cohort was 44 months, and the median follow-up time in the AAM cohort was 41 months. There were 3 CaM and 24 AAM who were lost to follow-up.
Pathologic evaluation and tissue microarray construction
Slides of the FFPE tissue from all RP specimens were reviewed by study pathologists to confirm the pathologic characteristics (TNM stage, Gleason score, margin status). The dominant tumor nodule, defined as the tumor with highest pathologic tumor stage, was selected from each case for construction of tissue microarrays (TMAs). TMAs were constructed using 0.6 mm cores from the FFPE blocks, with each sample represented in triplicate.
Fluorescence in situ hybridization (FISH) analysis of ERG rearrangement and PTEN deletion
Five μm–thick tissue sections from the TMA blocks were used for FISH analysis. For detection of ERG rearrangement, a dual-color break-apart interphase FISH assay was performed as previously described (14,25). Briefly, ERG rearrangement status was assessed using centromeric (BAC clone RP11-24A11 labeled red) and telomeric (BAC clone RP11-372O17 labeled green) probes (Figure 1). If >20% of tumor cells were found to have translocation or deletion, the tumor was considered to have an ERG rearrangement. For detection of PTEN deletion, a gene specific probe (BAC clone CTD-2047N14) and a reference probe located at 10q25.2 (RP11-431P18) were used (Figure 1). Deletion of PTEN was defined as fewer than two copies of the gene specific probe in the presence of two reference signals in >20% of the tumor nuclei. For detection of both ERG rearrangement and PTEN deletion, at least 200 tumor nuclei per case were evaluated using a fluorescence microscope (Olympus BX51; Olympus Optical, Tokyo, Japan).
Figure 1. Immunohistochemical Staining for ERG/SPINK1 and FISH for ERG Rearrangement and PTEN Deletion.
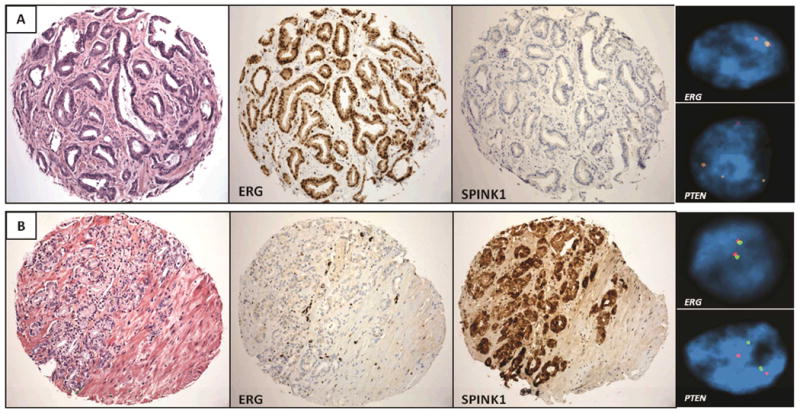
Panel A shows a prostatic adenocarcinoma which demonstrates positive ERG immunostaining and a corresponding ERG rearrangement by FISH. The tumor is negative for SPINK1 overexpression and shows a hemizygous deletion of PTEN.
Panel B shows a prostatic adenocarcinoma which demonstrates negative ERG immunostaining and no ERG rearrangement by FISH. The tumor shows SPINK1 overexpression and no deletion of PTEN by FISH.
Immunohistochemical analysis of ERG and SPINK1 overexpression
Immunohistochemical staining was applied using a commercially available antibody for SPINK1 (clone 4D4, 1:100 dilution, Abnova) and ERG (clone EPR 3864, 1:100 dilution, Epitomics) on the Discovery XT biomarker platform (Ventana Medical Systems, Inc.) (Figure 1). Semi-quantitative evaluation of cytoplasmic SPINK1 expression and nuclear ERG expression were separately performed. Staining of ≥5% of tumor cells was considered positive for each case.
SPOP mutation analysis
Using tissues cores from either fresh frozen material or archival FFPE blocks, samples of the same tumor nodule used for TMA construction were evaluated for SPOP mutations. DNA from fresh frozen material was extracted using phenol-chloroform and purified by ethanol precipitation method as previously described (26). DNA from archival FFPE material was extracted using the Qiagen Biorobot Universal system. High resolution melt analysis (HRM) followed by direct Sanger sequencing of putative SPOP somatic mutations was performed by standard methods following PCR amplification using specific primers. Sequences of the primers used for amplifying and sequencing SPOP have been recently described (21).
Statistical analysis
Chi-square or Fisher's exact tests were used to evaluate association between categorical variables. Wilcoxon Rank Sum Test was performed to compare continuous variables (e.g. age) between groups. For all statistical tests, a p-value <0.05 was considered statistically significant.
