Abstract
Two views of native α2-macroglobulin are revealed by electron microscopy of negatively stained samples; in one view the molecule resembles a padlock and in the other, a pair of lips. Interconversion of the two views upon tilting establishes that these are two different projected views of the same structure. Furthermore, the two views are related by a 45° rotation about their major axis because they interconvert when the specimens are tilted ±22.5°. Negatively stained molecules on Butvar films present a nearly equal distribution of the two views, whereas in frozen-hydrated samples the molecules almost exclusively are oriented in the lip view. Measurements from both views indicate that the α2-macroglobulin molecule is ~200 Å long and ~140 Å wide. Our results suggest that α2-macroglobulin is composed of two protomeric units, each in the shape of a twisted letter S. These units are joined together at their ends to form a complex with point group symmetry 222. The 45° interconversion angle between the lip and padlock views support this arrangement. Average images of unstained and stained lips are quite similar, indicating that the native structure is consistently preserved by the two electron microscopy procedures used in this investigation. This is substantiated by the interconversion between the lip and padlock views that occurs when the molecule is rotated 90° about its major twofold axis.
INTRODUCTION
α2-Macroglobulin (α2M3) isolated from human plasma is a large glycoprotein (Mr = 725,000) composed of four identical subunits (Mr = 180,000 for each subunit) (Sottrup-Jensen, 1989). A major reason for interest in the structure and function of α2M is its biological role as a scavenger of general proteases in the plasma of all vertebrates (Sottrup-Jensen, 1989).
Two characteristics shapes of the proteolyzed molecule have been seen by electron microscopy: an “H” shape (open form) and an ovate shape composed of two crescents facing each other with a central bar between them “(|)” (closed form) (Bloth et al., 1968; Barrett and Starkey, 1974; Starkey and Barrett, 1977; Schramm and Schramm, 1982; Tapon-Bretaudiere et al., 1985; Osada et al., 1986; Ikai et al., 1987; Bretaudiere et al., 1988; Gonias and Figler, 1989). Initially, the H and ovate shapes were presumed to represent the native and proteolyzed forms of α2M, respectively (Barrett and Starkey, 1974; Schramm and Schramm, 1982; Feldman et al., 1985). However, it has been shown recently that these two shapes are present only in samples containing the proteolyzed α2M, therefore suggesting that the two shapes represent two different projected views of the proteolyzed complex (Tapon-Bretaudiere et al., 1985; Bretaudiere et al., 1988). Tilting experiments were used to determine the relationship between the two views (Stoops et al., 1989). The H and ovate views of chymotrypsin-treated α2M interconverted if the molecule was tilted by 90° about the minor axis of the H view or the major axis of the ovate view. Thus, the interconversion between these shapes verified that the two views are orthogonal projections of the same structure.
These results raise questions about the structure of the native α2M and its subsequent conversion to the proteolyzed form. Based on electron microscopy studies of native α2M, we proposed that it has a padlock-like shape, the top and bottom arms of which open on proteolysis to give the H shape (Bretaudiere et al., 1988). Recently, Gonias and Figler (1989) proposed that the native structure has the shape of a doughnut to which two to four spheres are attached. This doughnut-shaped motif was also found by others who examined native α2M samples by electron microscopy (Tapon-Bretaudiere et al., 1985; Ikai et al., 1987). Gonias and Figler (1989) also proposed that, upon proteolysis of α2M, the spheres contract toward the central ring and then elongate to produce the H shape.
Because α2M consists of two identical protomers (Jensen and Sottrup-Jensen, 1986), it is reasonable to expect that projections of the molecule might reveal a twofold axis or mirror plane of symmetry. However, negatively stained samples of native α2M often consist of particles of varying shapes, many with no apparent symmetry. This suggests that the α2M structure may be easily perturbed by the electron microscopy preparation procedures. Our electron microscopy studies led us to conclude that the different shapes may have resulted from denaturation of the native structure (Bretaudiere et al., 1988). The heterogeneity of the particle shapes obtained by us (Bretaudiere et al., 1988) and others (Nishigai et al., 1985; Tapon-Bretaudiere et al., 1985; Ikai et al., 1987, 1988; Boisett et al., 1989; Gonias and Figler, 1989) and the lack of information concerning their relationships raise questions about the reliability of the projected views of the native structure that have been observed. This, in turn, casts doubt on proposals about structural changes that accompany proteolysis of the native complex (Feldman et al., 1985; Bretaudiere et al., 1988; Gonias and Figler, 1989).
