Abstract
Emerging evidence implicates cancer stem cells (CSCs) as primary determinants of the clinical behavior of human cancers, representing an ideal target for next-generation anticancer therapies. However CSCs are difficult to propagate in vitro, severely limiting the study of CSC biology and drug development. Here we report that growing cells from glioblastoma (GBM) cell lines on three dimensional (3D) porous chitosan-alginate (CA) scaffolds dramatically promotes the proliferation and enrichment of cells possessing the hallmarks of CSCs. CA scaffold-grown cells were found more tumorigenic in nude mouse xenografts than cells grown from monolayers. Growing in CA scaffolds rapidly promoted expression of genes involved in the epithelial-to-mesenchymal transition that has been implicated in the genesis of CSCs. Our results indicate that CA scaffolds have utility as a simple and inexpensive means to cultivate CSCs in vitro in support of studies to understand CSC biology and develop more effective anti-cancer therapies.
Keywords: brain tumor initiating cells, epithelial-to-mesenchymal transition, hyaluronic acid, microenvironment
1. Introduction
Cancer Stem cells (CSCs) constitute a minority subpopulation of tumor cells and are? characterized by the capacity of self-renewal, unlimited proliferation, and giving rise to tumor cells with a more differentiated phenotype [1–4]. Compared to the majority of cells in a tumor, CSCs are more malignant as evidenced by greater invasiveness, metastatic potential, and resistance to standard therapeutic interventions. These findings indicate that CSCs are primary determinants of tumor clinical behavior, making them attractive targets for more efficacious treatments. According to the current CSC or hierarchical model of cancer biology, drugs that can eradicate the CSC population in a tumor would likely be a highly effective therapy. It has been suggested that tumors with all CSCs removed would become a benign mass of cells that respond well to conventional chemotherapy [5]. However, development of drugs specific for CSCs is hindered by the difficulty in isolating and propagating these cells in vitro since they represent such a small proportion of the total cells in tumor tissue and in monolayer cultures of tumor cell lines, often less than 1% [6].
The most common method for isolating CSCs utilizes fluorescence-activated (FACS) or magnetic-activated (MACS) cell sorting of cells bound with antibodies specific for CSC surface markers (e.g., CD133, prominin-1) [6]. This approach requires costly antibody and dedicated equipment, and yet yields low numbers of viable cells. CSCs can also be isolated and propagated in vitro using serum-free, defined media as suspension cultures of tumorspheres [4]. However, tumorsphere growth is slow, requires large volumes of expensive specialized media and is frequently not successful due to the small percentage of CSC cells in the tumor of origin [4]. A major limitation of suspension cultures is the absence of a three dimensional (3D) environment required for cell-extracellular matrix interactions that facilitate proliferation and promote malignancy [7, 8]. Assays employing soft agar or agar microbeads have been used to grow isolated CSCs, but collection and subsequent analysis of cells is impaired by the high density and small pore sizes typical of polymerized agar [9].
We previously demonstrated that human glioblastoma (GBM) and hepatocellular carcinoma cell lines cultured on 3D chitosan-alginate (CA) scaffolds develop a more malignant phenotype, evidenced, in part, by increased tumorigenicity in nude mice, than those cultured as monolayers. We attributed the greater malignant potential of scaffold-grown cells to the presence of local structures in the CA matrix that mimic in vivo tumor niches [10, 11]. This conclusion is in accord with observations that the tumor microenvironment has a significant effect on the maintenance and self-renewal of CSCs [12], and with our previous report that CA scaffolds support the proliferation of human embryonic stem cells [13]. Here we investigated the ability of CA scaffolds to promote the proliferation and enrichment of the CSCs from GBM cell lines. We assessed CSC enrichment through CD133 flow cytometry and immunofluorescence, and further examined CSC growth kinetics through PCR analysis of GBM stem cell related genes. CSCs were confirmed through implantation into nude mice.
2. Materials and Methods
2.1. Cell lines and tissue culture
The human glioblastoma cell lines U-87 MG and U-118 MG were purchased from American Type Culture Collection (Manassas, VA) as was Minimum Essential Media (MEM). Cells were maintained according to manufacturer’s instructions in MEM containing 10% FBS (Atlanta Biologicals, Lawrenceville, GA) and 1% antibiotic–antimycotic (Invitrogen, Carlsbad, CA) at 37°C and 5% CO2 in a fully humidified incubator. Anti-human/mouse/rat CD133 primary antibodies (rabbit polyclonal to CD133) and FITC conjugated goat polyclonal secondary antibodies to rabbit IgG were purchased from Abcam (Cambridge, MA).
2.2. CA scaffold synthesis
Chitosan (practical grade, >75% deacetylated, MW = 190,000–375,000) and sodium alginate (alginic acid from brown seaweed) powders were purchased from Sigma-Aldrich (St. Louis, MO) and used without additional purification. CA scaffolds were prepared as previously reported [10, 11, 14]. Briefly, a solution of 4 wt% chitosan and 2 wt% acetic acid was mixed under constant stirring for 7 min to obtain a homogeneous solution. A 4 wt% alginate solution in deionized water was then added and mixed for 10 min, followed by constant mixing in a blender for 5 min to obtain a homogeneous CA solution. Approximately 3–4 ml of the CA solution was cast in each 24-well cell culture plate and frozen at −20°C overnight. The samples were then lyophilized, sectioned into 2 mm thick, 13 mm diameter disks, then crosslinked with 0.2 M CaCl2 for 10 min under vacuum, washed with deionized water several times to remove any excess salt, and sterilized in 70% (vol/vol) ethanol for 2 h under vacuum. The scaffolds were then washed three times with sterile PBS and placed on an orbital shaker for at least 12 h to remove any excess ethanol. To coat CA scaffolds with polycaprolactone (PCL), CA scaffolds were first dehydrated through two washes in excess tetrahydrofuran (THF). PCL was dissolved in THF at 10 mg/mL and added to dehydrated CA scaffolds for 2 hr to allow PCL to adsorb. CA scaffolds were then removed from PCL in THF and immediately dried with an air gun and washed in excess PBS overnight before culturing cells. A uniform PCL coating on CA scaffolds was confirmed using FTIR and SEM.