Results
ERG rearrangement, PTEN deletion, SPINK1 overexpression, and SPOP mutation in prostate cancer differ in African American vs. Caucasian men
ERG rearrangement was identified in 48/113 tumors (42.5%) of CaM. In AAM, however, ERG rearrangement was found in 29/105 tumors (27.6%; p=0.024). There was no significant difference in the mechanism of gene fusion between the two cohorts (translocation vs. translocation with deletion). Of note, ERG rearrangement by FISH and protein overexpression by immunohistochemistry were concordant in all cases. Hemizygous deletion of PTEN was seen in 19/96 tumors (19.8%) in CaM but only 7/101 tumors (6.9%) in African American men (p=0.011). SPINK1 overexpression was present in 9/110 tumors (8.2%) from CaM in contrast to 25/105 tumors (23.8%) from AAM (p=0.002). SPOP mutations were present in 8/78 (10.3%) prostate cancers in CaM in contrast to 4/88 (4.5%) prostate cancers in AAM; however, this difference was not statistically significant (p=0.230). In CaM, SPOP mutations involved the F133 (6 cases), F102 (1 case), and K129 (1 case) residues. SPOP mutations in AAM involved the F133 (2 cases), F102 (1 case), and Y87 (1 case) residues.
When adjusted for age, BMI, Gleason score, and pathologic stage, ERG rearrangement and SPINK1 overexpression remained significantly different between the two cohort (p=0.018 and p=0.008, respectively), and differences in PTEN deletion and SPOP mutation status approached statistical significance (p=0.061 and p=0.087, respectively). Table 2 summarizes the frequency of all of these molecular findings in the two cohorts.
Table 2. Prevalence of Molecular Aberrations in Prostate Cancer of AAM versus CaM.
| African American | Caucasian | p-value | Adjusted p-value* | |
|---|---|---|---|---|
| ERG rearrangement | 27.6% (29/105) | 42.5% (48/113) | 0.024 | 0.018 |
| PTEN deletion | 6.9% (7/101) | 19.8% (19/96) | 0.011 | 0.061 |
| SPINK1 overexpression | 23.8% (25/105) | 8.2% (9/110) | 0.002 | 0.008 |
| SPOP mutation | 4.5% (4/88) | 10.3% (8/78) | 0.230 | 0.087 |
Adjusted for age, BMI, Gleason score, and pathologic stage.
Association of molecular abnormalities with clinical and pathologic characteristics
When considering both ethnic groups combined, prostate cancers harboring PTEN deletions were found to be significantly associated with higher average age (p=0.001), higher Gleason score (p<0.001), higher pathological stage (p=0.003), and increased rate of biochemical recurrence (p=0.024). All other clinicopathologic parameters were statistically similar with respect to each molecular abnormality. Table 3 summarizes these findings.
Table 3. Association of Clinical and Pathologic Characteristics with Molecular Abnormalities (combined AAM and CaM cohorts).
| ERG rearrangement | PTEN deletion | SPINK1 overexpression | SPOP mutation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | P-value | Deleted | Wild type | P-value | Positive | Negative | P-value | Mutated | Wild type | P-value | |
| Age at Surgery (Mean ± SD) | 59.9 ± 7.0 | 60.7 ± 7.6 | 0.298 | 64.6 ± 7.4 | 59.5 ± 7.3 | 0.001 | 57.6 ± 8.2 | 60.9 ± 7.1 | 0.067 | 60.8 ± 7.2 | 60.3 ± 7.4 | 0.812 |
| BMI (Mean ± SD) | 26.9 ± 3.2 | 27.0 ± 4.8 | 0.622 | 26.4 ± 2.3 | 27.1 ± 4.6 | 0.628 | 26.0 ± 3.3 | 27.1 ± 4.5 | 0.190 | 27.0 ± 4.7 | 26.5 ± 3.8 | 0.899 |
| Gleason Score [no./total (%)] | 0.285 | <0.001 | 0.163 | 0.305 | ||||||||
| 6 | 13/37 (35) | 24/37 (65) | 1/35 (3) | 34/35 (97) | 9/36 (25) | 27/36 (75) | 0/29 (0) | 29/29 (100) | ||||
| 7(3+4) | 48/120 (40) | 72/120 (60) | 12/109 (11) | 97/109 (89) | 20/118 (17) | 98/118 (83) | 7/96 (7) | 89/96 (93) | ||||
| 7(4+3) | 9/39 (23) | 30/39 (77) | 5/35 (14) | 30/35 (66) | 4/39 (10) | 35/39 (90) | 4/34 (12) | 30/34 (88) | ||||
| 8 and 9 | 7/22 (32) | 15/22 (68) | 8/18 (44) | 10/18 (56) | 1/22 (5) | 21/22 (95) | 1/19 (5) | 18/19 (95) | ||||
| Pathological Stage [no./total (%)] | 0.139 | 0.003 | 1.000 | 0.259 | ||||||||
| T2 | 59/165 (35.8) | 106/165 (64.2) | 13/149 (8.7) | 136/149 (91.3) | 26/162 (16.0) | 136/162 (84.0) | 11/123 (8.9) | 112/123 (91.1) | ||||
| T3a | 16/38 (42.1) | 22/38 (57.9) | 8/33 (24.2) | 25/33 (75.8) | 6/38 (15.8) | 32/38 (84.2) | 0/28 (0) | 28/28 (100) | ||||
| T3b | 2/15 (13.3) | 13/15 (86.7) | 5/15 (33.3) | 10/15 (66.7) | 2/15 (13.3) | 13/15 (86.7) | 1/14 (7.1) | 13/14 (92.9) | ||||
| Margin Positivity [no./total (%)] | 8/218 (3.7) | 23/218 (10.5) | 0.311 | 2/197 (1.0) | 25/197 (12.7) | 0.541 | 4/215 (1.9) | 27/215 (12.6) | 0.793 | 1/166 (0.6) | 26/166 (15.7) | 0.440 |
