Two large, lipid-containing, double-stranded DNA (dsDNA) icosahedral viruses, each ∼1 GDa (1 × 109 daltons) and unlikely to be amenable to crystallization, have been studied by cryo-electron microscopy (cryo-EM) and three-dimensional image reconstruction. The first, the 1,850 Å diameter Chilo iridescent virus (CIV: genus Iridovirus, family Iridoviridae) infects the rice stem borer insect1. The second, the 1,900 Å diameter Paramecium bursaria chlorella virus, type 1 (PBCV-1: genus Chlorovirus, family Phycodnaviridae) infects certain unicellular, eukaryotic, chlorella-like green algae2. Despite differences in their pathogenicity and genome size, both viruses exhibit common structural features, suggesting a common evolutionary origin and therefore serve as a paradigm for other large viruses, such as African swine fever virus, frog virus-3 and many others (for example, see ref. 3). Both viruses have a layered structure consisting of a dsDNA–protein core, surrounded in turn by a lipid bilayer and one or more icosahedral capsid shells consisting of thousands of protein subunits4–7. Furthermore, the external capsid is essentially consistent with the quasi symmetry predictions of Caspar and Klug8. This is unexpected in view of the now well-documented departure from quasi symmetry in much smaller virusses such as polyoma9 or the inner core of the double-stranded RNA viruses (for example, see ref. 10).
CIV has a 209 kbp linear, circularly permuted (a linear molecule that maps as a circle) genome, whereas PBCV-1 has a 330 kbp linear, non-permuted genome with covalently closed, hairpin ends11,12. Some iridoviruses, like CIV, also contain fibers that protrude 350 Å or more from the outer surface of the capsid5,13 and this was confirmed in this study. In contrast to the iridoviruses, little is known about the chlorovirus structure. The chlorovirus capsids accommodate a wide range of genome sizes14,15 from 295 kbp16 to 380 kbp14. The PBCV-1 virion contains ∼50 proteins, with the 54 kDa surface glycoprotein, VP54, accounting for ∼40% of the total virion protein mass17. PBCV-1 and its close relatives are common in freshwater collected worldwide2,12. Like the tailed bacteriophages, PBCV-1 infects its host, Chlorella NC64A, by attaching to the cell wall, possibly through a unique vertex or a tail-like structure. The cell wall is then degraded at the point of attachment and viral DNA is released into the cell2,18. Analysis of the 330,740 bp PBCV-1 genome predicts that it contains 377 protein-encoding genes and 10 tRNA genes, including many interesting and unexpected genes12. To put the PBCV-1 genome size in perspective, the smallest free-living organism, a mycoplasma, encodes ∼470 proteins19.
Our cryo-EM studies of CIV and PBCV-1 show that both viruses have profiles characteristic of icosahedra viewed in different orientations. Most virions contain large, asymmetric masses distributed non-uniformly within them (Fig. 1a,d). CIV forms quasi crystalline, closely packed hexagonal arrays in which neighboring virions are separated by 400–600 Å due to a halo of fibers that surround the outer capsid. Image reconstructions of the two viruses, each at ∼26 Å resolution, show outer capsid structures composed of pseudo-hexagonal arrays of ‘capsomers’ (Fig. 1b,e). Close inspection demonstrates that each hexavalent capsomer is a trimeric structure (Fig. 2).
Fig. 1.