To reduce the variability induced by electron microscopy, Gonias and Figler (1989) cross-linked the α2M molecule with cis-dichlorodiammineplatinum, presumably to stabilize the complex prior to microscopy. Molecules of α2M treated in this way appeared to be mainly doughnut-shaped; however, considerable variability in the shapes and in the distribution and number of spheres around the central ring was observed.
In the present study we have taken precautions to minimize perturbation of the α2M structure as might be induced by conventional staining procedures. We have viewed molecules that appear as two predominant shapes: padlocks and lips. The latter shape, which has not been reported previously, is shown by tilting experiments to be related to the padlock view by a 45° rotation of the structure on its major axis. Images of frozen-hydrated α2M show that most molecules have the same lip appearance as seen in negative stain, indicating that the native structure of α2M is well preserved by both microscopy procedures. Furthermore, this conclusion is supported by image processing studies which demonstrate the close correspondence between the projections obtained by these two different methods.
MATERIALS AND METHODS
Protein preparation
α2M was purified using a modified version of the procedure of Imber and Pizza (1981). The purification procedure was followed by immunoaffinity chromatography to remove trace amounts of the “fast” form of α2M when necessary (Strickland et al., 1988). Based on the number of sulthydryl groups released during the reaction with excess trypsin (Steiner et al., 1985) the preparation was judged to be >98% active. We determined the amount of inactive α2M (α2M with its thioesters hydrolyzed) by using a sandwich ELISA (Larsson et al., 1989) to measure the amount of complex present and by analyzing the protein by SDS-PAGE and nondenaturing PAGE. The protein was then stored at 4°C. Assays performed at different times revealed that the preparation remained fully active throughout the electron microscopy experiments. Protein concentration was determined spectrophotometrically with (Jones et al., 1972).
Electron microscopy
α2M in 1 mM sodium phosphate (pH 7.2) was deposited on Butvar (Monsanto) or carbon-coated Butvar film on copper grids by the spray method (Oliver, 1973) with 0.25% methylamine tungstate used as a negative stain. To minimize denaturation effects, α2M was mixed with stain and deposited and dried on the grids within 10 sec. The method of Strickland and Bhattacharya (1984) was used to demonstrate that methylamine tungstate stain reacts negligibly with the thioester of α2M under these conditions. These experiments also showed that <5 mol% of the “fast” form of α2M is generated after exposure to the stain for 15 min. The concordance between the average images derived from the negative-stain and vitreous-ice data further supports this contention (see Results).
Specimens supported by carbon-coated Butvar films were examined in a JEOL JEM 1200 electron microscope at 100 kV with minimal irradiation microscopy (40 e/Å2). Micrographs were recorded on Kodak SO-163 film and developed in full-strength Kodak D19 for 12 min. Molecules deposited on the Butvar-coated grids were imaged by conventional irradiation procedures. Micrographs of negatively stained samples were recorded at × 75,000 with the objective lens at 0.1 µM underfocus. For cryomicroscopy, a 3-µl sample of α2M (0.1 mg/ml in 10 mM citrate, pH 6.0) was deposited on a glow-discharged, carbon-coated holey grid, the excess removed by blotting with filter paper, and the grid was then rapidly cooled in liquid ethane. Specimens were kept below −150°C in a Gatan cold holder and were examined in a Phillips Em 420 electron microscope operated at 80 kV and × 49 000 under minimal irradiation conditions (~20 e/Å2). Images were recorded at ~1.8 µM underfocus and the micrographs were developed as described above for the minimal exposure of the stained samples.