| Biochemical Recurrence [no./total (%)] | 7/191 (3.6) | 19/191 (9.9) | 0.383 | 7/171 (4.0) | 16/171 (9.3) | 0.024 | 5/188 (2.6) | 21/188 (11.1) | 0.562 | 2/141 (1.4) | 21/141 (14.9) | 0.743 |
Among CaM alone, prostate cancers harboring PTEN deletions were found to be significantly associated with higher average age (p=0.002), higher Gleason score (p=0.009), higher pathological stage (p=0.006), and increased rate of biochemical recurrence (p=0.034) (Supplemental Table S1). SPINK1 overexpression was associated with a lower Gleason score in the CaM cohort (p=0.016) (Supplemental Table S1). In the AAM cohort, all clinical and pathologic parameters were statistically similar with respect to each molecular abnormality (Supplemental Table S2).
Since the calendar years in which the AAM and CaM cases were accrued were disparate (2001-2011 and 2007-2009, respectively), statistical comparisons of the clinicopathological characteristics of patients accrued before 2007, from 2007-2009, and after 2009 were performed. No significant differences were observed with respect to pre-operative PSA, Gleason score, pathological stage, or the frequency of each of the molecular abnormalities (p>0.05 for all; data not shown).
ERG rearrangements and SPOP mutations are mutually exclusive, as are PTEN deletions and SPINK1 overexpression
ERG rearrangements and SPOP mutations were not seen together in any of the 178 tumors evaluable for both molecular alterations (p=0.009). PTEN deletion and SPINK1 overexpression were also mutually exclusive in the 195 tumors evaluable for both events (p=0.009). Furthermore, ERG rearrangement and SPINK1 overexpression were mutually exclusive in all but one of the 215 cases (p<0.001). No association was noted between ERG rearrangement and PTEN deletion, SPOP mutation and PTEN deletion, or SPOP mutation and SPINK1 overexpression (p>0.05). These findings are graphically depicted in Supplemental Figure S1.
Discussion
We investigated molecular differences in prostate cancer between two clinicopathologically similar cohorts of AAM and CaM treated at our institution. Our findings are concordant with recent studies showing that there is a significantly lower prevalence of ERG gene rearrangements in prostate cancers of AAM when compared to CaM (22–24,27). To our knowledge, this is the first study to evaluate ethnic differences in the prevalence of hemizygous PTEN deletions, SPINK1 overexpression, and SPOP mutation status in prostate cancer. These findings contribute to our understanding of biological differences in prostate cancer between AAM and CaM, building essential groundwork for the development of personalized cancer treatment regimens.
The discovery of recurrent gene rearrangements in prostate cancer involving androgen regulated genes (e.g. TMPRSS2) and ETS family genes (14) as well as more recent data from whole genome sequenced localized prostate cancers (26,28) has increased our understanding of the disease at the molecular level, identifying potentially diagnostic, prognostic, and therapeutic markers. Results from unscreened, population-based cohorts (e.g. Swedish Watchful Waiting Cohort) have suggested that untreated prostate cancer with ERG rearrangement runs a more aggressive clinical course than those without ERG rearrangement (29). In the setting of surgical or other interventions following diagnosis, the data are insufficient to make any reasonable conclusions. Yoshimoto M, et al later demonstrated that absence of ERG rearrangement and PTEN loss in prostate cancer is associated with a favorable outcome (30). Conversely, duplication of ERG rearrangement with interstitial deletion of sequences 5′ to ERG identified cases of fatal human prostate cancer in patients that had been conservatively managed (15). Regarding distinct molecular characteristics of prostate cancer among different ethnic/racial groups, two recent studies have assessed the difference in prevalence of ERG rearrangements between AAM and CaM (22–24). In the study by Magi-Galluzzi et al, ERG rearrangements were present in 50% of CaM versus 31% of AAM (p=0.07) and in the study by Rosen et al., ERG rearrangements were present in 41.9% of CaM versus 23.9% of AAM (p<0.0001). These findings are in concordance with our current study showing that ERG rearrangements are less frequent in prostate cancers in AAM (42.5% in CaM vs. 27.6% in AAM, p=0.024). When adjusted for age, BMI, Gleason score, and pathologic stage, the difference remained significant (p=0.018). Similar to the study by Magi-Galluzzi C et al, ERG rearrangement in our study did not correlate with other clinicopathologic parameters aside from ethnicity (22).
