Abstract
HuR is a ubiquitous nucleocytoplasmic RNA binding protein that exerts pleiotropic effects on cell growth and tumorigenesis. In this study, we explored the impact of conditional, tissue-specific genetic deletion of HuR on intestinal growth and tumorigenesis in mice. Mice lacking intestinal expression of HuR (Hur IKO mice) displayed reduced levels of cell proliferation in the small intestine and increased sensitivity to doxorubicin-induced acute intestinal injury, as evidenced by decreased villus height and a compensatory shift in proliferating cells. In the context of ApcMin mice, a transgenic model of intestinal tumorigenesis, intestinal deletion of the HuR gene caused a 3-fold decrease in tumor burden characterized by reduced proliferation, increased apoptosis and decreased expression of transcripts encoding anti-apoptotic HuR target RNAs. Similarly, Hur IKO mice subjected to an inflammatory colon carcinogenesis protocol (AOM-DSS administration) exhibited a 2-fold decrease in tumor burden. Hur IKO mice showed no change in ileal Asbt expression, fecal bile acid excretion or enterohepatic pool size that might explain the phenotype. Moreover, none of the HuR targets identified in Apc Min/+Hur IKO were altered in AOM-DSS treated Hur IKO mice, the latter of which exhibited increased apoptosis of colonic epithelial cells where elevation of a unique set of HuR-targeted pro-apoptotic factors was documented. Taken together, our results promote the concept of epithelial HuR as a contextual modifier of pro-apoptotic gene expression in intestinal cancers, acting independently of bile acid metabolism to promote cancer. In the small intestine, epithelial HuR promotes expression of pro-survival transcripts that support Wnt-dependent tumorigenesis, whereas in the large intestine epithelial HuR indirectly downregulates certain pro-apoptotic RNAs to attenuate colitis-associated cancer.
Keywords: Protein-RNA interaction, AU-rich RNA, mRNA stability, Post-translational regulation
Introduction
HuR is a ubiquitously expressed member of the Elav family of RNA binding proteins (1), whose regulation of target mRNA stability and/or translation reflects binding to cis-acting AU-rich elements within the 3′ untranslated region (UTR) of selected mRNAs (2). Transcriptome-wide analysis using RNA-protein crosslinking of unstimulated HeLa cells identified ~ 26,000 HuR binding sites, mainly localized in 3′ UTR regions but which also included intronic sites (3). Those studies further suggested that HuR interacts with more than 4800 genes, corresponding to approximately half of all HeLa transcripts (3). SiRNA knockdown of HuR demonstrated significantly decreased mRNA expression and concomitantly reduced protein synthesis of the majority of these candidate targets, placing HuR as a central player in a large network of factors that modulate posttranscriptional gene expression (3). This vast range of HuR targets identified in HeLa cells raises the question of whether there exists a cell-autonomous and/or developmental-stage specific hierarchy that modulates HuR-dependent pathways of gene expression in vivo and in particular how HuR might influence the development and progression of disease (4).
Several studies have correlated elevated cytoplasmic HuR expression with cancer initiation and progression in various types of human cancer cells, including lung (5), ovarian, breast, gastric, pancreatic and colon cancers (6–8). HuR analyzes in paired human normal and colon cancer revealed increased total HuR expression in tumor samples with increased cytoplasmic staining compared to the corresponding normal tissue (6). Similar findings of increased cytoplasmic HuR staining were observed in squamous esophageal cancer, where this pattern of expression was associated with decreased survival (9). These findings strongly suggest that HuR may play an important role in cancer initiation and progression and underscore the need for greater understanding of its cell- and tissue-specific roles in vivo.
Germline HuR deletion in mice leads to embryonic lethality as a result of impaired placental development (10), necessitating the use of conditional deletion strategies to examine the loss-of-function phenotype. However, global HuR deletion in adult mice also led to rapid lethality, associated with critical defects in hematopoietic progenitor cell production as well as defective intestinal stem cell dynamics and spontaneous villus atrophy (11). Those findings suggested that HuR plays a key role in intestinal epithelial growth and maintenance. More recent work used a conditional macrophage-specific HuR deletor line to reveal increased sensitivity to systemic inflammation and chemical colitis with enhanced secretion of pro-inflammatory cytokines (TNF, IL-6, IL-1b, IL-12) followed by progression to colitis-associated cancer (12).
Here we generated conditional intestine-specific HuR knockout mice (Hur IKO) and demonstrate that epithelial HuR is important for intestinal homeostasis. We generated compound Apc Min/+Hur IKO mice and demonstrate that intestinal HuR deletion attenuates small intestinal polyposis. We further show that Hur IKO mice exhibit 2-fold reduced colon tumor burden following azoxymethane and dextran sodium sulfate (AOM-DSS) administration. Both models reveal that intestinal HuR deletion protects against tumorigenesis by promoting apoptosis, but via regulation of cell-type specific transcripts.
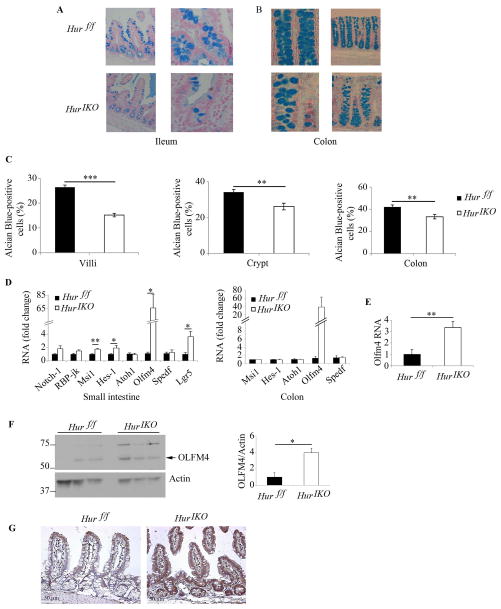
Figure 2. Conditional intestinal HuR deletion alters intestinal epithelial morphology.
Hur IKO mice show reduced number of goblet cells and increased expression of stem cell markers. Representative photographs of Alcian blue-stained sections of ileum (A) and colon (B) from Hur IKO and Hur f/f mice. C, The number of Goblet cells was scored by surveying 4 to 12 small intestinal villi, 2 to 8 small intestinal crypts and 3 to 9 colonic crypts. Results are represented as mean ± SE and expressed as percent of Alcian-blue-positive cells over total number of cells in villi and crypt respectively. (n= 4 to 7 mice/genotype) *** P<0.0001; ** P<0.01. D, Quantitative RT-PCR analysis of expression of intestinal stem cells markers. RNA was prepared from distal scraped mucosa. Data are represented as mean ± SE (n=6 animals/genotype) * P <0.05; ** P<0.001. E, Quantitative analysis of Olfm4 RNA expression in enterocytes isolated from Hur IKO and Hur f/f mice. Data represents mean ± SE (n=5 Hur f/f and 6 Hur IKO) ** P < 0.01. F, Scraped mucosa protein extracts from 3 separate Hur IKO and 3 separate controls, Hur f/f were separated on SDS-PAGE and probed with anti-OLFM4 antibody. OLFM4 expression was normalized to Actin and shown as OLFM4/Actin ratio (right panel) * P < 0.05. G, Immunohistochemical evaluation of OLFM4 shows increased expression in epithelial compartment from Hur IKO small intestine (Distal section) compared to Hur f/f controls.
Materials and Methods
Animals and experimental procedures
All mice protocols were approved by the Washington University Animal Studies Committee and conformed to criteria outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All mice were housed in a specific pathogen-free animal facility under standard 12:12-h light-dark cycle and were fed standard rodent Lab Chow (no. 5001, Purina Mills, St. Louis, MO) and tap water ad libitum. Conditional intestine-specific HuR deletor mice Hur f/f villin-Cre-ER T2 (Hur IKO) mice were generated by intraperitoneal injection of 35 day old mice with 1 mg tamoxifen (T5648, Sigma-Aldrich) for 5 consecutive days. Animals were sacrificed at 10–12 weeks of age unless stated otherwise. Hur-floxed (Hur f/f) littermates were used as controls. All animals were maintained in a C57BL/6J background. Apc min/+ Hur IKO mice were generated by crosses into Apc min/+ mice (Jax). Perinatal HuR deletion was performed by administering tamoxifen to dams of 2 days old suckling Apc min/+ Hur IKO mice. These animals are referred to as Apc min/+ Hur IKO (p). For induction of Doxorubicin injury, 8–10 week old mice were given a single intraperitoneal injection of 20 mg/kg body weight doxorubicin as described (13), (44583, Sigma-Aldrich). Animals were sacrificed 72 and 120 hr post-injection. For induction of colitis-associated cancer (CAC), 8 week old Hur f/f and Hur IKO mice were injected intraperitoneally with 10 mg/kg body weight azoxymethane (AOM) (A5486, Sigma-Aldrich). One week later, 2.5% dextran sodium sulfate (DSS) (MW 40000–50000 9011-18-1, Affymetrix Inc.) was provided in drinking water for 5 days, followed by 15 days of regular water. This cycle was repeated two more times; each time the DSS was provided for 7 days. Mice were sacrificed 12 weeks after the last DSS cycle. Tissues were collected and flash frozen for further RNA and protein analysis. Intestines were pinned and fixed in 10% formalin for scoring by screening with a Nikon SMZ800 dissecting microscope and photographed using a Photometrics CooLSNAPcf camera (Imaging Processing Services, Inc,). Measurements including width, length and total area were performed using Metavue software (Molecular Devices). Mice were injected intraperitoneally 2 hr before sacrifice with 200 μl BrdU solution (20 mg/ml) (B5002, Sigma-Aldrich).