2.3. Cell seeding on scaffolds
Cells were seeded onto PBS damp CA scaffolds in 12-well plates at 50,000 cells per scaffold in 50 µL of supplemented medium. Cells were allowed to attach to the scaffold for 1 h before adding 1 mL of medium to each well. For PCL and polystyrene (PS) scaffolds, 50,000 cells were seeded per scaffold following the manufacturer’s protocol. For 2D cultures, 12-well plates were inoculated with 1 mL medium containing 50,000 cells. Medium were replaced every 2 days or as required.
2.4. Cell proliferation analysis
Cell proliferation was determined using the Alamar Blue assay following the manufacturer’s protocol. Briefly, cells were washed with PBS before adding 1 mL of Alamar Blue solution (110 µg Resazurin per 1 mL medium) to each well. After continuing incubation for 1.5 h, the solution was transferred to a black-bottom 96-well plate to measure fluorescence. The cell number was calculated based on standard curves created for each cell line grown as monolayers. For time course determinations, cells were washed with D-PBS to remove Alamar Blue and returned to fresh medium.
2.5. Scanning electron microscopy
Samples in medium were fixed with 2.5% glutaraldehyde for 30 min at 37°C, followed by incubation in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer overnight at 4°C. After dehydration by serial washing in increasing ethanol concentrations (0%, 30%, 50%, 70%, 85%, 95%, 100%) with each wash performed twice, samples were critical point dried, sectioned, mounted, and sputter coated with platinum before imaging with a JSM-7000 SEM (JEOL, Tokyo, Japan).
2.6. Flow cytometry analysis
Cells were detached from scaffolds with Versene and washed into FACS buffer (2% FBS in D-PBS) at 1 million cells per mL. Cells were incubated with primary antibody (1:50) for 1 hr on ice, washed thrice with FACS buffer, and incubated with FITC conjugated secondary antibody (1:1000) for 30 min on ice and washed thrice with FACS buffer. Secondary only stained cells were used as a background control. Cells were analyzed on a FACSCanto flow cytometer (Beckton Dickinson, Franklin Lakes, NJ), and data processed using the FlowJo package (Tree Star, Ashland, OR).
2.7. Immunostaining
Cell cultured scaffolds were fixed overnight in 4% formaldehyde, embedded in paraffin, sectioned into 15 µm sections, and affixed to slides. Slides were deparaffinized with xylenes and rehydrated followed by antigen retrieval using a double boiler and Tris-based antigen retrieval buffer. Slides were blocked with 10% BSA for 2 hr, incubated with primary antibody (1:100) overnight at 4°C. For immunofluorescence imaging slides were washed thrice with 10% BSA before incubation with FITC-conjugated secondary antibody for 2 hr at 4°C (1:1000 dilution in 10% BSA). Slides were mounted using Prolong Gold antifade reagent containing DAPI as a counter stain to visualize cell nuclei. Images were obtained on an inverted fluorescent microscope (Nikon Instruments, Melville, NY) with the appropriate filters using a Nikon Ri1 Color Cooled Camera System and 60× Oil Objective Lens (Nikon Instruments, Melville, NY). For immunohistochemistry, slides were sequentially incubated with biotinylated secondary antibody, peroxidase-labeled avidin, and diaminobenzidine/hydrogen peroxide chromogen substrate and counterstained with hematoxylin before mounting.
2.8. PCR
Cells were detached from samples with versene and cell pellets stored at −80°C before RNA extraction using the Qiagen RNeasy kit (Qiagen, Valencia, CA) following the manufacturer’s protocol. Following reverse transcription (iScript cDNA synthesis kit, Bio-Rad, Hercules, CA), DNA transcripts were probed using BioRad iQ SYBR Green Supermix. A BioRad CFX96 Real-Time Detection System was used for PCR analysis and expression levels were normalized to GAPDH.
2.9. Tumorigenesis assay
Cells cultured on CA scaffolds for 15 days were detached using versene, and immunostained for CD133 as described above. CD133+ and CD133− cells were sorted into PBS containing 2% FBS using an Aria flow cytometer (Beckton Dickinson, Franklin Lakes, NJ). Cells (500 or 2000) were injected subcutaneously into the flanks of athymic nude mice (Charles River Labs, Wilmington, MA) of 6–8 weeks of age. All animal experiments were conducted in accordance with UW Internal Animal Care and Use Committee approved protocols. For unsorted cell tumor growth studies, scaffolds containing cells cultured for 10 days were implanted into the flanks of athymic nude mice. 2D cultured cells were implanted with growth factor reduced matrigel at the same cell number as cells on CA scaffolds. Tumors were measured using calipers and the volume was calculated using previously established methods [10, 11].