Regardless of ERG rearrangement's correlation with clinicopathologic features, ethnic differences in the prevalence of ERG rearrangements may have diagnostic implications, with urine-based screening tests currently under investigation (31,32). A recent review by Truong M et al., highlights ERG rearrangement transcripts as one of the more promising RNA markers for cancer detection in urine samples; a urine-based test which uses a combination of ERG rearrangement transcripts and prostate cancer antigen-3 (PCA3) has already been marketed for clinical use (33,34). Considering that our study and previous ones (22–24,27) have shown a decreased prevalence of ERG rearrangements or ERG expression in prostate cancer of AAM, such a urine-based diagnostic test will be less sensitive in this population and may not be as useful of a screening tool as it may be for prostate cancer in CaM.
PTEN, which encodes a phosphoinositide 3-phosphatase that negatively regulates the PI3K and mTOR signaling pathways, is a well-known tumor suppressor gene in many tumor types; in prostate cancer, mutations in PTEN have been found to be associated with higher Gleason score, a higher rate of metastasis, androgen independence, and an overall worse prognosis (15–18). In a more recent, large, nested case control study, decreased PTEN expression was shown to be associated with an increased risk of biochemical recurrence, independent of other clinicopathologic factors(35). Loss of PTEN results in elevated downstream activity in the PI3K and mTOR pathways, which have known therapeutic targets. Although therapeutic approaches to develop inhibitors targeting the PI3K-AKT pathway have failed in both pre-clinical and clinical trials for prostate cancer, there are newer AKT pathway inhibitors that show promise, such as AZD5363 (36). Specifically in prostate cancer cell lines, another recent study has shown that loss of PTEN and elevated AKT/mTOR activity are associated with sensitivity to ridaforolimus, a particular mTOR inhibitor under investigation (37). Although population-based mutational analyses on PTEN have been performed, there are few studies which have investigated ethnic differences in the prevalence of hemizygous loss of PTEN in specific tumors. One study by Winter JL et al., showed no significant racial differences in the expression of PTEN between invasive breast cancers in African American women and those in non-African American women (38). Similar to the racial disparities observed in prostate cancer, breast cancers in African American women are known to have a worse prognosis when compared to those in non-African Americans (39), but PTEN does not appear to play a major role in this disparity (38).
To our knowledge, our current study is the first to compare the prevalence of deletions in PTEN in prostate cancer between AAM and CaM. We found that hemizygous deletions in PTEN were less frequently present in our cohort of AAM compared to that observed in CaM (6.9% vs. 19.8%, p=0.011). However, when adjusted for age, BMI, Gleason score, and pathologic stage, the difference in prevalence was less pronounced and only trended toward significance (p=0.061). Our findings suggest that PTEN deletions may not be critical contributors to the increased incidence or mortality of prostate cancer in AAM, but larger studies with more power are warranted to confirm or refute our finding.
As expected, and consistent with prior literature, PTEN deletions in our study were significantly associated with a higher average age of patients, higher Gleason score, higher pathological stage, and increased rate of biochemical recurrence, though our analysis of biochemical recurrence is only a crude estimate that does not adjust for post-RP therapy and is limited by relatively short follow up time. These associations were statistically significant in CaM when analyzed alone, while no significant clinicopathologic associations with PTEN deletion were identified in AAM alone, possibly attributable to the low number of PTEN deletions in our AAM cohort. Although alterations in the mTOR/AKT and PI3K may still be present in AAM and prove to be therapeutic targets, they may be less frequently due to PTEN deletion than in CaM.
SPINK1 has structural similarity to epidermal growth factor (EGF) and has been demonstrated to activate the epidermal growth factor receptor (EGFR) on the surface of prostate cancer cells, leading to cell growth (20). Using a model of SPINK1-positive prostate cancer (22RV1 cells), Ateeq et al. showed that monoclonal antibodies to either SPINK1 or EGFR (cetuximab) could slow the growth of SPINK1-positive tumors by over 60% and 40%, respectively, suggesting that it may be a reasonable therapeutic target (40). Moreover, SPINK1 overexpression has identified an aggressive subtype of ETS negative prostate cancer, validated in different cohorts (19). SPINK1 and ERG rearrangements have been found to be mutually exclusive in other studies as well (41,42), similar to our current findings. A study by Leinonen et al. found SPINK1 overexpression to be present in 10% of prostate cancers and also found it to be associated with an aggressive form of the disease, although mutual exclusivity with ERG rearrangements were not observed in this particular study (43).