Image averaging
Image processing was performed on a VAX 11/785 (Digital Equipment Corp.) and an IRIS workstation (Silicon Graphics Inc.), both running the UNIX operating system. Our SUPRIM image processing system was used to analyze images. Eighteen regions from six micrographs were digitized with an Eikonix Model 78/99 camera. Three micrographs each from frozen-hydrated and stained samples were digitized as 1536 × 1536 × 12 bit arrays with a pixel size of 5 Å. Particle images were interactively extracted from the displayed data in a 60 × 60 pixel window; 150 images of frozen-hydrated particles and 236 images of negatively stained particles were extracted. The negatively stained particles were visually sorted into two sets: 71 exhibiting the lip appearance and 165 exhibiting the padlock shape. All froen-hydrated particles had the lip appearance. These three sets of images were separately aligned with representative reference images by auto- and cross-correlation procedures (Frank, 1980). Three or four iterations of alignment and averaging were performed for each group of particles. Each successive iteration used the previous average as a reference. The alignment process was repeated until changes in the positions of individual images were judged to be insignificant.
After alignment, each particle was masked to eliminate spurious background noise (Frank and Verschoor, 1984), and this data was analyzed with the multivariate statistical technique of correspondence analysis (Van Heel and Frank, 1981). Particle images of each type were organized into clusters by hierarchical ascendent classification (Benzecri, 1982) based on the first 10 eigenvector coordinates. A dissimilarity index was computed for each image (Ward, 1963). Based on the dissimilarity indices of the nodes, a level was selected, the hierarchical tree was cut, and average images corresponding to each “branch” of the tree at that level were constructed.
The resolution limits of the averaged data sets were estimated from calculations of the spectral signal-to-noise ratios (Unser et al., 1987), the phase residuals (Frank et al., 1981), and the Fourier ring correlations (Saxton and Baumeister, 1982; Van Heel and Stoffler-Meilicke, 1985). The reported error ranges are at the 95% confidence limit for the spectral signal-to-noise method, and two standard deviations for the phase residual and Fourier ring correlation methods. The values of the phase residual and ring correlation resolutions represent the means for 10 separate comparisons of average images generated by random division of the image data into two subsets. Unser et al. (1989) showed the necessity for comparing a number of different averages to arrive at a statistically correct estimate for the resolution.
Fine details in the averages were enhanced with a high-pass Butterworth filter (Gonzalez and White, 1987). A low-pass Fermi filter (Frank et al., 1985) was applied at the resolution limit determined by the spectral signal-to-noise ratio method. Final images were displayed as continuous-tone, pseudo-color representations, overlayed with contours, and printed on a Mitsubishi C650 color printer.
Analysis of tilted and untilted interconverting images
Identical areas from images of stained samples that had been tilted ±22.5° were digitized. Those particle images that appeared either as padlocks or lips at a 22.5° tilt were extracted along with the corresponding image at a −22.5° tilt and displayed with enhanced contrast on a high-resolution video monitor. Images were recorded on a Matrix ProColor film recorder. Twenty eight pairs of lips and padlocks views of molecules with their major axes parallel to the tilt axis were sorted visually into two sets. Each set was aligned against the respective average obtained from the previous averaging procedures. Images were recorded and displayed as described above.