3. Results
3.1. CD133 expression in CA scaffold cultured cells
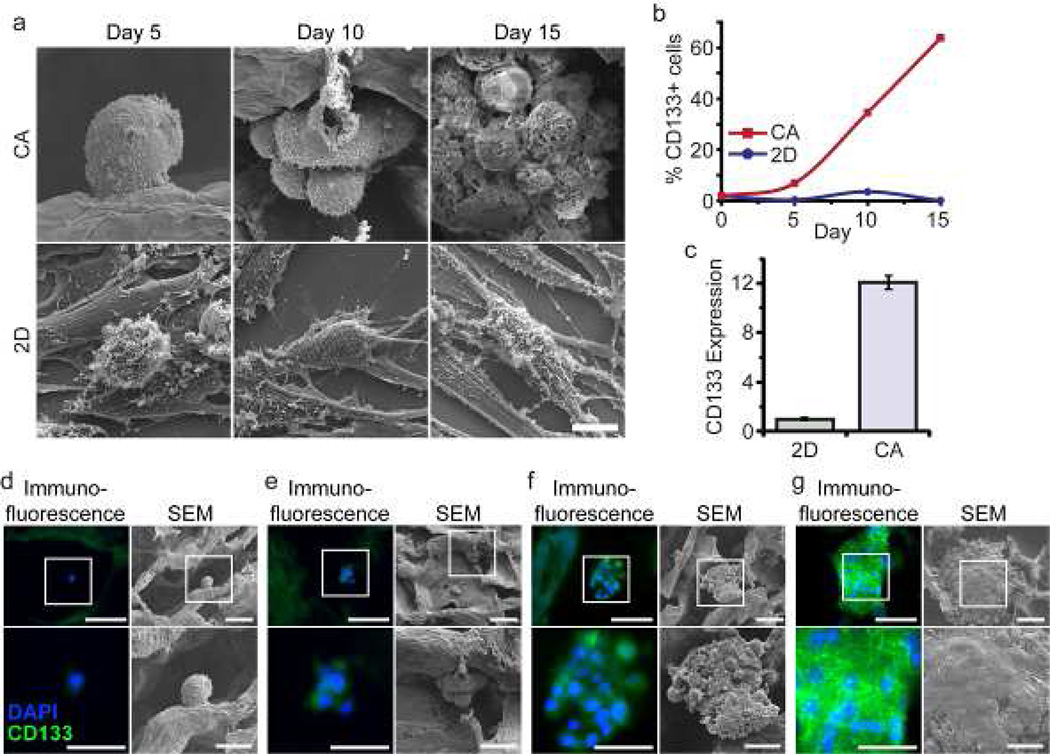
Cells were seeded directly in 6-well plates containing cover slips or 12-well plates containing CA scaffolds at 50,000 cells per sample in fully supplemented culture media. After 5, 10, and 15 days of culture, cells were fixed, dehydrated, and super-critically dried for SEM imaging. Human U-118 MG GBM cells grown on CA scaffolds display pronounced differences in morphology and expression of CD133, a marker of GBM CSCs [15–17], than cells grown as monolayers. In accord with our earlier report [10], scanning electron microscopy (SEM) revealed that growth on CA scaffolds produced aggregations of spherical- or ovoid-shaped cells (tumor spheroids) while growth in monolayer yielded sheets of flat, epithelioid cells with numerous, extended processes (Fig. 1a). As illustrated in Figure 1b, growth on CA scaffolds for 15 days was accompanied by a 1226-fold increase in the population of CD133 immunopositive U-118 MG cells and an increase in fraction of CD133+ cells in GBM cell population from 1.5% to 62.7%. In contrast, the fraction of CD133+ cells in monolayer cultures remained unchanged (~1%) in the same time period. Notably, CD133 mRNA abundance in U-87 MG grown in CA scaffolds was 12-fold higher than grown in monolayers (Fig. 1c), suggesting that scaffolds stimulate de novo CD133 gene expression rather than the acquisition of a spherical morphology exposing occult CD133 protein on the cell surface. Finally, examination of U-118 MG CD133 immunopositivity in fixed, paraffin-embedded sections of scaffolds revealed that solitary cells expressed no detectable CD133 protein (Fig. 1d), while cells that formed clusters showed apparent CD133 immunostaining, and the intensity of the immunostaining increased with the size of the tumor cell clusters (Fig. 1e-g). These results suggest that CD133− cells were unable to grow or grow very slowly whereas CD133+ cells preferentially grew into large clusters in the CA scaffolds, resulting in CSC enrichment.
Figure 1.
Growth of CD133+ GBM cells on CA scaffolds. a) SEM images comparing the morphology and proliferation of human U-118 MG GBM cells cultured on 3D CA scaffolds and 2D monolayers over 15 days. Scale bar corresponds to 10 µm. b) Comparison of the change in fraction of CD133+ cells in U-118 MG cell population grown for 15 days on CA scaffolds or as monolayers. Immunopositivity for CD133 was determined by flow cytometry. c) CD133 mRNA content determined by real time PCR in U-87 MG GBM cells grown for 10 days on CA scaffolds compared to that of monolayer cultures. CD133 mRNA content was normalized to the monolayer condition. d-g) Immunostaining for CD133 (green) and SEM imaging of CA-scaffold cultured U-118 MG GBM cells at day 10. The boxed regions in the top-row images correspond to the areas of the bottom images. Blue color reflects DAPI counter-staining of nuclei. d) Solitary U-118 MG cells generally showed no CD133 staining. e) Small clusters of U-118 MG cells showed faint CD133 staining. f and g) Intensity of CD133 staining increases as clusters of U-118 MG cells grow larger. Scale bars for panels d-g correspond to 50 µm for the upper row and 25 µm for the lower row.