Our study is the first to show that SPINK1 overexpression in a particular tumor type correlates with African American ethnicity. In our study, prostate cancers from AAM showed SPINK1 overexpression in 23.8% of cases compared with 8.2% of prostate cancers in CaM (p=0.002), and this difference remained statistically significant after adjusting for age, BMI, Gleason score, and pathologic stage (p=0.008). In the context of the aforementioned literature, which has demonstrated that SPINK1 overexpression is associated with more aggressive prostate cancers, our study suggests that SPINK1 overexpression may be one molecular aberrancy that plays a role in the increased incidence and/or mortality observed in AAM with prostate cancer, although we emphasize that our study was not designed to demonstrate association of these molecular alterations with clinical outcomes. Furthermore, any targeted therapies to SPINK1 that could develop in the future, as proposed by Ateeq B et al. (40), potentially may benefit more AAM than CaM with prostate cancer.
More recently, whole genome and exome sequencing of prostate cancer has elucidated novel recurrent mutations in prostate cancer such as SPOP, MED12, and FOXA1 (25,26). The most common non-synonymous somatic mutation involves SPOP, which encodes the substrate-binding subunit of a cullin-based E3 ubiquitin ligase (44,45). This recurrent mutation defines a new molecular subtype of ETS-negative prostate cancer (21). After having sequenced the SPOP gene in more than 300 primary prostate cancers and metastases, all SPOP mutations affected conserved residues in the structurally defined substrate binding cleft (21). Recent work in breast cancer has shown that SPOP directly interacts with a p160 steroid resistant coactivator, SRC-3, part of a family of proteins which are overexpressed in numerous human cancers; they are associated with poor clinical outcomes and resistance to therapy and are considered to be potential therapeutic targets (45,46). The interaction of SPOP and SRC-3 in breast cancer promotes cullin 3-dependent ubiquitination and proteolysis, thereby supporting SPOP's role as a potential tumor suppressor (47).
In prostate cancer cell lines, SPOP mutants have been shown to be unable to interact with SRC-3 protein or promote its ubiquitination and subsequent degradation, suggesting that SPOP plays a critical tumor suppressor role in prostate cancer and supporting the potential of SRC-3 as a therapeutic target in prostate cancer (48). In our study, SPOP mutations were less frequently seen in prostate cancers from AAM than from CaM, although this difference did not reach statistical significance (p=0.230). When adjusted for age, BMI, Gleason score, and pathologic stage, however, this difference approaches statistical significance (p=0.087). Although further work is needed in order to fully elucidate the biological and prognostic significance of SPOP mutations in prostate cancer in vivo, as well as the therapeutic potential of SRC-3, our findings suggest that SPOP mutations are less likely to play a significant role in prostate cancer in AAM compared to CaM.
Our study certainly is not devoid of limitations. First, our cohorts contained a relatively modest number of patients from a single institution; larger studies are needed to validate our findings. In addition, there were some cases for which there was missing data on the molecular alterations, highlighted in Supplemental Figure S1, which was attributable to missing or insufficient tissue on the TMAs (for ERG rearrangement, SPINK1 overexpression, and PTEN deletion) or insufficient tissue for DNA extraction (for SPOP mutation analysis). Second, while two techniques were used to assess ERG rearrangement and were concordant, only one technique was used for the other molecular alterations, limiting our ability to confirm these molecular changes. Third, we also must emphasize that our study was not designed or powered to assess clinical outcomes of our patient cohorts, and, therefore, any conclusions as to whether the molecular alterations in these patients have prognostic value would be premature. Assessment of biochemical recurrence in our cohorts was performed only to show rough consistency with prior literature on PTEN and its association with worse outcomes(15–18). In addition, the median length of follow up time in both cohorts was relatively short (41 months and 44 months in AAM and CaM, respectively) with a large number of AAM lost to follow up (24 patients). Lastly, we did not obtain socioeconomic data on our patient population, which has been shown to contribute to prostate cancer outcomes(2). However, given that all patients in our study were treated at a single academic tertiary care facility, that all patients had pre-existing health insurance, and that >70% of patients in each cohort had private insurance coverage suggest that our patients had comparable access to oncologic care. The comparable (and mostly low) pathologic stages of the tumors between both cohorts at radical prostatectomy also suggest that screening and early diagnosis of prostate cancer occurred in both groups.