Determination of bile acid pool size and fecal bile acid output
Bile acid pool size was determined from the total content of the entire small intestine, gallbladder and liver (14). Total bile acid mass was determined enzymatically (431–15001, Wako Chemicals) and fecal bile acid output was determined from stool collected from individually housed mice for 72 h (14, 15). Where indicated, mice of different genotypes were fed a diet containing 2% cholestyramine (C4650, Sigma-Aldrich) for 10 days, and fecal bile acid excretion determined.
Immunohistochemical analysis
Immunohistochemical analysis for H&E, HuR, OLFM4, BrdU, TUNEL and Alcian Blue/PAS were conducted on formalin-fixed paraffin embedded tissues. Sections (4 μM) were stained with the following antibodies: anti-HuR primary Antibody 1:200 (SC 5261, Santa Cruz Biotechnology); rabbit anti-OLFM4 1:100 (ab96280, Abcam Inc.) rat anti-BrdU 1:300 (Accurate Chemical & Scientific Corp). Apoptosis was analyzed by TUNEL staining following the ApopTag Peroxidase In situ Oligo Ligation (ISOL) Apoptosis Detection Kit (S7200, Chemicon International). Intestinal proliferation was determined by scoring full longitudinal sections of crypts and reported as the number of BrdU positive cells normalized to the total number of cells per crypt. Similarly the apoptotic index evaluates the number of TUNEL-positive cells normalized to total number of cells in crypt and villi. Goblet cells were visualized using Alcian blue/periodic acid-Schiff’s reagent (AB/PAS) and counterstained with hematoxylin. For each animal the number of Goblet cells was determined as percentage of total number of cells per crypt. Similarly, the number of Alcian blue/PAS–stained Goblet cells per villus was counted and expressed as percentage of the total villus cell number. For scoring cell position, each crypt was divided in half and cells were numbered sequentially from crypt base to crypt-villus junction, with cell position one being occupied by the first cell at the base of each half crypt (13). Villus length and crypt depth were evaluated on Hematoxylin-eosin (H&E)-stained sections using Axio Imager software on images captured with an Axio Imager A1 and an AxioCam MRC 5 high-resolution camera (Carl Zeiss Microimaging).
Electron microscopy
2 mm segments of the jejunum were flushed with PBS followed by immersion in 3% Glutaraldehyde (EM grade); 1% formaldehyde (EM grade) in 0.1 M sodium cacodylate (pH 7.4); 2 mM CaCl2; and 2 mM MgCl2. Specimens were stained in 2% aqueous uranyl acetate, and viewed on a Hitachi H-600 electron microscope (Hitachi High Technologies). Three animals per genotype were examined to obtain representative images.
Protein extraction and Western blotting
Scraped mucosa was homogenized in tissue lysis buffer containing 20 mM Tris (pH 8); 0.15 M NaCl; 2 mM EDTA; 1 mM sodium vanadate; 0.1 M sodium fluoride; 50 mM β-glycerophosphate; 5% glycerol, 2x protease inhibitor (Roche Applied Science); 1% Triton; and 0.1% SDS. Homogenate (60 μg protein) were resolved by 10% SDS-PAGE, transferred to PVDF membrane, and probed with mouse anti-HuR antibody (3A2)(Sc5261 Santa Cruz Biotechnology Inc.), mouse anti-Cyclin D1 (Mouse MAb, NeoMarkers, Fremont, CA), rabbit anti-C-Myc (Sc-764, Santa Cruz Biotechnology Inc.), rabbit anti-OLFM4 (ab96280 Abcam Inc.) and a rabbit anti-β-catenin antibody (9582, Cell Signaling Technology). Equal loading was verified using a rabbit anti-α actin antibody (A2066, Sigma-Aldrich) or rabbit anti-Hsp40 (SPA-400, Assay designs).
RNA extraction and quantitative PCR analysis
Total RNA was isolated from scraped mucosa (proximal, middle, distal intestine and colon) using TRIzol (Ambion), and used to synthesize cDNA using High Capacity cDNA Reverse transcription kit (Applied Biosystems). Quantitative PCR was conducted in triplicate on an ABI Step One Plus Detection System (Applied Biosystems) using SYBR GreenER qPCR SuperMix (Invitrogen).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 4.0 (GraphPad Software, Inc.) and Microsoft Excel. Data are reported as mean ± SEM. All data were subjected to one-way ANOVA and Mann-Whitney test. All data underwent two-tailed Student t-testing as deemed appropriate. For all comparisons, statistical significance was set at a p value of <0.05.
Results
Conditional intestinal HuR deletion alters intestinal epithelial proliferation
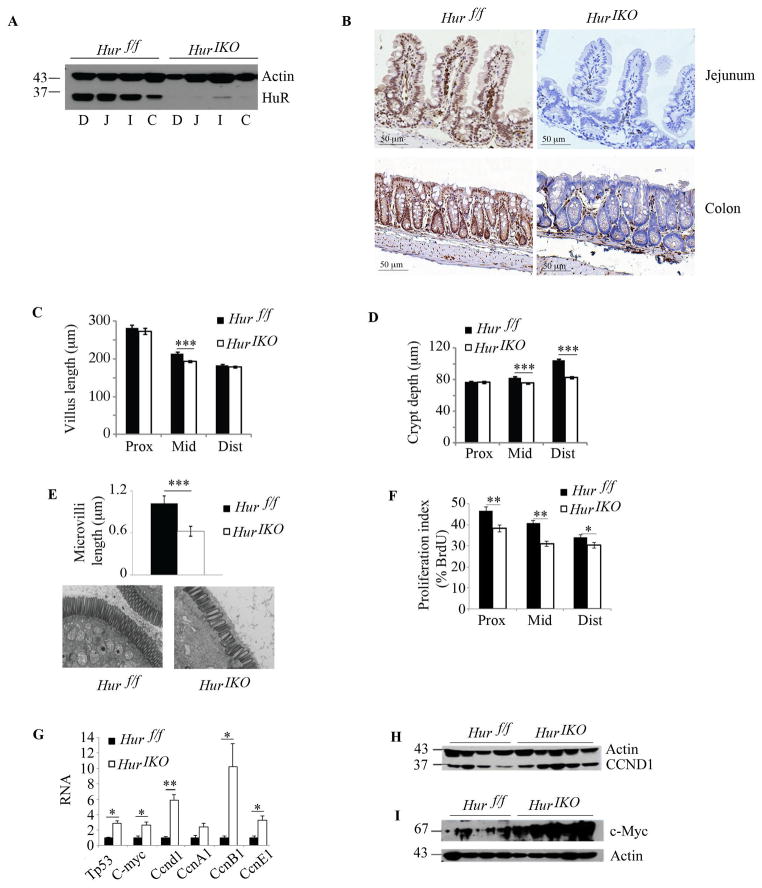
HuR expression was attenuated throughout the small intestine and colon in 10 –12 weeks old Hur IKO mice with ~ 90% reduction in protein expression compared to Hur f/f littermate controls (Figure 1A). Immunohistochemical analysis showed HuR expression in nuclei of crypt, mature villus and stromal cells of Hur f/f mice, with epithelial deletion in Hur IKO mice (Figure 1B). Hur IKO mice exhibited shorter villi and crypts (Figure 1C–D) and reduced microvillus length (Figure 1E). Hur IKO mice also exhibited reduced proliferation, evidenced by decreased BrdU-positive epithelial cells throughout the entire small intestine (Figure 1F). There was no change in intestinal apoptosis by genotype (data not shown). In order to understand the pathways that might be altered in this phenotype, we surveyed expression of RNAs encoding factors involved in cell growth and survival. We demonstrated upregulation of Tp53, C-myc, Ccnd1, CcnB1 and CcnE1 mRNAs in Hur IKO mice (Figure 1G) and a corresponding increase of CCND1 and C-Myc protein compared to Hur f/f mice (Figure 1H–I). Together these observations suggest that intestinal epithelial HuR deletion is associated with alterations in the expression of genes regulating proliferation and cell cycling programs.