3.2. Characterization of CD133+ CA scaffold cultured cells
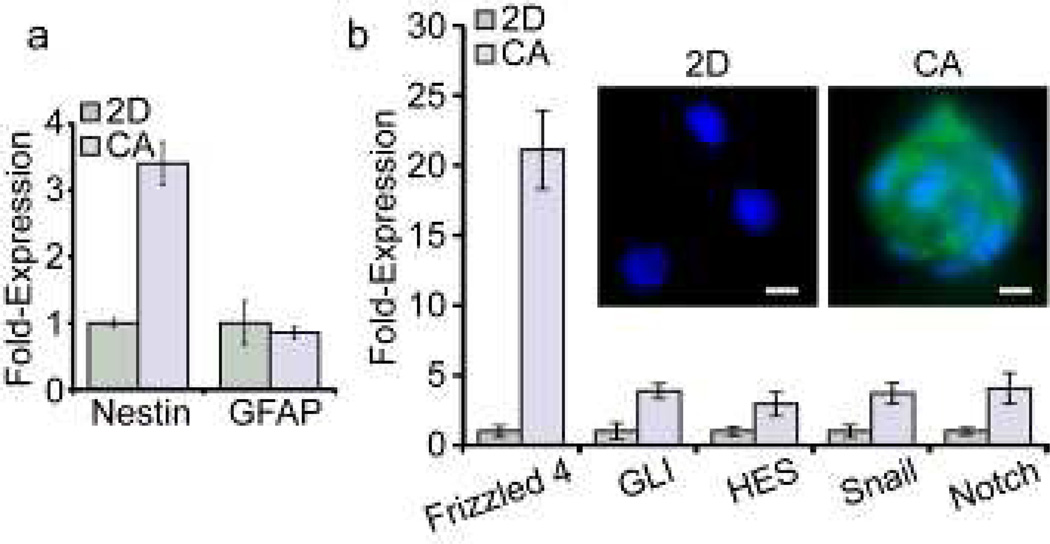
Considerable evidence indicates that transformation of normal neural stem cells underlies the genesis of GBM and is accompanied by the aberrant expression of genes that promote the normal development of neural cells [18]. We therefore compared the expression of mRNA for a panel of genes found in normal neural progenitor cells in scaffold and monolayer cultures of U-118 MG. Ten days after cell seeding, cells harvested from scaffolds showed a 3.5-fold higher abundance of mRNA for nestin, a cytoskeletal protein specific to neural progenitor cells [18], compared to monolayer cells (Fig. 2a). In contrast, scaffold-grown cells showed no increase in mRNA expression for GFAP, a cytoskeletal protein that replaces nestin as neural progenitors differentiate into mature glial cells [18]. As shown in Figure 2b, growth on scaffolds was accompanied by elevated expression of mRNA for other neural development genes that have been implicated in the genesis of GBM [19–22], including Frizzled 4 of the WNT signaling pathway, GLI and Snail of the hedgehog pathway, and HES of the Notch pathway. These data strongly indicate that growth of U-118 MG on scaffolds is accompanied by elevated expression of a host of genes characteristic of undifferentiated GBM CSCs.
Figure 2.
CA scaffold-grown U-118 MG GBM cells express characteristic neural stem cell markers, and exhibit phenotypic characteristics of CSCs. a) mRNA content determined by real time PCR revealed that relative to 2D monolayer cultures, cells grown on CA scaffolds show elevated expression of the neural progenitor intermediate filament nestin while the level of mRNA for GFAP, the intermediate filament of mature glia is unchanged. b) Comparison of expression of mRNA of genes associated with normal neural cell development and the genesis of GBM in cells grown on CA scaffolds relative to cells grown as monolayers. Immunofluorescence images in inset shows that enhanced protein expression (green) accompanied elevation of mRNA level for frizzled-4. Scale bars are 10 µm.