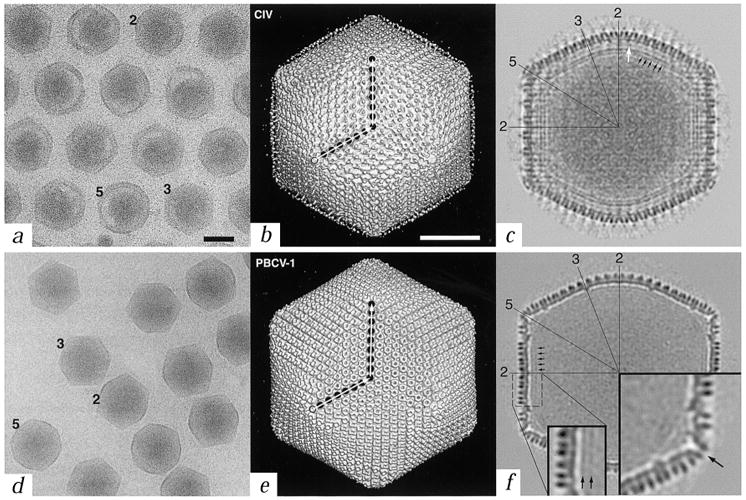
Comparison of CIV and PBCV-1 virions. a, d, Micrographs of vitrified samples of CIV and PBCV-1 virions (black bar = 1000 Å). Representative particles oriented with their two-, three-, or five-fold axes nearly parallel to the view direction are identified with numbers. Long, thin fibers emanate from the surface of CIV virions and maintain a regular intervirion spacing (a). b, e, Shaded-surface representations of the three-dimensional reconstructions of CIV and PBCV-1, each viewed down an icosahedral three-fold axis (white bar = 500 Å). Each capsid consists of twelve pentavalent and either 1,460 (CIV) or 1,680 (PBCV-1) hexavalent ‘capsomers’, arranged with T = 147 (h = 7, k = 7 in CIV) and T = 169d (h = 7, k = 8 in PBCV-1) icosahedral, quasi-equivalent lattices. The T (triangulation) number, which describes the geometrical arrangement of capsomers on a two-dimensional lattice, is given by the relationship T = h2+hk+k2, where h and k are integers that define lattice points8. The d (for dextro) in T = 169d indicates that the PBCV-1 capsid belongs to a right-handed, skew class of T lattice8. Filled circles mark the positions of capsomers along h and k directions (open circle is the origin of h, k lattice). Only the proximal end of each fiber in CIV is visible, presumably because the fibers are inherently flexible resulting in the distal portions smearing out in the reconstruction process. c, f, Central sections of the reconstruction density maps as viewed along a two-fold axis (same magnification as panels (b) and (e)). A lipid bilayer exists beneath the capsid shell in both virions (black arrows in (c) and (f) and in inset in (f)), although it is less prominent in PBCV-1. The PBCV-1 bilayer is more easily detected in images of disrupted virions, prepared by vitrification or negative staining (data not shown). Numerous connections, which constitutes an additional shell, occur between the outer shell and the bilayer in CIV (white arrow in (c)). Several icosahedral two-, three-, and five-fold symmetry axes are identified in both sections. Large inset in (f) is a view of the region near a pentavalent capsomer and reveals an axial channel (arrow). Both insets in (f) are at 2× magnification compared to (b) (c) (e), and (f).
Fig. 2.
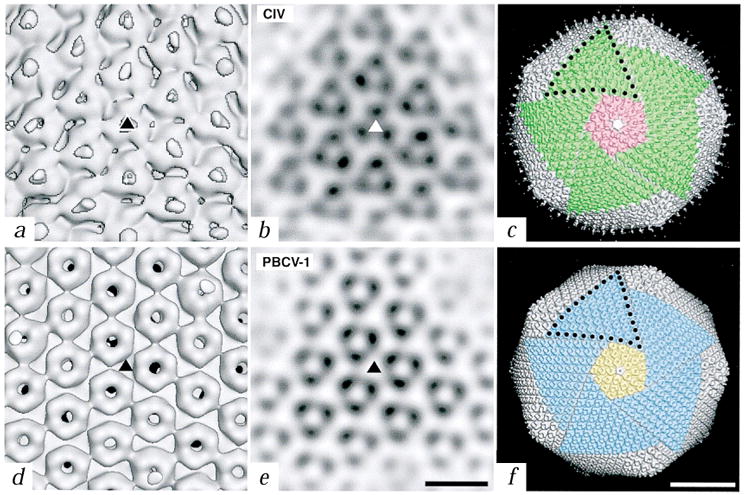
Close-up views of the trimers of CIV and PBCV-1. a, d, Shaded-surface representations of the reconstructions viewed down an icosahedral three-fold axis (triangles). Fibers in CIV are centered over the trimers whereas the trimers in PBCV-1 have a central, concave depression and axial channel. b, e, Planar sections through the reconstructed density maps viewed along an icosahedral three-fold axis (triangles). (Bar, 100 Å for panels (a) (b) (d), and (e)). c, f, Comparison of the symmetron facets of CIV (c) and PBCV-1 (f). Five trisymmetrons are highlighted in each reconstruction (green in CIV and blue in PBCV-1) and a single pentasymmetron is colored pink in CIV and yellow in PBCV-1. A pentavalent capsomer (uncolored) lies at the center of each pentasymmetron. Ten capsomers in CIV and eleven in PBCV-1 form the edge of each trisymmetron (black dots). Bar, 500 Å.