RESULTS
a. Electron Microscopy
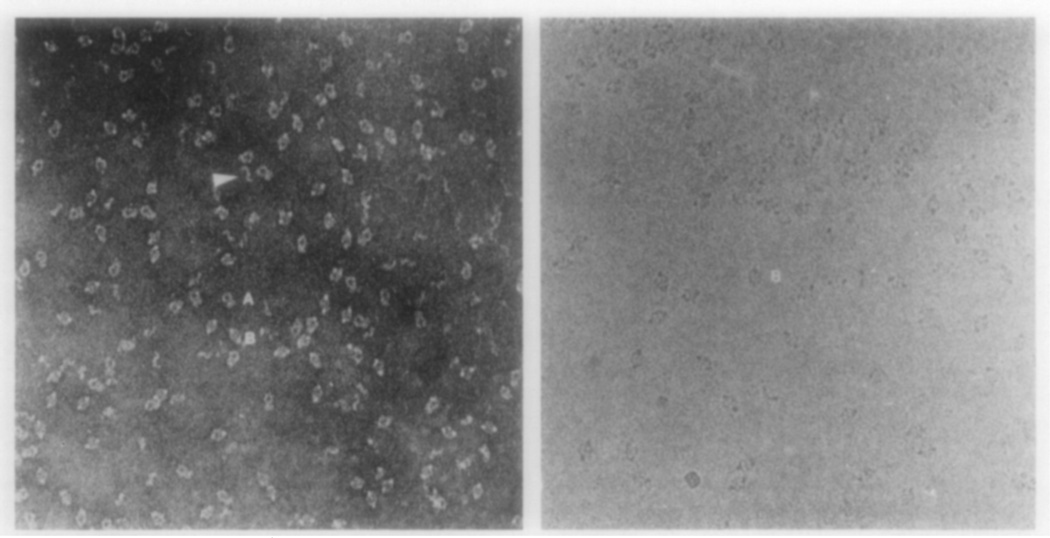
A field of α2M molecules on Butvar film (Fig. 1, left) consists primarily of particles displaying two characteristic shapes: a padlock (the particle labeled A) and a pair of lips (the particle labeled B), hereafter referred to as “lip.” A corresponding field of frozen-hydrated molecules suspended in a layer of ice (Fig. 1, right) consists exclusively of the lip-shaped particles. Both particle types have approximately the same major dimension (200 Å).
FIG. 1.
Electron micrographs of α2M stained with methylamine tungstate (left) or frozen-hydrated (right) (× 175,000). (A) and (B) denote the padlock and lip views, respectively. The two views are nearly equally represented in the negatively stained sample, whereas the lip view is almost exclusively seen in the frozen-hydrated sample. The arrow points out a possible dimer, which presumably results from the dissociation of the complex.
These two distinct shapes initially were thought to represent projections of different α2M structures. However, interconversion of the two shapes occurs when the samples are tilted ±45° (Fig. 2) and this demonstrates that the lips and padlocks represent different projected views of the same structure. Since the lip view is seen almost exclusively in frozen-hydrated samples and the padlock view predominates for molecules adsorbed to nitrocellulose film (Bretaudiere et al., 1988), these findings show that surface interactions may induce a preferential orientation of the molecules. For example, some molecules appear to have strong interactions at the air-water interface in specimens prepared for cryoelectron microscopy (Dubochet et al., 1988). Because proteins are known to interact with nitrocellulose film, α2M may also adsorb in a preferred orientation. However, a nearly equal distribution of the lip and padlock views and intermediate views thereof are seen on the Butvar film, suggesting that there is little, if any, interaction between the α2M and this support film. Similar results (three distinct projected views) were obtained for ornithine decarboxylase on Butvar films (Stoops et al., 1991). The interconversion of the two projected views demonstrates that the three-dimensional structure of α2M is well preserved in the stain on the Butvar film. The lack of appreciable interactions between the molecule and the Butvar film may contribute to the preservation of the native structure. For instance, when methylamine tungstate-stained α2M was supported by nitrocellulose, more than half of the particles appeared to have nonuniform shapes (Bretaudiere et al., 1988).