Figure 1. Conditional intestinal HuR deletion (HuR IKO) alters intestinal epithelial proliferation.
A, Expression analysis of HuR following tamoxifen-induced HuR deletion in small intestine and colon. Scraped mucosal protein extracts from 10 –12 weeks-old Hur IKO and control, Hur f/f were separated on SDS-PAGE and probed with anti-HuR antibody. Duodenum (D), Jejunum (J), ileum (I), colon (C). Actin was used as loading control. B, Immunohistochemical evaluation of small intestine (jejunum) and colon confirms epithelium-specific HuR deletion as compared with littermate control animals Hur f/f. C–D, Villi length and crypt depth were compared between Hur IKO and controls Hur f/f mice. Eight to 10 mice per genotype were evaluated. On average, 30 villi and 14 crypts per animal were examined. Data represent mean ± SE, *** P<0.0001. E, Microvillus length was evaluated by electron microscopy and reported as mean ± SE, *** P<0.0001. Representative photographs for each genotype are shown below. F, Evaluation of BrdU incorporation across the entire small intestine. Proliferation index is expressed as percentage of BrdU-positive epithelial cells over the total number of cells per crypt. Data represent mean ± SE (n=8–10 mice/genotype), ** P<0.001; * P<0.05. 20 crypts were evaluated per animal. G, Cell growth related gene expression was evaluated by quantitative RT-PCR. Data normalized to Gapdh RNA expression are represented as mean ± SE (n=4 to 6 animals/genotype) * P <0.05; ** P <0.001. H and I, Western blot analysis of cyclin D1 (CCND1) and c-Myc, respectively, in small intestine scraped mucosa from Hur IKO and Hur f/f control mice. Actin is shown as loading control.
Conditional intestinal HuR deletion alters intestinal epithelial morphology
Alcian blue staining revealed reduced goblet cells in the small intestine (Figure 2A, C) and colon (Figure 2B, C) of Hur IKO mice. These changes were associated with a 2-fold increase in Musashi1 (Msi1) and Hes-1 mRNA in Hur IKO mice (Figure 2D), both of which have been implicated in regulating mammalian intestinal development (16, 17). The findings with Msi1 mRNA expression were unexpected, since HuR has been demonstrated to stabilize Msi1 RNA and promote its translation (18). However, because of the key role of Notch signaling in intestinal differentiation (19) we analyzed the effect of Hur deletion on the expression of both Notch1 receptor and of RBP-jk, its downstream target. Both RNAs showed a trend towards increased expression in Hur IKO mice (Figure 2D), suggesting that Notch dependent regulation cannot be invoked as an explanation of the upregulation of Msi1 and Hes-1 expression in Hur IKO animals, implying that yet other pathways must also be in play. Hur IKO mice also showed a >60-fold increase in mRNA abundance of Olfm4 in small intestine (and a similar trend in colon), with a ~3-fold induction of Lgr5 RNA (Figure 2D). The induction of Olfm4 mRNA was also confirmed in isolated enterocytes from Hur IKO mice (Figure 2E). The induction of Olfm4 expression in Hur IKO mice was further confirmed by verifying increased OLFM4 protein in Hur IKO small intestine (Figure 2F, G). These findings together suggest that intestinal HuR deletion alters intestinal homeostasis and Lgr5-dependent pathways of differentiation, most likely through Notch-independent mechanisms (20).
Aged Hur IKO mice exhibit normal intestinal proliferation and no alteration of cell growth-related factors
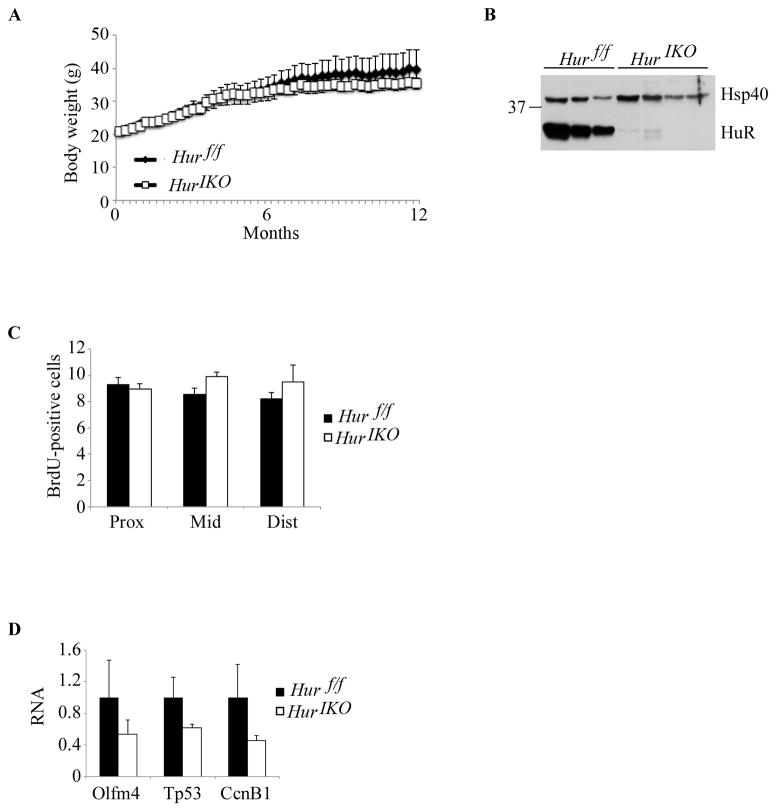
Since the effect of epithelial HuR deletion in young (~12 week old) mice were rather subtle, we asked whether prolonged HuR deletion of over one year might lead to a more dramatic phenotype. However, this turned out not to be the case. Aged Hur IKO mice grew comparably to Hur f/f controls (Figure 3A). Also, despite evidence that intestinal HuR protein expression was still attenuated (Figure 3B), small intestinal proliferation was no longer reduced in aged Hur IKO mice (Figure 3C). Furthermore, the expression of RNAs encoding factors involved in intestinal homeostasis, cell growth and survival—all of which were up-regulated in young mice (Figure 1G)—were unchanged at one year (Figure 3D). These findings suggest that there is a temporal component to the adaptive effects of intestinal epithelial HuR deletion in modulating compensatory pathways involved in intestinal integrity.
Figure 3. One year-old Hur IKO mice shows normal intestinal epithelial proliferation.
A, Average body weight of aged show-fed Hur IKO and Hur f/f mice. B, HuR conditional knockout was confirmed by Western blot of intestinal scraped mucosa from 3 individual Hur f/f and 4 Hur IKO mice. Hsp40 was used as loading control. C, Quantitation of BrdU incorporation in the small intestine. Data are represented as mean ± SE (n=4 animals/genotype). D, Quantitative RT-PCR analysis of Olfm4 and cell growth related factors. RNA was extracted from distal scraped mucosa. Data are represented as mean ± SE (n= 4 animals/genotype).