The outer diameters of the viral capsids range from 1,615 and 1,650 Å along the two- and three-fold axes to 1,850 and 1,900 Å along the five-fold axes for CIV and PBCV-1, respectively (Fig. 1c,f). The capsomers of CIV have a triangular profile (∼80 Å in diameter and ∼75 Å high) and the fibers are associated with and project radially from the center of the hexavalent but not pentavalent capsomers. Capsomers of PBCV-1 are doughnut-shaped (∼70 Å in diameter and ∼75 Å high) and most contain axial channels of ∼17 Å in diameter. The capsomers of both viruses interconnect at their bases to form contiguous, 20–25 Å thick icosahedral shells. The protruding portion of each hexavalent capsomer extends ∼50 Å above the surface of the shells (Fig. 2a,d).
Density corresponding to a lipid bilayer, ∼40 Å thick, that follows the contour of the outer capsid is visible inside both viruses, although it is more prominent in CIV (Fig. 1c,f). In CIV, the bilayer appears to be tethered by connections with an additional shell of density that lies between the outer layer of capsomers and the lipid bilayer (white arrow; Fig. 1c). Such a second shell is not apparent in PBCV-1 (Fig. 1f). Occasional disruptions of the membrane might reflect the presence of transmembrane proteins. The centers of the bilayers occur at radii 637 and 703 Å at the two-fold axes and radii 729 and 756 Å at the five-fold axes in CIV and PBCV-1, respectively (Fig. 1c,f). These values are consistent with mass measurements that indicate each virus consists of ∼9% lipid11,17,20. Like the outer shell, the inner capsid of CIV consists of trimeric structures and these are rotated ∼60° about their axes relative to those in the outer layer (data not shown).
Assuming all hexavalent capsomers in PBCV-1 are identical chemically, the outer capsid of PBCV-1 contains 5,040 copies of VP54, associated as 1,680 trimers, consistent with earlier biochemical measurements17. The similar size of the PBCV-1 pentavalent and hexavalent capsomers suggests that the pentavalent capsomers are likely to be oligomers of a different structural protein. CIV encapsidates its genome and 26 viral proteins within the outer and inner capsid shells that are composed, respectively, of 51.4 kDa major capsid proteins P50 and P′50 (refs 21,22). P50 and P′50 are identical proteins in sequence; however, P50 exists as a non-covalent trimer whereas P′50 is linked by disulfide bonds in each trimer21. Hence, there are 4,380 copies (= 1,460 × 3) each of P50 and P′50. Composition of the pentavalent capsomers is unknown for both viruses, but, in CIV these capsomers are clearly larger than those in PBCV-1 (Fig. 2c,f).
Inspection of the outer capsid surfaces of CIV and especially of PBCV-1, shows the presence of large pentagonal and triangular facets (Fig. 2c,f). Such facets were first recognized and described (named pentasymmetrons and trisymmetrons) by Wrigley23, who examined negatively stained preparations of disrupted Sericesthis iridescent virus, and an additional type of facet, ‘groups of nine’, was later described in adenovirus24. Each of the twelve pentasymmetrons at the vertices of CIV and PBCV-1 contains 31 capsomers, of which one is pentavalent and the remaining 30 are trimers that occupy hexavalent positions in the capsids. Both virus capsids contain twenty trisymmetrons; each of which contains 55 and 66 trimers in CIV and PBCV 1, respectively. All trimers within each type of trisymmetron are arranged in similar orientations. This produces a two-dimensional, triangular ‘crystal’ and hence gives each triangular facet its characteristic, near planar morphology that leads to the overall icosahedral morphology of the assembled capsid. The CIV and PBCV-1 trimers differ by ∼60° rotation about the trimer axis (Fig. 2b,e). Despite the lack of direct evidence for the assembly pathway of any large, icosahedral, dsDNA-containing virus, the existence of characteristic facets suggests that they may be mature forms of self-assembled, intermediate substructures that facilitate efficient morphogenesis of large icosahedral capsids24.