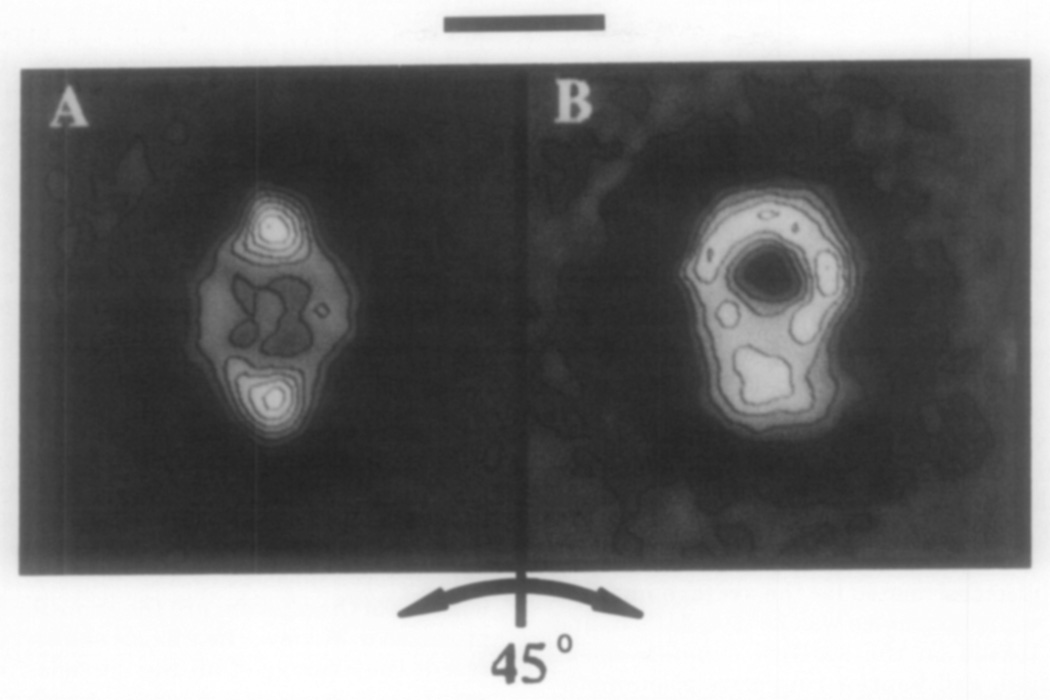
FIG. 2.
Computer averaged images of 28 α2M molecules shown to interconvert between the lip (A) and the padlock (B) views obtained from the specimen tilted ±22.5°. The major axis of the tilted images are 10–15% shorter than the average images in Fig. 3. The shortening of this dimension resulted from the fact that the major axis of the molecule was not exactly parallel to the tilt axis of the stage. The untilted particles in this data set had a major axis of 200 Å. The scale bar corresponds to 100 Å.
Based on the interconversion of the padlock with the lip that took place when the structure was rotated 45°, we propose that the lip and padlock shapes are related by a 45° rotation of the structure on its major axis (Fig. 2). Interconversions between orthogonal projections have been demonstrated with ornithine decarboxylase (Stoops et al., 1991), and observed with the proteolyzed form of α2M (Stoops et al., 1989) using the Butvar support film. The lack of any major interaction between the support film and the molecules affords the opportunity to obtain some structures oriented with their lip view perpendicular to the beam when the stage is tilted 22.5° (Stoops et al., 1991). Thus, the padlock view of the molecule is revealed when the stage is rotated −22.5°.
Some α2M molecules have the shape of a question mark (Fig. 1, denoted by the arrow). This structure may arise from dissociation of α2M at a low concentration (25 µg/ml) in the stain, resulting in the formation of dimers. The fact that this shape was not observed in the frozen-hydrated fields may be explained by the higher protein concentration employed (0.1 mg/ml) and rules out the possibility that this shape represents a contaminating protein in the α2M preparation. The dimer, held together by two disulfide bonds (Jensen and Sottrup-Jensen, 1986), is the protomeric unit in the complex. A structure comprised of two protomers that face each other might give rise to the padlock view.