HuR IKO mice exhibit increased injury following doxorubicin administration
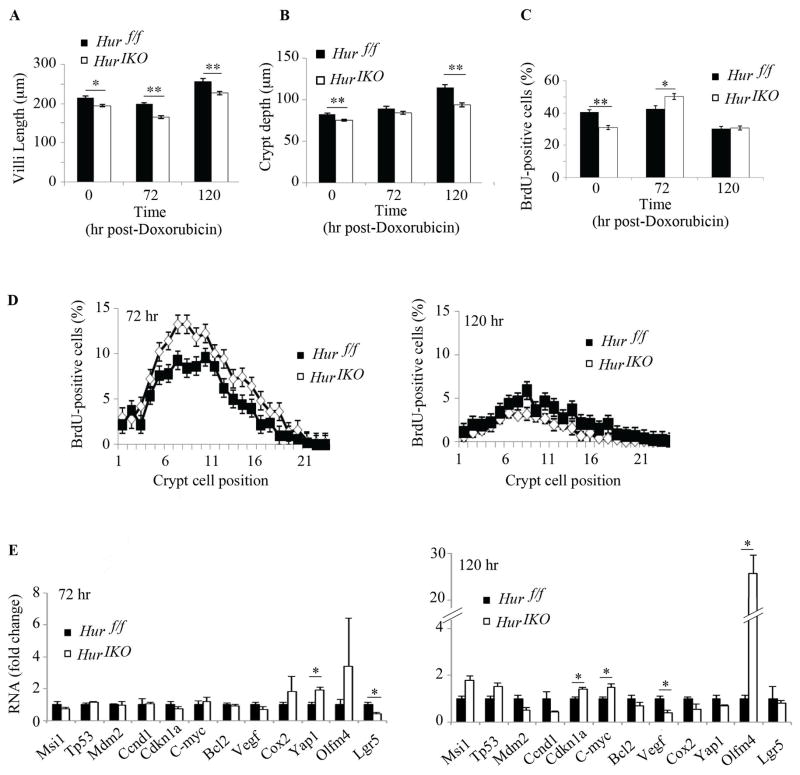
The findings to this point suggest that intestinal HuR deletion results in subtle alterations in basal small intestinal homeostatic regulation. We next examined the response of Hur IKO mice to an acute chemical injury, in which intestinal morphology was examined at 72 (peak of injury) and 120 h (regenerating phase) following doxorubicin injection (13). The findings reveal subtle yet significant increased injury in Hur IKO mice, with decreased villus length compared to baseline (and significantly shorter than Hur f/f villi at all time points, Figure 4A). Hur IKO mice showed a significant increase in compensatory proliferation at the peak damage phase (Figure 4C) associated with a greater number of proliferating cells (positions +4 and above, i.e transit-amplifying cells, Figure 4D), suggesting expansion of crypt progenitor cells. 72h after injury, there was a significant increase in mRNA encoding the growth-promoting factor Yap1 in Hur IKO mice (21) followed by a decline at 120h (Figure 4E). Expression of mRNA encoding Lgr5 was significantly reduced at 72h in Hur IKO mice (Figure 3E). We observed increased mRNA abundance of Olfm4, Cdkn1a and C-myc and a significant decrease in Vegf RNA at 120h in Hur IKO mice (Figure 4E). These findings, coupled with the increased compensatory proliferation observed in Hur IKO mice suggest that loss of epithelial HuR increases sensitivity to acute intestinal injury likely through multiple adaptive pathways.
Figure 4. HuR IKO mice exhibit increased injury following doxorubicin administration.
Time-dependent changes in villus length and crypt depth following cytotoxic small intestinal injury. For all measurements, numbers of animals per genotype and per time points are as followed: 72 hr: n=5 animals/genotype; 120 hr: 3 animals/genotype. Evaluations shown here are from the mid small intestine. A, Villus length was measured 72 and 120 hr following doxorubicin injection. (20–40 villi were measured at 72 and 120 hr post-doxorubicin injection). Measurements represent mean ± SE, * P <0.05; ** P <0.001. B, crypt depth was evaluated in 10 to 20 crypts from mice sacrificed 72 hr post-doxorubicin injection and in 15–20 crypts from mice sacrificed 120 hr post-doxorubicin. Crypt depth is significantly reduced in Hur IKO mice compared to Hur f/f mice. Data are represented as mean ± SE, ** P < 0.001. C, BrdU incorporation was evaluated 72 and 120 hr post-doxorubicin injection. Ten and twenty crypts per animals were analyzed at 72 and 120 hr post-doxorubicin respectively. Proliferative cells counts are expressed as number of BrdU-positive cells per crypt (mean ± SE) * p <0.05; ** P< 0.0001. D, The position of proliferative cells in the crypt was recorded during the damage phase (72hr) and during repair phase (120hr). Data are represented as percent of BrdU-positive cells at each position relative to total number of cell in the crypts. E, Quantitative RT-PCR analysis of mid-jejunum mRNA expression at peak damage and repair phases. Data were normalized to Gapdh mRNA expression and reported as mean ± SE (n= 3 animals/genotype), * P<0.05.
Intestinal HuR deletion attenuates polyposis in Apc min/+ mice
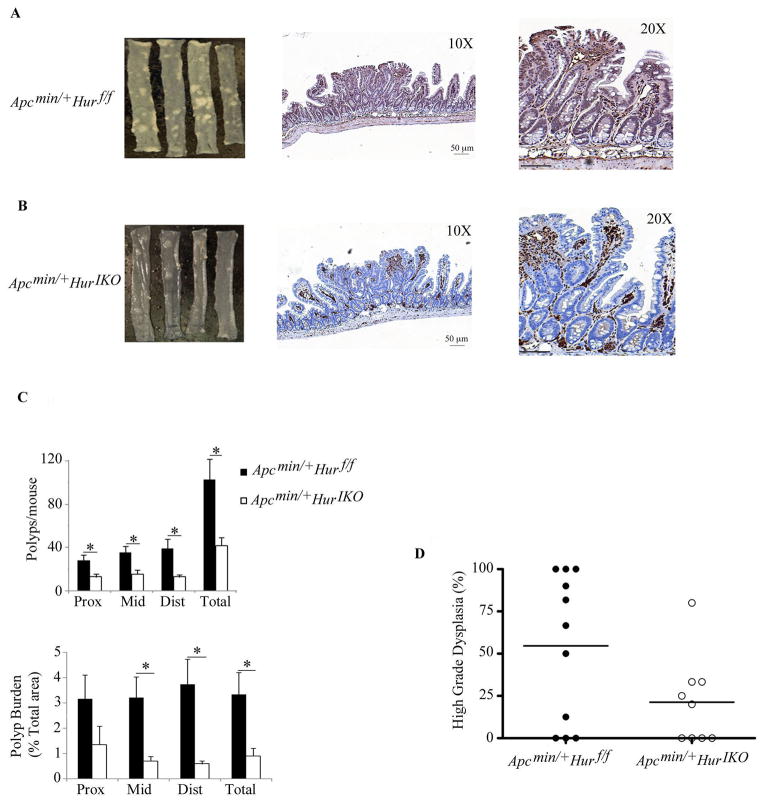
The aforementioned results showing altered expression of cell cycle genes and an increased proliferative response to acute injury in Hur IKO mice raised the question of whether intestinal HuR deletion would promote or attenuate intestinal tumorigenesis. To address this question we initiated conditional HuR deletion in 5 week old compound Apc min/+ Hur IKO mice and examined spontaneous intestinal polyposis at 103 days i.e. after 68 days of HuR deletion. Using this protocol, we observed reduced polyp formation in Apc min/+ Hur IKO mice compared to controls (Figure 5A, B), with immunohistochemical validation of epithelial-specific HuR expression (Figure 5B). Apc min/+ Hur IKO mice exhibited a 60% reduction in tumor number (mean of 42 polyps/mouse Apc min/+ Hur IKO vs 102 polyps/mouse in Apc min/+ Hur f/f) and a 70% decrease in tumor burden (estimated from total area) across the entire small intestine (Figure 5C). The tumors in Apc min/+ Hur IKO mice were also significantly smaller (Figure 5C). Colonic polyp burden was also significantly reduced (~ 3-fold) in Hur IKO mice (data not shown). Further analysis revealed a trend towards decreased high-grade dysplasia in polyps from Apc min/+ Hur IKO mice (Figure 5D). These observations collectively suggest that deletion of intestinal epithelial HuR attenuates tumorigenesis. We further investigated whether perinatal deletion of epithelial HuR would alter tumor initiation. In this approach, we undertook conditional intestinal HuR deletion in 2 day-old Apc min/+ Hur IKO mice and analyzed tumor burden 100 days later. Surprisingly, perinatal HuR-deleted animals showed no difference in tumor burden (or dysplasia) compared to control animals (Supplemental Figure 1C–D) indicating that early postnatal epithelial HuR is not essential for small intestinal adenoma initiation.
Figure 5. Intestinal HuR deletion attenuates polyposis in Apcmin/+ mice.
A–B, Left panels, representative gross morphology of polyps through the entire small intestine from Apc min/+ Hur f/f (A) and Apc min/+ Hur IKO mice (B). Middle and right panels, HuR-stained sections of small intestine showing HuR deletion restricted to the epithelial cell subpopulation. Scale bars 50 μm. C, Top panel, absence of HuR is associated with reduced polyp number per mouse and reduced polyp area in each third of small intestine (lower panel) (proximal (Prox), middle (Mid) and distal (Dist); (n=10 mice/genotype) * P< 0.05. D, Trend to reduced high-grade dysplasia in polyps from Apc min/+ Hur IKO mice (P<0.06).