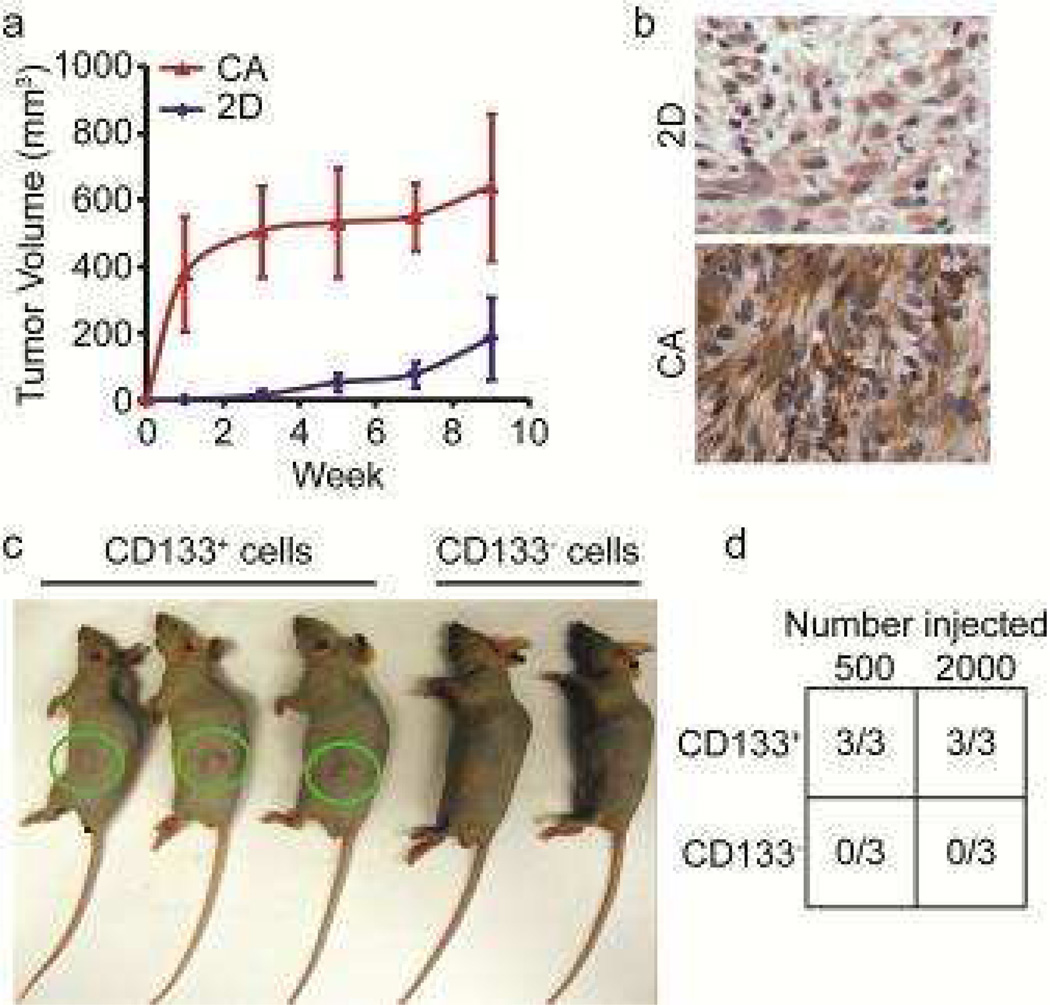
CSCs are characterized by their ability to readily form tumors in nude mice [1–3], where CSCs and not non-stem cancer cells drive tumor formation [23]. CD133+ and CD133− cells from U-118 MG cells cultured in CA scaffolds for 10 days were separated using FACS, and implanted subcutaneously into the flanks of nude mice at either 200 or 2000 cells per mouse. As shown in Figure 3a, subcutaneous flank tumors were detectable earlier and grew to a larger size in animals inoculated with U-118 MG cells grown on scaffolds compared to those grown as monolayers. The tumors from scaffold-grown cells also expressed a higher level of CD133 (Fig. 3b), indicating that the enhanced tumorigenicity of cells from scaffolds reflected expression and maintenance of the CSC phenotype. Notably, tumors were readily formed in all animals 9 weeks after receiving injection of 500 or 2,000 CD133+ cells harvested from scaffolds while no tumors formed in animals injected with CD133− cells harvested from scaffolds (Figs. 3c and 3d). This more stringent test of tumorigenicity provides additional evidence that scaffold-grown CD133+ cells possess the hallmark properties of CSCs.
Figure 3.
CA scaffold-grown U-118 MG GBM cells exhibit phenotypic characteristics of CSCs. a) Size of flank tumors in nude mice injected subcutaneously with 50,000 cells harvested from CA scaffolds or 2D monolayers. b) Tumors grown from scaffold-grown cells display greater immunopositivity for CD133 (brown) than tumors grown from 2D monolayer cells. c) Flank tumors in nude mice injected with 500 CD133+ or CD133− U-118 MG GBM cells sorted by FACS after culture in CA scaffolds. The tumors (green circles) were evident only in mice injected with CD133+ cells. d) Tumorigenicity in nude mice injected subcutaneously with either 500 or 2,000 FACS-isolated CD133+ or CD133− cells grown on CA scaffolds for 10 days. Tumors grew only in all mice implanted with CD133+ cells.