Such large, icosahedral viruses package their genomes inside capsid shells that are surprisingly precise and highly symmetric structures. Striking similarities in the capsid structures of CIV and PBCV-1, such as the existence of trimeric capsomers and planar facets, hint that the assembly of large icosahedral viruses may follow analogous pathways. Indeed, many large dsDNA viruses contain closely related, ∼50kDa major capsid proteins that associate as trimers25. In addition, adenovirus and bacteriophage PRD1, among the largest viruses studied to date by cryo-EM, have T = 25 capsids consisting of major capsid proteins that associate as trimers26,27. Phylogenetic analyses indicate the DNA polymerases from phycodnaviruses are near the root of all eukaryotic DNA polymerase delta proteins28, which suggests these viruses have a long evolutionary history. Hence, structural information from CIV and PBCV-1 may reveal patterns that exist in other viruses in the same manner as the tertiary structure of proteins and can be used to map phylogenetic trees.
Methods
Purified samples of CIV were prepared by homogenizing 50 third-instar, highly infected Galleria mellonella larvae in 110 ml of deionized water in a Potter homogenizer at 4 °C. After centrifugation at 2,500 g for 10 min, the homogenate was layered on an ECTEOLA cellulose column. Virions were collected immediately after the void volume and concentrated by centrifugation at 23,000 g for 30 min. The bluish pellet was resuspended in deionized water, brought to a concentration of 20mg ml-1, and stored at 4 °C. PBCV-1 samples were prepared as described29. For electron microscopy, 3.5 μl aliquots of purified virus were prepared and vitrified as described30 and images were recorded 2–2.5 μm underfocus in a Philips CM200 FEG microscope at a nominal magnification of 38,000× with an electron dose of 22e- Å-2. All steps of image processing, three-dimensional reconstruction, resolution assessment and data visualization were performed essentially as described31. Three-dimensional reconstructions of CIV and PBCV-1 were computed to 26 Å resolution from 460 and 356 boxed particle images, respectively, as described32,33. Comparison with an independent reconstruction of PBCV-1 (data not shown) showed that the correlation coefficient remained above 0.5 out to 27 Å resolution. The absolute hand of the PBCV-1 T = 169d skew lattice was determined from eight pairs of micrographs recorded at 0° and -5° tilt in the microscope and at ∼2.2 μm underfocus as described34. The average correlation coefficients comparing raw images to model projections were 0.297 ± 0.098 and 0.157 ± 0.076 for the dextro and laevo data sets, respectively. In addition, only 5 of the 189 images of the tilted specimens were designated as laevo and these did not statistically differ from a dextro choice.
Acknowledgments
We thank J. C. Veyrunes and D. Burbank for their help in purifying CIV and PBCV-1, respectively, R. Ashmore for programming, and A. Friedman, M. Graves, L. Lane, A. Simpson, S. Walker, and W. Zhang for helpful discussions. This work was supported in part by NIH grants to T.S.B., M.G.R. and J.V.E., and NSF Grand Challenge and Shared Instrumentation grants to T.S.B. and M.G.R. A grant from the Lucille P. Markey Foundation and reinvestment funds from the office of the University Executive Vice President for Academic Affairs have benefited T.S.B. and M.G.R.