In an era where precision therapy of prostate cancer is rapidly changing, molecular characterization of both localized and metastatic prostate tumors will help stratify which men will benefit from active surveillance, surgery, targeted therapy, and hormonal and/or chemoradiation therapy. Already, recent studies have shown that ethnicity is an important factor in the progression of prostate cancers under active surveillance (12,49), suggesting perhaps that prostate cancers among different races should be managed differently. It has also been shown previously that prostate cancers with ERG rearrangement have a worse outcome under active surveillance(29), highlighting its potential importance in influencing therapeutic management. Furthermore, a study by Bismar et al. suggests that molecular aberrancies in PTEN, ERG, and SPINK1 may be involved in the development of castration-resistant prostate cancer, emphasizing their clinical importance (42). Our current study highlights the significant differences that exist at the molecular level when prostate cancers from clinicopathologically similar AAM and CaM are compared. We have demonstrated that ERG gene rearrangement, PTEN deletion, and SPOP mutation have a lower prevalence in prostate tumors of AAM and likely play a lesser role in incidence or mortality differences. In contrast, SPINK1 overexpression, a molecular aberrancy that has been found to be associated with more aggressive prostate cancer (19,41,50), is more prevalent in AAM, suggesting that it plays a more important role in the disease within this ethnic group. As our study was not designed to assess clinical outcomes in association with these molecular alterations, larger studies with more detailed clinical outcome data will be needed in order to determine if any of these molecular differences at least partially explain the disparities in incidence and mortality between these two ethnic groups. In addition, future work on whole genome/exome sequencing of prostate cancer will help us to better characterize potential therapeutic targets in prostate tumors among different ethnic groups.
Supplementary Material
Translational Relevance.
African Americans have a higher incidence of prostate cancer and higher mortality from the disease than that observed in Caucasians. Although socioeconomic factors may contribute to these differences, underlying genetic differences are believed to play a role as well. In the current study, we highlight significant differences in ERG gene rearrangement, PTEN deletion, SPINK1 overexpression, and SPOP mutation status in prostate cancer of African- American men compared to Caucasian men. Our findings suggest biologic differences between prostate cancers from these two ethnic groups, with ERG rearrangement, PTEN deletion, and SPOP mutation less frequent in African American men and SPINK1 overexpression more frequent. In view of forthcoming new molecular diagnostic modalities and targeted therapies for prostate cancer, molecular classification of this disease is germane; understanding ethnic differences in this disease will allow for optimizing screening methods and selecting appropriate treatment plans.
Acknowledgments
Funding Support: NIH (EDRN) U01-CA111275 (M.A.R., J.M.M.), NCI R01-CA125612-05A1 (M.A.R.), NIH U01-CA113913-08 (A.K.T.), Prostate Cancer Foundation (M.A.R.), Prostate Cancer Foundation Young Investigator Award (C.E.B.), Urology Care Foundation Research Scholar Award (C.E.B.), Weill Cornell Medical College Department of Pathology & Laboratory Medicine Translational Research Program
Footnotes
Conflicts of Interest: A patent has been issued to Weill Medical College of Cornell University on SPOP mutations in prostate cancer; C.E.B and M.A.R are listed as co-inventors.
Presented in part at the 101st Annual Meeting of the United States and Canadian Academy of Pathology in Vancouver, BC, March 2012
References
- 1.Ries LAG, Eisner MP, Kosary CL, Hankey BF, Miller BA, Clegg L, Mariotto A, Feuer EJ, Edwards BK, editors. SEER Cancer Statistics Review, 1975-2002. National Cancer Institute; Bethesda, MD: http://seer.cancer.gov/csr/1975_2002/, based on November 2004 SEER data submission, posted to the SEER web site 2005. [Google Scholar]
- 2.Freedland SJ, Isaacs WB. Explaining racial differences in prostate cancer in the United States: sociology or biology? Prostate. 2005;62:243–52. doi: 10.1002/pros.20052. [DOI] [PubMed] [Google Scholar]
- 3.Alexander G, Brawley O. Prostate cancer treatment outcome in blacks and whites: a summary of the literature. Semin Urol Oncol. 1998;16:232–4. [PubMed] [Google Scholar]
- 4.Powell IJ, Banerjee M, Novallo M, Sakr W, Grignon D, Wood DP, et al. Prostate cancer biochemical recurrence stage for stage is more frequent among African-American than white men with locally advanced but not organ-confined disease. Urology. 2000;55:246–51. doi: 10.1016/s0090-4295(99)00436-7. [DOI] [PubMed] [Google Scholar]
- 5.Platz EA, Rimm EB, Willett WC, Kantoff PW, Giovannucci E. Racial variation in prostate cancer incidence and in hormonal system markers among male health professionals. J Natl Cancer Inst. 2000;92:2009–17. doi: 10.1093/jnci/92.24.2009. [DOI] [PubMed] [Google Scholar]