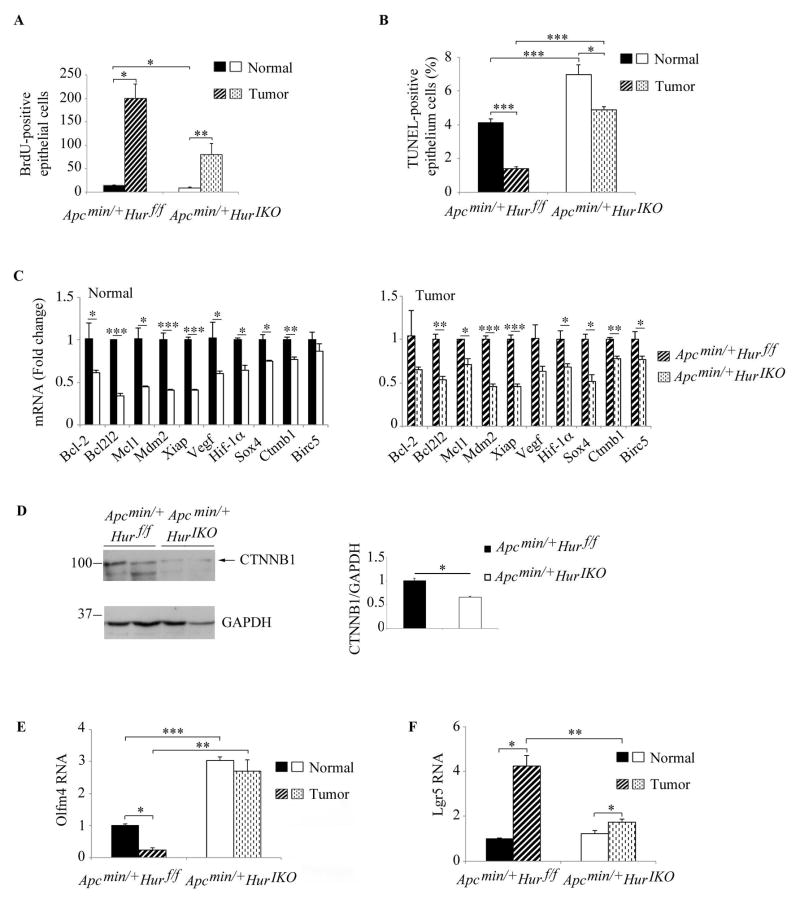
We next turned to an examination of the potential pathways by which intestinal HuR deletion modifies the tumor susceptibility phenotype. Intestinal tissue (both normal, uninvolved and polyp) from Apc min/+ Hur IKO mice demonstrated a two-fold reduction in proliferation compared to control (Figure 6A). In addition, TUNEL staining revealed a significant (~1.8 fold) increased apoptosis in normal tissues from Apc min/+ Hur IKO mice compared to Apc min/+Hur f/f controls (Figure 6B). Apoptosis was significantly elevated (~ 5 fold) in Hur IKO polyps compared to Hur f/f polyps (Figure 6B). These data suggest that small intestine epithelial HuR influences both enterocyte proliferation and apoptosis, which in turn attenuates tumor susceptibility.
Figure 6. Intestinal HuR deletion attenuates proliferation and increases apoptosis in normal and polyp tissues in Apcmin/+ mice.
A, Proliferative cells were counted and expressed as means ± SE of BrdU-positive cells (n= 2–5 mice/genotype; 2–4 40X fields were surveyed in Apc min/+ Hur f/f mice and 3–10 40X fields in Apc min/+ Hur IKO mice) * P< 0.05; ** P < 0.01. B, Apoptotic cells were visualized by TUNEL staining and counted. Counts are expressed as percent of TUNEL-positive cells. Data represent mean ± SE (n=10/genotype; 20 40X fields per animal and genotype were analyzed) * P<0.05; *** P < 0.0001. C, mRNA expression of apoptosis-related genes. RNAs extracted from full thickness normal and polyp tissues were analyzed by quantitative PCR and expression normalized to Gapdh mRNA. Data were compared to RNAs isolated from Apc min/+ Hur f/f mice and reported as means ± SE (n= 4 animals/genotype) * P <0.05; ** P <0.01; *** P <0.001. D, Full thickness protein extracts were separated on SDS-PAGE and β-catenin protein (CTNNB1) expression analyzed by Western blot. GAPDH was used for normalization as loading control. E–F, Quantitative RT-PCR analysis of Olfm4 and Lgr5 expression. Data are represented as mean ± SE, * P<0.05; ** P<0.01; *** P<0.001.
Numerous studies in cancer cell lines have shown that RNAs encoding several anti-apoptotic factors are targets of HuR and that RNA binding promotes mRNA stability and increases mRNA expression (11, 22–27). Accordingly, we hypothesized that HuR deletion may be associated with decreased expression of these candidates RNAs, which might in turn lead to activation of apoptotic pathways. Quantitative PCR analysis of anti-apoptotic RNA (reported targets of HuR) revealed reduced expression of Bcl2l2, Xiap, Hif-1α mRNAs in both normal and tumor tissues from Apc min/+ Hur IKO mice compared to Apc min/+ Hur f/f animals (Figure 6C). Apc min/+ Hur IKO mice also demonstrated reduced β-catenin RNA and a corresponding decrease in protein expression (Ctnnb1) (Figure 6C, D). The downstream β-catenin target, Survivin (Birc5) RNA was significantly downregulated in Hur IKO polyps (Figure 6C), consistent with attenuated activation of Wnt-dependent signaling.
Based on findings (above, Figures 2, 4) that expression of Olfm4 mRNA was upregulated in Hur IKO small intestine, we analyzed Olfm4 RNA expression in normal and polyp tissue from Apc min/+ Hur IKO mice. These findings revealed a 3-fold increase in Olfm4 mRNA in normal tissue from Apc min/+ Hur IKO compared to Hur f/f mice (Figure 6E). Olfm4 mRNA expression was decreased 5-fold in Hur f/f tumor, but was unchanged in polyp tissue from Apc min/+ Hur IKO mice, further magnifying the differential induction of Olfm4 mRNA by genotype (Figure 6E). In addition, the expression of Lgr5 mRNA was increased 4 fold in Hur f/f polyp compared to normal tissue. By contrast, Lgr5 mRNA was induced 1.4 fold in Hur IKO tumors with attenuated mRNA expression compared to Hur f/f tumor (Figure 6F). These findings collectively suggest that in a genetic background of spontaneous intestinal tumorigenesis (ie Apc Min/+) epithelial HuR may regulate actively proliferating stem cells, and in turn function to promote growth via induction of anti-apoptotic factors.
Intestinal HuR deletion does not influence ASBT expression or bile acid homeostasis
Among the candidate intestinal mRNAs for which HuR has been implicated in regulating expression is the ileal sodium-dependent bile acid transporter (Asbt), with in vitro data suggesting that intestinal epithelial HuR may stabilize Asbt mRNA, increasing its expression and promoting bile acid absorption and recycling (28). This possibility is important since studies have linked altered bile acid metabolism with colorectal cancer (29, 30). However, we found no changes in ASBT gene expression or any parameter of enterohepatic bile acid metabolism in Hur IKO mice (Supplemental Figure 2).
Intestinal HuR deletion protects against AOM-DSS cancer associated colitis
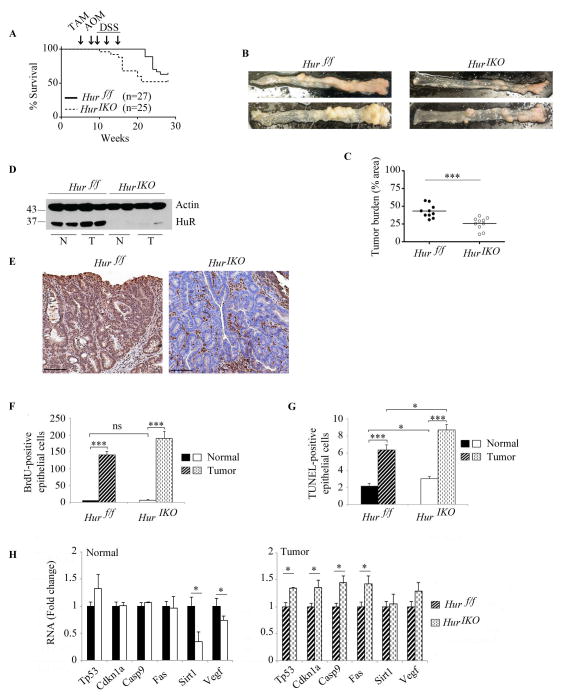
We extended the phenotypic characterization of intestinal HuR deletion to yet another model of tumorigenesis, namely colitis associated cancer. These studies were prompted in part by work indicating the key role of inflammatory cytokines in oncogenesis, including many cytokine mRNAs identified as targets of HuR (31, 32). In addition, other work demonstrated that myeloid-specific HuR expression modulates intestinal inflammation and carcinogenesis (12). Those findings demonstrated that myeloid-lineage specific HuR deletion dramatically increased susceptibility to colitis associated cancer, while myeloid transgenic HuR overexpression was protective (12). We deleted HuR in 35 day old mice and exposed the Hur IKO mice to AOM followed by 3 cycles of DSS. Hur IKO mice began to die after the first cycle of DSS (1 animal out of 25) (Figure 7A). Five out of 25 (20%) Hur IKO mice died after the third cycle of DSS. At the time of sacrifice (12 weeks after the last cycle of DSS) 48 % (12/25) of the treated Hur IKO mice died, a 52% survival rate. By contrast, 11% (3/27) of Hur f/f controls died 6 weeks after the last cycle DSS (Figure 7A). At time of sacrifice, 10/27 Hur f/f mice died, a 63% survival rate. These findings suggest that Hur IKO mice might be more sensitive to DSS exposure since they appear to die earlier than Hur f/f controls, despite a similar overall survival rate for the entire experiment (ie 52% vs 63%).