b. Image Averaging
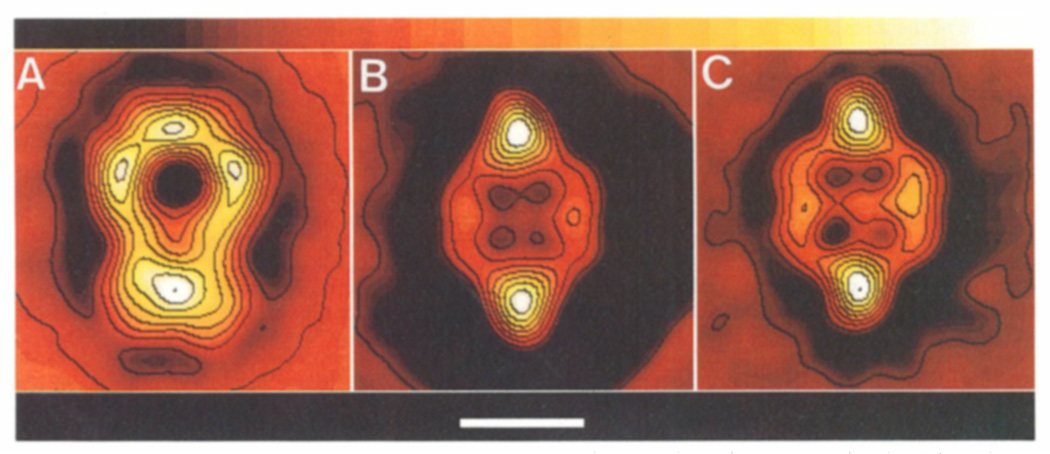
The average images for the padlock (n = 165) and lip (n = 71) views of stained molecules and the lip view (n = 150) of unstained molecules, where n is the number of particles averaged in each set, are shown in Figs. 3A-3C, respectively. The longest dimension in all views is ~200 Å. The width of the lip in both samples is 138 Å, and the width of the padlock loop is 142 Å. The padlock view exhibits the highest density of protein (stain exclusion) at its base and has three other dense regions distributed around the hole that lies on the vertical axis in the upper half of the structure (Fig. 3A). The lip has an oval shape with its maximum density on the vertical axis at both the top and bottom of the structure. The center of the lip has a square-shaped, low-density region ~60 Å wide, consisting of four depressions of even lower protein density.
FIG. 3.
Average images from the stained α2M molecules derived from the carbon-coated Butvar film in padlock (A) and lip (B) views and from unstained molecules in the lip view (C). The bar corresponds to 100 Å, the color bar at the top of the figure shows the progression of protein density from low (dark) to high (white). Image processing of the molecules on the Butvar film was made difficult because of the variable orientations between the padlock and lip views.
The resolution estimated for the average image of negatively stained samples was 35 ± 4, 41 ± 3, and 28 ± 3 Å (based on the spectral signal-to-noise, phase residual, and Fourier ring correlation methods, respectively; see Materials and Methods). The corresponding values of the resolution for the average image of the unstained sample were similar, being 34 ± 7, 38 ± 4, and 25 ± 2 Å. Similar differences in resolution using these criteria have been reported previously (Unser et al., 1987, 1989). Accordingly, all averages were low-pass filtered to 35 A (Fig. 3).
Correspondence analysis nearly equally segregated the padlock images into four major clusters. Images in the different clusters primarily varied in the compactness of density in the upper loop and bottom region as observed previously (Bretaudiere et al., 1988). Similar analysis of stained α2M in the lip view divided the images into two nearly equal-sized clusters differing primarily in the density distribution within the central, square-shaped region. Correspondence analysis of the lip images from unstained specimens gave three clusters of nearly equal membership. The three clusters were mainly distinguished by the appearance of the square-shaped region as was found for stained molecules. The average obtained from images in one cluster appeared similar to the average shown in Fig. 3C; the other clusters of images showed a more asymmetric distribution of density in the square-shaped region. The reason for variability in these clusters is unknown, but variability may reflect small differences in the orientation of the structure on the support surface. Since the overall shape, size, and major features of the clusters are the same for a particular projected α2M view, the images shown in Fig. 3 represent the average of all the particles in each data set.
DISCUSSION
Previous electron microscopy studies of native α2M exhibited a wide variety of projected views. Consequently, proposed models for the α2M structure are being reevaluated. In the present study, the interconversion between the lip and the padlock views (Fig. 2), the stability of the α2M molecule in the presence of stain (see Materials and Methods), and the close correspondence between the different average images obtained from stained and frozen-hydrated samples (Fig. 3) strongly suggest that the native structure of α2M has been well preserved. Because each of the two to four clusters found by correspondence analysis for each view closely resembles the global average, systematic variations in the lip or padlock views are small. On the basis of these observations, we believe that the padlock and lip views represent projected images of the same structure.