3.3. Effect of CA scaffold microenvironment on CSC enrichment
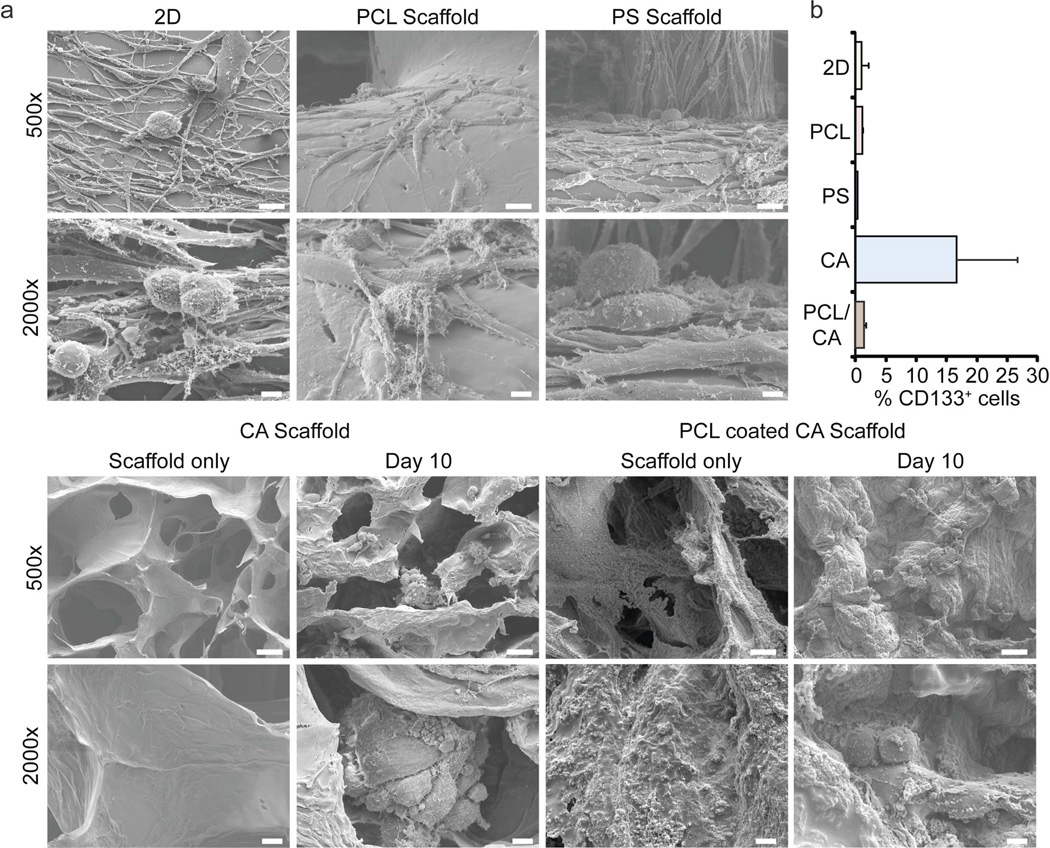
To assess if the enrichment of GBM CSCs is simply a consequence of growth in any 3D structure, we compared morphology and CD133 immunopositivity of the human glioma line U-87 MG cultured for 10 days on commercially available polycaprolactone (PCL) and polystyrene (PS) scaffolds with cells grown on CA scaffolds and as monolayers. As shown in Figure 4a, the morphology of U-87 MG cells grown on the commercial scaffolds differed little from that of monolayer cells, in contrast to the clusters of spherical or ovoid cells on the CA scaffolds. Additionally, cells cultured on CA scaffolds coated with a thin layer of PCL did not develop the clusters of spherical cells that were inherent to CA scaffold culture. These data suggest that it was not simply the 3D structure of the scaffolds that promoted formation of the tumor spheroids; rather, it was likely a combination of the 3D and chemical structure of the scaffolds. Moreover, near 20% of cells on the CA scaffold were immunopositive for CD133 after 10 days of culture compared to only 1–2% of CD133+ cells grown on the other substrates (Fig. 4b).
Figure 4.
Proliferation of CD133+ U-118 MG CSC GBM cells is promoted by the physicochemical environment of the CA scaffolds. a) SEM images of cells cultured in 2D plates, commercially available polycaprolactone (PCL) and polystyrene (PS) scaffolds, CA scaffolds, and PCL coated CA scaffolds. Scale bars: 25 µm at 500× and 5 µm at 2000×. b) Fraction of CD133+ cells determined by flow cytometry after growth for 10 days on 2D plates, PCL, PS or CA scaffolds.
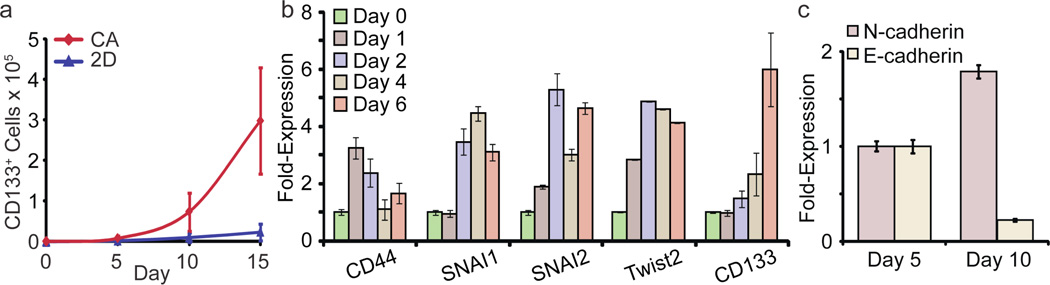
The greater proliferation of U-87 MG CSCs on CA scaffolds compared to monolayer culture is illustrated in Figure 5a. After a delay of 5 days, U-87 MG CSCs grew rapidly on CA scaffolds through day 15. By 15 days after cell seeding the CSC fraction increased from 0.3% to 42%; the total number of U-87 MG CD133+ CSCs on CA scaffolds increased 2188-fold while those grown as monolayers showed little increase. These results indicate that the proliferation and enrichment of CD133+ cells on the CA scaffold reflects the chemistry of the substrate as well as its geometry and that culture on CA scaffolds is a facile method of producing large numbers of CSC for subsequent study.
Figure 5.
Proliferation of CD133+ U-87 MG CSC GBM cells on CA scaffolds is accompanied by expression of genes that mediate EMT. a) Growth curves for CD133+ cells on CA scaffolds and in monolayer cultures. CD133+ cell number was assessed by Alamar blue and flow cytometry. b) Change in mRNA contents for EMT related genes and CD133 during growth on CA scaffolds for 6 days. c) Expression of N-cadherin and E-cadherin mRNA in scaffold-grown cells at day 5 and 10 determined by real time PCR.