Figure 7. Intestinal HuR deletion protects against AOM-DSS cancer associated colitis.
A, Kaplan-Meier survival curves of Hur IKO and Hur f/f after exposure to AOM-DSS (n=27 Hur f/f and 25 Hur IKO mice). Arrows indicate Tamoxifen-induced Hur knockout (week 5) followed by one injection of AOM (week 8) and three consecutive exposures to DSS (weeks 9, 12 and 15). Log-rank test (Mantel-Cox) was not significant (P value 0.137). B, Gross morphology photographs of colon from Hur f/f and Hur IKO 20 weeks after exposure to AOM-DSS. C, Pinned colons were analyzed under dissecting microscope and surveyed for total polyp number and size relative to total colon area. Hur IKO mice show a significantly reduced tumor burden, *** P<0.001. D, Western blot analysis of HuR in protein extracts prepared from full thickness normal (N) and tumor (T) tissues isolated from Hur f/f and Hur IKO mice treated with AOM-DSS. Note the increased expression of HuR in tumor isolated from control animals. E, Representative HuR staining of polyps in colon from Hur f/f and Hur IKO mice. Note the complete absence of HuR in epithelial cells with staining confined to the stroma. Scale bars 50 μm. F, Hur IKO mice treated with AOM-DSS show no alteration of cell proliferation. BrdU-positive epithelial cells were counted and data are represented as total number of BrdU-positive cells per high magnification power (40X). On average 16 fields were evaluated per mouse. (n=3 to 4 mice per genotype). Data represent mean ± SE, *** P<0.001. G, Hur IKO mice show increased apoptosis 20 weeks after AOM-DSS administration. Apoptotic cells were visualized by TUNEL staining and counted. Data represent total number of TUNEL-positive epithelial cells per high-resolution field (40X magnification). Six to 10 fields were analyzed per mouse. (n=3 to 5 mice per genotype). Data represent mean ± SE, * P<0.05; *** P< 10 −10. H, Following AOM-DSS exposure, Hur IKO mice showed elevated expression of a subset of RNAs encoding pro-apoptotic factors. RNA was extracted from normal and tumor tissues. RNA expression was evaluated by quantitative PCR and normalized to Gapdh RNA. Data are represented as mean ± SE (n=3 to 5 per genotype) * P <0.05.
Nevertheless, at the time of sacrifice (ie 161 days after HuR deletion) the surviving Hur IKO mice manifested a reduced colonic tumor burden compared to Hur f/f mice (Figure 7B and C). Western blot analysis of colon tissue verified attenuated HuR expression in Hur IKO mice (Figure 7D). There was a subtle but reproducible increase in HuR protein expression in Hur f/f tumor tissue compared to normal (Figure 7D), consistent with the increased HuR expression previously reported in colorectal and other cancers (6, 7). Immunohistochemical analysis of HuR expression in Hur IKO mice revealed staining restricted to stromal cells (Figure 7E). Epithelial proliferation was increased in tumor tissue compared to normal colon in both genotypes but there was no difference in tumor proliferation by genotype (Figure 7F). By contrast Hur IKO mice showed increased TUNEL-positive epithelial cells compared to AOM-DSS-treated Hur f/f controls (Figure 7G). These findings suggest that increased apoptosis in tumors from Hur IKO mice may contribute to the attenuated tumor burden in colitis associated cancer.
To investigate the pathways accounting for the increased apoptosis with HuR deletion, we analyzed the expression of pro- and anti-apoptotic HuR RNA targets (11, 22–27). Two RNAs encoding factors with anti-apoptotic activity, Sirt1 and Vegf both of which have been demonstrated to undergo HuR-dependent modulation in other settings (33–35), were downregulated in normal tissue from Hur IKO mice (Figure 7H). Vegf-A RNA was also recently verified as an HuR target in mouse macrophages (36). Other pro-apoptotic factors (Tp53, Cdkn1a, Casp9 and Fas) showed unchanged RNA expression in normal tissues from Hur IKO vs Hur f/f mice, but these mRNAs were significantly upregulated in Hur IKO tumor tissues (Figure 7H). The expression of these RNAs is also presented by genotype, comparing mRNA levels in tumor versus normal tissue (Supplemental Figure 3). These observations together suggest that in the background of AOM-DSS induced colitis-associated cancer, HuR deletion promotes tumor apoptosis by activation of pathways including the p53-dependent intrinsic apoptotic pathway as well as Casp9, Cdkn1a and Fas.
Discussion
The central findings of this study are that conditional, intestinal epithelial specific HuR deletion in adult mice produces a subtle and temporally-dependent growth phenotype, with an exaggerated response to cytotoxic injury and attenuation of both spontaneous intestinal polyposis and colitis-associated cancer. The findings collectively highlight the role of HuR in modulating tissue and cell-specific pathways of small intestinal and colonic epithelial homeostasis and also the role of epithelial HuR expression in the initiation and progression of intestinal tumorigenesis. Several elements of these overarching conclusions merit additional discussion.
Among the key observations was that conditional intestinal epithelial-specific HuR deletion in adult mice is not critical for survival. Earlier studies using tamoxifen-inducible Rosa26/Cre mediated HuR deletion in young adult (8 weeks old) mice demonstrated marked abnormalities in the gastrointestinal tract within two to four days of (global) HuR deletion and lethality within 10 days (11). In particular, those studies demonstrated villus atrophy, decreased crypt proliferation and goblet cell loss, findings reminiscent of those observed in the current study (11). However, a major distinction in the observations from the current study and those earlier findings is that intestinal atrophy with villus blunting in Hur IKO mice was more modest and not progressive, compared to the global knockout model. These findings suggest that the gross morphological disruption of both small intestine and colonic mucosal architecture following global HuR deletion may reflect the loss of stromal HuR expression in addition to the loss of enterocyte-specific HuR expression. It is worth noting that epithelial-specific HuR deletion virtually eliminated HuR protein expression (Figure 1A, B) from scraped mucosal samples of small intestine and full thickness colonic extracts (both of which would include epithelial and stromal compartments), suggesting that even a small population of mesenchymal or stromal cells expressing HuR must play a crucial role in epithelial maintenance. The exaggerated response to acute cytotoxic small intestinal injury mediated by doxorubicin exposure underscores the role of HuR in maintaining intestinal homeostasis by controlling stem cell proliferation and subcellular lineage. Among the pathways and mediators implicated in these adaptive processes, we observed upregulation of TP53 as observed earlier in the global HuR deletion experiments (11). However, we did not observe a change in Mdm2 expression in small intestine or colon of Hur IKO mice (data not shown) as noted in early time points (48h) following global HuR deletion. By contrast, we observed increased expression of Musashi 1 in the small intestine of Hur IKO mice while no changes were observed at 48h following global HuR deletion (11). Our observations contrast with the changes noted in the setting of global HuR deletion and strongly suggest that there is a temporal as well as a cell-specific (ie spatial) component to the adaptive pathways and phenotypes observed following intestinal HuR deletion.