References
- 1.Fukaya M, Nasu S. Appl Entomol Zool. 1966;1:69–72. [Google Scholar]
- 2.Van Etten J, Lane L, Meints R. Microbiol Rev. 1991;55:586–620. doi: 10.1128/mr.55.4.586-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy FA, et al. Virus Taxonomy. Springer-Verlag Wien; New York: 1995. pp. 92–94. [Google Scholar]
- 4.Stoltz DB. J Ultrastruct Res. 1971;37:219–239. doi: 10.1016/s0022-5320(71)80052-7. [DOI] [PubMed] [Google Scholar]
- 5.Stoltz DB. J Ultrastruct Res. 1973;43:58–74. doi: 10.1016/s0022-5320(73)90070-1. [DOI] [PubMed] [Google Scholar]
- 6.Darcy F, Devauchelle G. In: Animal Virus Structure Vol 3, Perspectives in Medical Virology. Nermut MV, Steven AC, editors. Elsevier; Amsterdam: 1987. pp. 407–420. [Google Scholar]
- 7.Williams T. Adv Virus Res. 1996;46:345–412. doi: 10.1016/s0065-3527(08)60076-7. [DOI] [PubMed] [Google Scholar]
- 8.Caspar DLD, Klug A. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 9.Rayment I, Baker TS, Caspar DLD, Murakami WT. Nature. 1982;295:110–115. doi: 10.1038/295110a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimes JM, et al. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 11.Devauchelle G, et al. Curr Top Micro Immun. 1985;116:37–48. doi: 10.1007/978-3-642-70280-8_3. [DOI] [PubMed] [Google Scholar]
- 12.Van Etten JL, Meints RH. Ann Rev Microbiol. 1999;53:447–494. doi: 10.1146/annurev.micro.53.1.447. [DOI] [PubMed] [Google Scholar]
- 13.Guérillon J, Barray S, Devauchelle G. Arch Virol. 1982;73:161–170. doi: 10.1007/BF01314724. [DOI] [PubMed] [Google Scholar]
- 14.Rohozinski J, Girton LE, Van Etten JL. Virology. 1989;168:363–369. doi: 10.1016/0042-6822(89)90277-8. [DOI] [PubMed] [Google Scholar]
- 15.Yamada T, Higashiyama T, Fukuda T. Appl Environ Microbiol. 1991;57:3433–3437. doi: 10.1128/aem.57.12.3433-3437.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landstein D, Burbank DE, Xia Y, Meints RH, Van Etten JL. Virology. 1995;214:413–420. doi: 10.1006/viro.1995.0051. [DOI] [PubMed] [Google Scholar]
- 17.Skrdla MP, Burbank DE, Xia Y, Meints RH, Van Etten JL. Virology. 1984;135:308–315. doi: 10.1016/0042-6822(84)90188-0. [DOI] [PubMed] [Google Scholar]
- 18.Meints RH, Lee K, Burbank DE, Van Etten JL. Virology. 1984;138:341–346. doi: 10.1016/0042-6822(84)90358-1. [DOI] [PubMed] [Google Scholar]
- 19.Fraser CM, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 20.Kelly DC, Vance DE. J Gen Virol. 1973;21:417–423. doi: 10.1099/0022-1317-21-2-417. [DOI] [PubMed] [Google Scholar]
- 21.Cerutti M, Devauchelle G. Virology. 1985;145:123–131. doi: 10.1016/0042-6822(85)90207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stohwasser R, Raab K, Schnitzler P, Janssen W, Darai G. J Gen Virol. 1993;74:873–879. doi: 10.1099/0022-1317-74-5-873. [DOI] [PubMed] [Google Scholar]
- 23.Wrigley NG. J Gen Virol. 1969;5:123–134. doi: 10.1099/0022-1317-5-1-123. [DOI] [PubMed] [Google Scholar]
- 24.Burnett RM. J Mol Biol. 1985;185:125–143. doi: 10.1016/0022-2836(85)90187-1. [DOI] [PubMed] [Google Scholar]
- 25.Tidona CA, Schnitzler P, Kehm R, Darai G. Virus Genes. 1998;16:59–66. doi: 10.1023/a:1007949710031. [DOI] [PubMed] [Google Scholar]
- 26.Stewart PL, Fuller SD, Burnett RM. EMBO J. 1993;12:2589–2599. doi: 10.1002/j.1460-2075.1993.tb05919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butcher SJ, Bamford DH, Fuller SD. EMBO J. 1995;14:6078–6086. doi: 10.1002/j.1460-2075.1995.tb00298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villarreal LP. In: Origin and evolution of viruses. Domingo E, Webster R, Holland J, editors. Academic Press; San Diego: 1999. pp. 391–420. [Google Scholar]
- 29.Van Etten JL, Burbank DE, Xia Y, Meints RH. Virology. 1983;126:117–125. doi: 10.1016/0042-6822(83)90466-x. [DOI] [PubMed] [Google Scholar]
- 30.Olson NH, Baker TS. Ultramicrosc. 1989;30:281–298. doi: 10.1016/0304-3991(89)90057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belnap DM, et al. J Mol Biol. 1996;259:249–263. doi: 10.1006/jmbi.1996.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuller SD, Butcher SJ, Cheng RH, Baker TS. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- 33.Baker TS, Cheng RH. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 34.Belnap DM, Olson NH, Baker TS. J Struct Biol. 1997;120:44–51. doi: 10.1006/jsbi.1997.3896. [DOI] [PubMed] [Google Scholar]