3.4. Analysis of CA scaffold-induced CSC enrichment
The delay that precedes the rapid proliferation of CD133+ CSCs observed above suggests that changes in gene expression are necessary for cancer cells to proliferate on CA scaffolds. Emerging evidence indicates that the pathway that mediates the epithelial-to-mesenchymal transition (EMT) in cancer can also promote reversion of non-CSC tumor cells to CSCs [24, 25]. EMT is mediated by signaling cascades induced by the interaction of the transmembrane glycoprotein CD44 with extracellular matrix [26, 27]. Activation of CD44 signaling is also associated with enhanced proliferation, invasion, and chemoresistance in cancer cells [27]. We examined the expression of CD44 and other genes that participate in EMT in scaffold-grown U-87 MG cells. As shown in Figure 5b, CD44 mRNA content was elevated within a day of culture on scaffolds. Elevation of mRNA for Twist2, Snai1 and Snai2, genes that participate in EMT [16, 24], was subsequently detected beginning at day 2. Overexpression of CD44 induced the overexpression of Snai1, Snai2, and Twist2 through downstream signaling which resulted in overexpression of CD133 between days 4–6. Another hallmark of EMT, enhanced expression of the cell adhesion molecule N-cadherin accompanied by suppression of E-cadherin, occurred between days 5 and 10 (Fig. 5c). In toto, these findings strongly suggest that activation of at least some elements that participate in EMT accompany the increased proliferation of CD133+ CSCs on CA scaffolds.
4. Discussion
We have shown that culturing GBM and other cancer cells on CA scaffolds promotes the proliferation of cells expressing the hallmarks of CSCs. After an initial lag, there was an exponential increase in the number of CD133+ cells growing on CA scaffolds, reaching ~70% after 15 days. The stemness of these CD133+ cells was verified using the gold-standard tumor formation assay where cells are implanted into nude mice at low inocula and without Matrigel. Only CD133+ cells from CA scaffolds grew tumors whereas CD133− cells showed no tumor formation. Additionally, cells cultured in CA scaffolds displayed increased expression of stem cell related genes such as Nestin, Frizzled 4, GLI, HES, Snail, and Notch.
The acquisition of the CSC phenotype appears to be specific to the chemistry of the CA scaffolds. Cells cultured in other 3D scaffolds showed no increase in CD133 immunopositivity. Importantly, cells cultured in PCL coated CA scaffolds, in which the 3D structure of the scaffold was conserved but the surface chemistry changed, did not show tumor spheroid formation nor CD133+ cell enrichment. Therefore, it was both the structural and chemical environment of the CA scaffolds that promoted rapid enrichment of CSCs. In fact, CD44, a transmembrane glycoprotein, was found to be upregulated in CA scaffold cultured cells at early time points and likely promoted the downstream expression of CD133 through SNAI1, SNAI2, and Twist2. The enrichment of CSCs was found to be mediated by expression of genes involved in EMT, and most importantly generated a cadherin switch.
It is intriguing that cells cultured in CA scaffolds were able to revert to a more stem-like state and displayed the properties of CSCs. The fact that non-CSCs can become CSCs under the right microenvironmental cues implies that CSC-targeted therapies may not be as effective as once thought [28–30]. If non-CSCs in the tumor microenvironment can become CSCs, then cells targeted by anti-CSC therapies could simply be replaced by non-CSCs through genes involved in EMT. Therefore, it will be crucial to learn if there are only a subset of non-CSCs that can become CSCs, or if it is a property of all cancer cells. These CA scaffolds can provide the medium for these studies. Additionally, these CA scaffolds can be used to probe the specific microenvironmental cues (cellular and non-cellular [10, 11, 31–33]) that promote the conversion of non-CSCs into CSCs to generate effective microenvironmental therapies.
5. Conclusions
We have demonstrated that culture of human glioblastoma cell lines on 3D porous CA scaffolds promotes the growth and enrichment of the CD133+ CSC population. We found that cells exposed to the chemical and structural microenvironment provided by the CA scaffolds reverted to a more stem-like state through CD44 signaling and EMT. This finding could have broad implications in developing cancer stem cell specific therapies, as these treatments might not provide the desired therapeutic efficacy if non-CSCs can become CSCs. Significantly, this method of culture rapidly produces large numbers of CSCs that can easily be collected for subsequent analysis without the use of defined media. CA scaffolds will facilitate the study of CSC biology and provide a new in vitro model for the discovery of more efficacious anti-cancer therapies.