Among the most highly altered mRNAs detected following intestinal HuR deletion, we observed a striking increase in olfactomedin4 (Olfm4) expression in both small intestine and colon. Olfm4 encodes a glycoprotein implicated in promoting cell proliferation as well as cell adhesion and migration (Reviewed in (37)). Of particular relevance to the current study, Olfm4 overexpression was detected in 90% of human colon cancer tumors compared to normal, uninvolved tissue with increased expression in a variety of other solid tumors (38). However, yet other studies observed that Olfm4 also functions as a tumor suppressor and impairs metastasis in an orthotopic model of metastatic mouse melanoma, suggesting that the role of Olfm4 in cancer may be cell specific (39). We observed increased Olfm4 mRNA expression both at baseline and also in the adapting small intestine following doxorubicin mediated injury, raising the important question of whether this transcript is a direct or indirect target of HuR and what (if any) effects are observed on the expression of its cognate protein product. A survey of the RNA array data sets from gain- and loss-of-HuR knockdown in HeLa and RKO cells did not report changes in Olfm4 RNA (3, 6). However, computational analysis of Par-Clip HuR RNA targets revealed a preference for AU-rich motifs and both mouse and human Olfm4 3′ UTR contain canonical HuR motifs (UUUUUUU, UUUGUUU, UUUAUUU) as well as preferred AU-rich motifs (AUUUUUUA, AUUUAUU) raising the possibility that Olfm4 may be an HuR RNA target. However, while Olfm4 mRNA was upregulated in intestine from young (12 week old) Hur IKO mice, there was no change in aged Hur IKO mice, suggesting that there may be indirect, temporally regulated, mechanisms of Olfm4 regulation in the setting of HuR deletion.
The loss of goblet cells observed in Hur IKO mice, coupled with the increased proliferation observed at 72h in response to doxorubicin injury (Figure 3D), raised the possibility that Hur IKO mice might be more tumor prone in the background of altered Wnt signaling (ie in the Apc Min/+ background). However, this turned out not to be the case. The findings demonstrated reduced small intestinal tumor burden in Hur IKO mice, in association with reduced proliferation and increased apoptosis. These findings are consistent with earlier observations from global HuR deletion where decreased number of progenitor cells was also coupled with decreased proliferation and increased apoptosis (11). The observation that tumor burden in perinatal Apc min/+ HuR IKO mice was indistinguishable from control mice suggest that perinatal small intestinal epithelial HuR is not essential for adenoma initiation. Further studies focusing on mesenchymal/stromal HuR deletion will be required to examine the contribution of non-epithelial intestinal HuR in tumorigenesis and the cross talk between stromal and epithelial compartments in tumor initiation and progression (40–42).
The current findings also begin to address our understanding of the role of intestinal HuR in colonocyte homeostasis and in colitis-associated cancer. HuR has been implicated in stabilizing Asbt mRNA, increasing its expression postnatally in rodents (43). However, we found no change in Asbt mRNA or protein expression and no changes in fecal bile acid output or enterohepatic pool size in Hur IKO mice, under either basal conditions or with increased fecal bile acid losses. These findings make it extremely unlikely that any of the intestinal phenotypes observed in Hur IKO mice are in any way related to altered bile acid metabolism.
We next turned to an examination of the role of intestinal HuR expression in the setting of colitis-associated cancer. Those studies were prompted by the results of studies in which transgenic overexpression of HuR in macrophages and myeloid cells attenuated colon inflammation while myeloid-specific HuR deletion enhanced endotoxemia and exaggerated the severity of colitis-associated cancer (12). The current findings demonstrate that HuR IKO mice manifest attenuation of colitis-associated cancer. The molecular mechanisms likely include reduced expression of the anti-apoptotic factors Vegf and Sirt1 in normal tissue, as well as enhanced expression of pro-apoptotic factors involved in the p53-dependent and death receptor pathways (Tp53, Casp9, Fas). The decreased expression of Vegf and Sirt1 mRNAs in Hur IKO mice may be a direct effect since HuR is reported to stabilize these RNAs (33, 36, 44). In addition, previous studies have shown a positive correlation between cytoplasmic accumulation of HuR and Vegf proteins with increased angiogenesis and tumor size (45). The mechanisms accounting for the upregulation of candidate pro-apoptotic factors (Capsase 9, Vegf and TP53) are likely to include indirect and possibly multicomponent interactions in vivo, as alluded to above. However such suggestions will require further validation, particularly because of the well recognized discordance between the stabilizing effects of HuR on target mRNA half-life in cell lines versus the effects noted upon transgenic overexpression or targeted HuR deletion in-vivo (12).
In summary, the current studies illustrate the important role of enterocyte-specific HuR expression in small intestinal epithelial homeostasis and in two models of tumorigenesis. The finding that intestinal HuR deletion is protective in these two distinct models of intestinal tumorigenesis implies the need for greater understanding of the pathways involved in order to exploit these observations in a therapeutic context. Such understanding will need to include approaches to dissect the molecular mechanisms involved in a tissue- and cell-type dependent manner and should take into consideration the important temporal role of epithelial HuR in preserving intestinal integrity. That being said, our findings imply that targeting epithelial HuR within a critical time window might be an approach to attenuate intestinal tumor progression.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by grants (HL-38180, DK-56260 and DDRCC DK52574, particularly murine and imaging cores, to NOD), an American Society of Colon and Rectal Surgeons Fellowship award (GM), an American Society of Colon and Rectal Surgeons Research Foundation Career Development Award (SD) and an NCI Centers for Translational Research on Energetics and Cancer (TREC) U54CA155496 Pilot Project Award (SD), and HL-49094 to TH.
The authors are grateful to Kym Carter for outstanding technical support.
Footnotes
The authors declare that there are no conflicts of interest to report.
Author contributions: Study design: AG, VB, NOD; Performed experiments: AG, VB, GM, CK, YX, SK, JL; Key Reagent Provision: SC, TH; Data interpretation and analysis: AG, VB, GM, CK, SC, TH, IN, SD, NOD; Wrote manuscript: AG, VB, NOD with input from all the authors.
References
- 1.Lu JY, Schneider RJ. Tissue distribution of AU-rich mRNA-binding proteins involved in regulation of mRNA decay. The Journal of biological chemistry. 2004;279(13):12974–9. doi: 10.1074/jbc.M310433200. [DOI] [PubMed] [Google Scholar]
- 2.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58(2):266–77. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, et al. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43(3):340–52. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Srikantan S, Gorospe M. HuR function in disease. Frontiers in bioscience. 2012;17:189–205. doi: 10.2741/3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaxall BC, Dwyer-Nield LD, Bauer AK, Bohlmeyer TJ, Malkinson AM, Port JD. Differential expression and localization of the mRNA binding proteins, AU-rich element mRNA binding protein (AUF1) and Hu antigen R (HuR), in neoplastic lung tissue. Mol Carcinog. 2000;28(2):76–83. [PubMed] [Google Scholar]
- 6.Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, et al. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22(46):7146–54. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 7.Lopez de Silanes I, Lal A, Gorospe M. HuR: post-transcriptional paths to malignancy. RNA biology. 2005;2(1):11–3. doi: 10.4161/rna.2.1.1552. [DOI] [PubMed] [Google Scholar]
- 8.Burkhart RA, Pineda DM, Chand SN, Romeo C, Londin ER, Karoly ED, et al. HuR is a post-transcriptional regulator of core metabolic enzymes in pancreatic cancer. RNA biology. 2013;10(8):1312–23. doi: 10.4161/rna.25274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C, Xue G, Bi J, Geng M, Chu H, Guan Y, et al. Cytoplasmic expression of the ELAV-like protein HuR as a potential prognostic marker in esophageal squamous cell carcinoma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(1):73–80. doi: 10.1007/s13277-013-1008-4. [DOI] [PubMed] [Google Scholar]