- 6.Bennett CL, Price DK, Kim S, Liu D, Jovanovic BD, Nathan D, et al. Racial variation in CAG repeat lengths within the androgen receptor gene among prostate cancer patients of lower socioeconomic status. J Clin Oncol. 2002;20:3599–604. doi: 10.1200/JCO.2002.11.085. [DOI] [PubMed] [Google Scholar]
- 7.Gaston KE, Kim D, Singh S, Ford OH, Mohler JL. Racial differences in androgen receptor protein expression in men with clinically localized prostate cancer. J Urol. 2003;170:990–3. doi: 10.1097/01.ju.0000079761.56154.e5. [DOI] [PubMed] [Google Scholar]
- 8.Ross R, Bernstein L, Judd H, Hanisch R, Pike M, Henderson B. Serum testosterone levels in healthy young black and white men. J Natl Cancer Inst. 1986;76:45–8. [PubMed] [Google Scholar]
- 9.Gapstur SM, Gann PH, Kopp P, Colangelo L, Longcope C, Liu K. Serum androgen concentrations in young men: A longitudinal analysis of associations with age, obesity, and race: The CARDIA male hormone study serum androgen concentrations in young men. Cancer Epidemiol Biomarkers Prev. 2002:1041–7. [PubMed] [Google Scholar]
- 10.Kim HS, Moreira DM, Jayachandran J, Gerber L, Bañez LL, Vollmer RT, et al. Prostate biopsies from black men express higher levels of aggressive disease biomarkers than prostate biopsies from white men. Prostate Cancer Prostatic Dis. 2011;14:262–5. doi: 10.1038/pcan.2011.18. [DOI] [PubMed] [Google Scholar]
- 11.Grisanzio C, Werner L, Takeda D, Awoyemi BC, Pomerantz MM, Yamada H. Genetic and functional analyses implicate the NUDT11, HNF1B, and SLC22A3 genes in prostate cancer pathogenesis. Proc Natl Acad Sci USA. 2012;109:11252–7. doi: 10.1073/pnas.1200853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iremashvili V, Soloway MS, Rosenberg DL, Manoharan M. Clinical and demographic characteristics associated with prostate cancer progression in patients on active surveillance. J Urol Elsevier Inc. 2012;187:1594–9. doi: 10.1016/j.juro.2011.12.082. [DOI] [PubMed] [Google Scholar]
- 13.Kwabi-Addo B, Wang S, Chung W, Jelinek J, Patierno SR, Wang BD, et al. Identification of differentially methylated genes in normal prostate tissues from African American and Caucasian men. Clin Cancer Res. 2010;16:3539–47. doi: 10.1158/1078-0432.CCR-09-3342. [DOI] [PubMed] [Google Scholar]
- 14.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science (80-) 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 15.Shen MM, Abate-Shen C. Pten inactivation and the emergence of androgen-independent prostate cancer. Cancer Res. 2007;67:6535–8. doi: 10.1158/0008-5472.CAN-07-1271. [DOI] [PubMed] [Google Scholar]
- 16.Pourmand G, Ziaee AA, Abedi AR, Mehrsai A, Alavi HA, Ahmadi A, et al. Role of PTEN gene in progression of prostate cancer. Urol J. 2007;4:95–100. [PubMed] [Google Scholar]
- 17.Reid AHM, Attard G, Ambroisine L, Fisher G, Kovacs G, Brewer D, et al. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br J Cancer. 2010;102:678–84. doi: 10.1038/sj.bjc.6605554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Su J, DingZhang X. PTEN deletion and heme oxygenase-1 overexpression cooperate in prostate cancer progression and are associated with adverse clinical outcome. J. 2011;224:90–100. doi: 10.1002/path.2855. [DOI] [PubMed] [Google Scholar]
- 19.Tomlins SA, Rhodes DR, Yu J, Varambally S, Mehra R, Perner S, et al. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell. 2008;13:519–28. doi: 10.1016/j.ccr.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paju A, Hotakainen K, Cao Y, Laurila T. Increased expression of tumor-associated trypsin inhibitor, TATI, in prostate cancer and in androgen-independent 22Rv1 cells. Eur Urol. 2007;52:1670–9. doi: 10.1016/j.eururo.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 21.Barbieri C, Baca S, Lawrence M. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magi-Galluzzi C, Tsusuki T, Elson P, Simmerman K, LaFargue C, Esgueva R, et al. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate. 2011;71:489–97. doi: 10.1002/pros.21265. [DOI] [PubMed] [Google Scholar]
- 23.Rosen P, Pfister D, Young D, Petrovics G, Chen Y, Cullen J, et al. Differences in frequency of ERG oncoprotein expression between index tumors of Caucasian and African American patients with prostate cancer. Urology Elsevier Inc. 2012;80:749–53. doi: 10.1016/j.urology.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powell IJ, Dyson G, Land S, Ruterbusch J, Bock CH, Lenk S, et al. Genes associated with prostate cancer are differentially expressed in african american and European american men. Cancer Epidemiol Biomarkers Prev. 2013;22:891–7. doi: 10.1158/1055-9965.EPI-12-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perner S, Demichelis F, Beroukhim R. TMPRSS2: ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–41. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 26.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosquera JM, Mehra R, Regan MM, Perner S, Genega EM, Bueti G, et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res. 2009;15:4706–11. doi: 10.1158/1078-0432.CCR-08-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baca S, Prandi D, Lawrence M. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–77. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demichelis F, Fall K, Perner S, Andrén O, Schmidt F, Setlur SR, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–9. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 30.Yoshimoto M, Joshua AM, Cunha IW, Coudry Ra, Fonseca FP, Ludkovski SR, et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008;21:1451–60. doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 31.Young A, Palanisamy N, Siddiqui J, Wood DP, Wei JT, Chinnaiyan AM, et al. Correlation of urine TMPRSS2:ERG and PCA3 to ERG+ and total prostate cancer burden. Am J Clin Pathol. 2012;138:685–96. doi: 10.1309/AJCPU7PPWUPYG8OH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomlins SA, Aubin SMJ, Siddiqui J, Lonigro RJ, Sefton-Miller L, Miick S, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3:94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Truong M, Yang B, Jarrard DF. Toward the detection of prostate cancer in urine: a critical analysis. J Urol. 2013;189:422–9. doi: 10.1016/j.juro.2012.04.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salami SS, Schmidt F, Laxman B, Regan MM, Rickman DS, Scherr D, et al. Combining urinary detection of TMPRSS2:ERG and PCA3 with serum PSA to predict diagnosis of prostate cancer. Urol Oncol. 2013;31:566–71. doi: 10.1016/j.urolonc.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaux A, Peskoe SB, Gonzalez-Roibon N, Schultz L, Albadine R, Hicks J, et al. Loss of PTEN expression is associated with increased risk of recurrence after prostatectomy for clinically localized prostate cancer. Mod Pathol. 2012;25:1543–9. doi: 10.1038/modpathol.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamoureux F, Zoubeidi A. Dual inhibition of autophagy and the AKT pathway in prostate cancer. Autophagy. 2013;9 doi: 10.4161/auto.24921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Squillace RM, Miller D, Wardwell SD, Wang F, Clackson T, Rivera VM. Synergistic activity of the mTOR inhibitor ridaforolimus and the antiandrogen bicalutamide in prostate cancer models. Int J Oncol. 2012;41:425–32. doi: 10.3892/ijo.2012.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winter JL, Stackhouse BL, Russell GB, Kute TE. Measurement of PTEN expression using tissue microarrays to determine a race-specific prognostic marker in breast cancer. Arch Pathol Lab Med. 2007;131:767–72. doi: 10.5858/2007-131-767-MOPEUT. [DOI] [PubMed] [Google Scholar]
- 39.American Cancer Society. Cancer Facts & Figures for African Americans 2013-2014. Atlanta: American Cancer Society; 2013. [Google Scholar]
- 40.Ateeq B, Tomlins S. Therapeutic targeting of SPINK1-positive prostate cancer. Sci Transl Med. 2011;3:1–18. doi: 10.1126/scitranslmed.3001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lippolis G, Edsjö A, Stenman UH, Bjartell A. A high-density tissue microarray from patients with clinically localized prostate cancer reveals ERG and TATI exclusivity in tumor cells. Prostate Cancer Prostatic Dis. 2013;16:145–50. doi: 10.1038/pcan.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bismar T, Yoshimoto M, Duan Q. Interactions and relationships of PTEN, ERG, SPINK1 and AR in castration-resistant prostate cancer. 2012;60:645–52. doi: 10.1111/j.1365-2559.2011.04116.x. [DOI] [PubMed] [Google Scholar]
- 43.Leinonen K, Tolonen T. Association of SPINK1 expression and TMPRSS2: ERG fusion with prognosis in endocrine-treated prostate cancer. Clin Cancer. 2010;16:2845–51. doi: 10.1158/1078-0432.CCR-09-2505. [DOI] [PubMed] [Google Scholar]
- 44.Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagai Y, Kojima T, Muro Y, Hachiya T, Nishizawa Y, Wakabayashi T, et al. Identification of a novel nuclear speckle-type protein, SPOP. FEBS Lett. 1997;418:23–6. doi: 10.1016/s0014-5793(97)01340-9. [DOI] [PubMed] [Google Scholar]
- 46.Xu J, Wu RC, O'Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615–30. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li C, Ao J, Fu J, Lee DF, Xu J, Lonard D, et al. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011;30:4350–64. doi: 10.1038/onc.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geng C, He B, Xu L, Barbieri CE, Eedunuri VK, Chew SA, et al. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc Natl Acad Sci USA. 2013;110:6997–7002. doi: 10.1073/pnas.1304502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abern M, Bassett M, Tsivian M. Race is associated with discontinuation of active surveillance of low-risk prostate cancer: Results from the Duke Prostate Center. Prostate Cancer Prostatic Dis. 2012;16:85–90. doi: 10.1038/pcan.2012.38. [DOI] [PubMed] [Google Scholar]
- 50.Bismar T, Yoshimoto M, Vollmer R. PTEN genomic deletion is an early event associated with ERG gene rearrangements in prostate cancer. BJU Int. 2011;107:477–85. doi: 10.1111/j.1464-410X.2010.09470.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