Acknowledgements
This work was supported by NIH grant R01CA172455. FK acknowledges support from an NIH training grant (T32CA138312) and an American Brain Tumor Association Fellowship in Honor of Susan Kramer. SJF acknowledges support from the Egtvedt Fellowship. We acknowledge the use of the SEM at the Dept. of Materials Science and Engineering, the confocal and optical microscopes at Keck Microscopy Imaging Facility, and the flow cytometers at the Dept. of Immunology at the University of Washington. We acknowledge laboratory assistance from Ms. Allison M. Lewis and Ms. Samara K. Sytsma in preparing CA scaffolds. We thank Prof. Benjamin S. Glick for submitting the RFP plasmid to AddGene.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.O'Brien CA, Kreso A, Jamieson CH. Cancer stem cells and self-renewal. Clin Cancer Res. 2010;16:3113–3120. doi: 10.1158/1078-0432.CCR-09-2824. [DOI] [PubMed] [Google Scholar]
- 2.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 4.Zhou BBS, Zhang HY, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov. 2009;8:806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly SE, Di Benedetto A, Greco A, Howard CM, Sollars VE, Primerano DA, et al. Rapid selection and proliferation of CD133+ cells from cancer cell lines: chemotherapeutic implications. PLoS One. 2010;5:e10035. doi: 10.1371/journal.pone.0010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ, et al. Engineering tumors with 3D scaffolds. Nat Methods. 2007;4:855–860. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 8.Rao W, Zhao S, Yu J, Lu X, Zynger DL, He X. Enhanced enrichment of prostate cancer stem-like cells with miniaturized 3D culture in liquid core-hydrogel shell microcapsules. Biomaterials. 2014;35:7762–7773. doi: 10.1016/j.biomaterials.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith BH, Gazda LS, Conn BL, Jain K, Asina S, Levine DM, et al. Three-dimensional culture of mouse renal carcinoma cells in agarose macrobeads selects for a subpopulation of cells with cancer stem cell or cancer progenitor properties. Cancer Res. 2011;71:716–724. doi: 10.1158/0008-5472.CAN-10-2254. [DOI] [PubMed] [Google Scholar]
- 10.Kievit FM, Florczyk SJ, Leung MC, Veiseh O, Park JO, Disis ML, et al. Chitosan-alginate 3D scaffolds as a mimic of the glioma tumor microenvironment. Biomaterials. 2010;31:5903–5910. doi: 10.1016/j.biomaterials.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung M, Kievit FM, Florczyk SJ, Veiseh O, Wu J, Park JO, et al. Chitosan-alginate scaffold culture system for hepatocellular carcinoma increases malignancy and drug resistance. Pharm Res. 2010;27:1939–1948. doi: 10.1007/s11095-010-0198-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borovski T, De Sousa EMF, Vermeulen L, Medema JP. Cancer stem cell niche: the place to be. Cancer Res. 2011;71:634–639. doi: 10.1158/0008-5472.CAN-10-3220. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Leung M, Hopper R, Ellenbogen R, Zhang M. Feeder-free self-renewal of human embryonic stem cells in 3D porous natural polymer scaffolds. Biomaterials. 2010;31:404–412. doi: 10.1016/j.biomaterials.2009.09.070. [DOI] [PubMed] [Google Scholar]
- 14.Florczyk SJ, Kim DJ, Wood DL, Zhang M. Influence of processing parameters on pore structure of 3D porous chitosan-alginate polyelectrolyte complex scaffolds. J Biomed Mater Res A. 2011;98:614–620. doi: 10.1002/jbm.a.33153. [DOI] [PubMed] [Google Scholar]
- 15.Ricci-Vitiani L, Pallini R, Biffoni M, Todaro M, Invernici G, Cenci T, et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468:824–828. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- 16.Fan X, Khaki L, Zhu TS, Soules ME, Talsma CE, Gul N, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 18.Westphal M, Lamszus K. The neurobiology of gliomas: from cell biology to the development of therapeutic approaches. Nat Rev Neurosci. 2011;12:495–508. doi: 10.1038/nrn3060. [DOI] [PubMed] [Google Scholar]
- 19.Jin X, Jeon HY, Joo KM, Kim JK, Jin J, Kim SH, et al. Frizzled 4 regulates stemness and invasiveness of migrating glioma cells established by serial intracranial transplantation. Cancer Res. 2011;71:3066–3075. doi: 10.1158/0008-5472.CAN-10-1495. [DOI] [PubMed] [Google Scholar]
- 20.Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizzo P, Miao H, D'Souza G, Osipo C, Song LL, Yun J, et al. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res. 2008;68:5226–5235. doi: 10.1158/0008-5472.CAN-07-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 23.Lathia JD, Gallagher J, Myers JT, Li M, Vasanji A, McLendon RE, et al. Direct in vivo evidence for tumor propagation by glioblastoma cancer stem cells. PLoS One. 2011;6:e24807. doi: 10.1371/journal.pone.0024807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouyang G, Wang Z, Fang X, Liu J, Yang CJ. Molecular signaling of the epithelial to mesenchymal transition in generating and maintaining cancer stem cells. Cell Mol Life Sci. 2010;67:2605–2618. doi: 10.1007/s00018-010-0338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Misra S, Heldin P, Hascall VC, Karamanos NK, Skandalis SS, Markwald RR, et al. Hyaluronan- CD44 interactions as potential targets for cancer therapy. Febs J. 2011;278:1429–1443. doi: 10.1111/j.1742-4658.2011.08071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toole BP. Hyaluronan-CD44 interactions in cancer: paradoxes and possibilities. Clin Cancer Res. 2009;15:7462–7468. doi: 10.1158/1078-0432.CCR-09-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells - what challenges do they pose? Nat Rev Drug Discov. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 31.Florczyk SJ, Liu G, Kievit FM, Lewis AM, Wu JD, Zhang M. 3D porous chitosan-alginate scaffolds: a new matrix for studying prostate cancer cell-lymphocyte interactions in vitro. Adv Healthc Mater. 2012;1:590–599. doi: 10.1002/adhm.201100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phan-Lai V, Kievit FM, Florczyk SJ, Wang K, Disis ML, Zhang M. CCL21 and IFNgamma recruit and activate tumor specific T cells in 3D scaffold model of breast cancer. Anticancer Agents Med Chem. 2014;14:204–210. doi: 10.2174/18715206113136660375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phan-Lai V, Florczyk SJ, Kievit FM, Wang K, Gad E, Disis ML, et al. Three-dimensional scaffolds to evaluate tumor associated fibroblast-mediated suppression of breast tumor specific T cells. Biomacromolecules. 2013;14:1330–1337. doi: 10.1021/bm301928u. [DOI] [PMC free article] [PubMed] [Google Scholar]