- 10.Katsanou V, Milatos S, Yiakouvaki A, Sgantzis N, Kotsoni A, Alexiou M, et al. The RNA-binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development. Mol Cell Biol. 2009;29(10):2762–76. doi: 10.1128/MCB.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh M, Aguila HL, Michaud J, Ai Y, Wu MT, Hemmes A, et al. Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. The Journal of clinical investigation. 2009;119(12):3530–43. doi: 10.1172/JCI38263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yiakouvaki A, Dimitriou M, Karakasiliotis I, Eftychi C, Theocharis S, Kontoyiannis DL. Myeloid cell expression of the RNA-binding protein HuR protects mice from pathologic inflammation and colorectal carcinogenesis. The Journal of clinical investigation. 2012;122(1):48–61. doi: 10.1172/JCI45021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dekaney CM, Gulati AS, Garrison AP, Helmrath MA, Henning SJ. Regeneration of intestinal stem/progenitor cells following doxorubicin treatment of mice. American journal of physiology Gastrointestinal and liver physiology. 2009;297(3):G461–70. doi: 10.1152/ajpgi.90446.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarz M, Russell DW, Dietschy JM, Turley SD. Marked reduction in bile acid synthesis in cholesterol 7alpha-hydroxylase-deficient mice does not lead to diminished tissue cholesterol turnover or to hypercholesterolemia. Journal of lipid research. 1998;39(9):1833–43. [PubMed] [Google Scholar]
- 15.Setchell KD, Lawson AM, Tanida N, Sjovall J. General methods for the analysis of metabolic profiles of bile acids and related compounds in feces. Journal of lipid research. 1983;24(8):1085–100. [PubMed] [Google Scholar]
- 16.Lan SY, Yu T, Xia ZS, Yuan YH, Shi L, Lin Y, et al. Musashi 1-positive cells derived from mouse embryonic stem cells can differentiate into neural and intestinal epithelial-like cells in vivo. Cell biology international. 2010;34(12):1171–80. doi: 10.1042/CBI20100108. [DOI] [PubMed] [Google Scholar]
- 17.Ueo T, Imayoshi I, Kobayashi T, Ohtsuka T, Seno H, Nakase H, et al. The role of Hes genes in intestinal development, homeostasis and tumor formation. Development. 2012;139(6):1071–82. doi: 10.1242/dev.069070. [DOI] [PubMed] [Google Scholar]
- 18.Vo DT, Abdelmohsen K, Martindale JL, Qiao M, Tominaga K, Burton TL, et al. The oncogenic RNA-binding protein Musashi1 is regulated by HuR via mRNA translation and stability in glioblastoma cells. Molecular cancer research: MCR. 2012;10(1):143–55. doi: 10.1158/1541-7786.MCR-11-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kazanjian A, Shroyer NF. NOTCH Signaling and ATOH1 in Colorectal Cancers. Current colorectal cancer reports. 2011;7(2):121–7. doi: 10.1007/s11888-011-0090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dehmer JJ, Garrison AP, Speck KE, Dekaney CM, Van Landeghem L, Sun X, et al. Expansion of intestinal epithelial stem cells during murine development. PloS one. 2011;6(11):e27070. doi: 10.1371/journal.pone.0027070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barry ER, Morikawa T, Butler BL, Shrestha K, de la Rosa R, Yan KS, et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493(7430):106–10. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdelmohsen K, Lal A, Kim HH, Gorospe M. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle. 2007;6(11):1288–92. doi: 10.4161/cc.6.11.4299. [DOI] [PubMed] [Google Scholar]
- 23.Masuda K, Abdelmohsen K, Kim MM, Srikantan S, Lee EK, Tominaga K, et al. Global dissociation of HuR-mRNA complexes promotes cell survival after ionizing radiation. The EMBO journal. 2011;30(6):1040–53. doi: 10.1038/emboj.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci U S A. 2003;100(14):8354–9. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, et al. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. The Journal of clinical investigation. 2001;108(11):1657–65. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donahue JM, Chang ET, Xiao L, Wang PY, Rao JN, Turner DJ, et al. The RNA-binding protein HuR stabilizes survivin mRNA in human oesophageal epithelial cells. Biochem J. 2011;437(1):89–96. doi: 10.1042/BJ20110028. [DOI] [PubMed] [Google Scholar]
- 27.von Roretz C, Lian XJ, Macri AM, Punjani N, Clair E, Drouin O, et al. Apoptotic-induced cleavage shifts HuR from being a promoter of survival to an activator of caspase-mediated apoptosis. Cell death and differentiation. 2013;20(1):154–68. doi: 10.1038/cdd.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen F, Shyu AB, Shneider BL. Hu antigen R and tristetraprolin: counter-regulators of rat apical sodium-dependent bile acid transporter by way of effects on messenger RNA stability. Hepatology. 2011;54(4):1371–8. doi: 10.1002/hep.24496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernstein H, Bernstein C, Payne CM, Dvorak K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J Gastroenterol. 2009;15(27):3329–40. doi: 10.3748/wjg.15.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kosters A, Karpen SJ. Bile acid transporters in health and disease. Xenobiotica; the fate of foreign compounds in biological systems. 2008;38(7–8):1043–71. doi: 10.1080/00498250802040584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grivennikov SI. Inflammation and colorectal cancer: colitis-associated neoplasia. Seminars in immunopathology. 2013;35(2):229–44. doi: 10.1007/s00281-012-0352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grivennikov SI, Karin M. Inflammatory cytokines in cancer: tumour necrosis factor and interleukin 6 take the stage. Annals of the rheumatic diseases. 2011;70 (Suppl 1):i104–8. doi: 10.1136/ard.2010.140145. [DOI] [PubMed] [Google Scholar]
- 33.Sakuma T, Nakagawa T, Ido K, Takeuchi H, Sato K, Kubota T. Expression of vascular endothelial growth factor-A and mRNA stability factor HuR in human meningiomas. Journal of neuro-oncology. 2008;88(2):143–55. doi: 10.1007/s11060-008-9559-8. [DOI] [PubMed] [Google Scholar]
- 34.Goldberg-Cohen I, Furneauxb H, Levy AP. A 40-bp RNA element that mediates stabilization of vascular endothelial growth factor mRNA by HuR. The Journal of biological chemistry. 2002;277(16):13635–40. doi: 10.1074/jbc.M108703200. [DOI] [PubMed] [Google Scholar]
- 35.Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25(4):543–57. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang SH, Lu YC, Li X, Hsieh WY, Xiong Y, Ghosh M, et al. Antagonistic function of the RNA-binding protein HuR and miR-200b in post-transcriptional regulation of vascular endothelial growth factor-A expression and angiogenesis. The Journal of biological chemistry. 2013;288(7):4908–21. doi: 10.1074/jbc.M112.423871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grover PK, Hardingham JE, Cummins AG. Stem cell marker olfactomedin 4: critical appraisal of its characteristics and role in tumorigenesis. Cancer metastasis reviews. 2010;29(4):761–75. doi: 10.1007/s10555-010-9262-z. [DOI] [PubMed] [Google Scholar]
- 38.Koshida S, Kobayashi D, Moriai R, Tsuji N, Watanabe N. Specific overexpression of OLFM4(GW112/HGC-1) mRNA in colon, breast and lung cancer tissues detected using quantitative analysis. Cancer science. 2007;98(3):315–20. doi: 10.1111/j.1349-7006.2006.00383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park KS, Kim KK, Piao ZH, Kim MK, Lee HJ, Kim YC, et al. Olfactomedin 4 suppresses tumor growth and metastasis of mouse melanoma cells through downregulation of integrin and MMP genes. Molecules and cells. 2012;34(6):555–61. doi: 10.1007/s10059-012-0251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeindl-Eberhart E, Brandl L, Liebmann S, Ormanns S, Scheel SK, Brabletz T, et al. Epithelial-mesenchymal transition induces endoplasmic-reticulum-stress response in human colorectal tumor cells. PloS one. 2014;9(1):e87386. doi: 10.1371/journal.pone.0087386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Stevens PD, Liu J, Yang H, Wang W, Wang C, et al. PHLPP is a Negative Regulator of RAF1 that Reduces Colorectal Cancer Cell Motility and Prevents Tumor Progression in Mice. Gastroenterology. 2014 doi: 10.1053/j.gastro.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geng Y, Chandrasekaran S, Agastin S, Li J, King MR. Dynamic Switch Between Two Adhesion Phenotypes in Colorectal Cancer Cells. Cellular and molecular bioengineering. 2014;7:35–44. doi: 10.1007/s12195-013-0313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christie DM, Dawson PA, Thevananther S, Shneider BL. Comparative analysis of the ontogeny of a sodium-dependent bile acid transporter in rat kidney and ileum. Am J Physiol. 1996;271(2 Pt 1):G377–85. doi: 10.1152/ajpgi.1996.271.2.G377. [DOI] [PubMed] [Google Scholar]
- 44.Ido K, Nakagawa T, Sakuma T, Takeuchi H, Sato K, Kubota T. Expression of vascular endothelial growth factor-A and mRNA stability factor HuR in human astrocytic tumors. Neuropathology: official journal of the Japanese Society of Neuropathology. 2008;28(6):604–11. doi: 10.1111/j.1440-1789.2008.00926.x. [DOI] [PubMed] [Google Scholar]
- 45.Yoo PS, Mulkeen AL, Cha CH. Post-transcriptional regulation of vascular endothelial growth factor: implications for tumor angiogenesis. World J Gastroenterol. 2006;12(31):4937–42. doi: 10.3748/wjg.v12.i31.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.