The doughnut (Nishigai et al., 1985; Tapon-Bretaudiere et al., 1985; Ikai et al., 1987, 1988; Gonias and Figler, 1989) and other motifs (Nishigai et al., 1985; Tapon-Bretaudiere et al., 1985) are difficult to explain on the basis of the results presented here. It is unlikely that the doughnut represents yet another projection of the native α2M molecule since the lip-to-padlock interconversion seems to preclude a doughnut-shaped top view that is orthogonal to both the lip and padlock views. We do observe the doughnut shape when α2M is stained with uranyl acetate or uranyl formate. Because the doughnut shape is not observed in samples stained with methylamine tungstate (pH 7.2) or in frozen-hydrated samples, it may be that the low pH of the uranyl salt solutions (~4–5) perturbs the α2M structure. In addition, we have not observed the doughnut view in α2M supported on Butvar film and stained with methylamine tungstate. This and previous studies (Stoops et al., 1991) suggest that molecules can assume random orientations on Butvar films, and thus, the doughnut shape ought to be observed if it truly represents a projected view of the native α2M structure.
This study supports our previous proposal regarding the assembly of the native structure of α2M (Bretaudiere et al., 1988) and offers additional insight about the shape and arrangement of the two protomers in the complex. We propose that the protomer has the shape of a twisted letter S such that the top half of the structure is rotated approximately 90° with respect to its bottom half. A projection of this structure could give the question mark view seen in Fig. 1. We propose that the joining of the ends of the two protomers gives rise to the α2M structure which has point group symmetry 222.
This study establishes a reliable methodology for imaging the structure of α2M by electron microscopy and demonstrates that the padlock and lip views are two distinct but related projections of the same structure. A three-dimensional reconstruction of α2M is needed to understand better the spatial relationship between these projections. Knowledge of the three-dimensional structures of the native and proteolyzed forms of α2M might also identify structural changes that accompany the transformation of the native molecule to the proteolyzed form.
Acknowledgments
The authors thank Pamela P. Powell for editorial assistance and Annie M. Rose for manuscript preparation. This work was supported in part by Grants GM-39536 (to J.-P.B., and now to J.K.S.), HL-42886 to J.K.S., HL-30200 to D.K.S., and GM-33050 to T.S.B. from the National Institutes of Health, United States Public Health Service. D.K.S. is the recipient of a Research Career Development Award (HL-02113).
Footnotes
A preliminary report of this study was presented at the 74th Annual Meeting of the Federation of American Societies for Experimental Biology held in New Orleans, Louisiana on June 4–7, 1990 (STOOPS, J. K., OLSON, N., BAKER, T., BRETAUDIERE, J.-P., AND STRICKLAND, D. K. [1990] FASEB J. 4, A2160).
Abbreviations used: α2M, α2-macroglobulin; SDS, sodium do-decyl sulfate; PAGE, polyacrylamide gel electrophoresis.
Note added in proof. Random orientations of α2M have been obtained in vitreous ice revealing the padlock as well as the lip shape. In addition, a figure 8 shape has been observed, this further supporting the proposal that the molecule has point group symmetry 222.
REFERENCES
- Barrett AJ, Starkey PM. Bayer-Symp. 1974;5:72–77. [Google Scholar]
- Benzecri J-P. Cahiers de l’Analyse des Donnees. 1982;7:209–218. [Google Scholar]
- Bloth B, Chesebro B, Svehag S-E. J. Exp. Med. 1968;127:749–756. doi: 10.1084/jem.127.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisett N, Taveau J-C, Pochan F, Tardieu A, Barry M, Lamy JN, Delain E. J. Biol. Chem. 1989;264:12,046–12,052. [PubMed] [Google Scholar]
- Bretaudiere J-P, Bretaudiere JT, Stoops JK. Proc. Natl. Acad. Sci. USA. 1988;85:1437–1441. doi: 10.1073/pnas.85.5.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Chang M-J, Homo J-C, Lepault J, McDowell AW, Schultz P. Q. Reu. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Feldman SR, Gonias SL, Pizzo SV. Proc. Natl. Acad. Sci. USA. 1985;82:5700–5704. doi: 10.1073/pnas.82.17.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. In: Topics in Current Physics. Hawkes PW, editor. Berlin: Springer-Verlag; 1980. pp. 187–221. [Google Scholar]
- Frank J, Verschoor A. J. Mol. Biol. 1984;178:696–698. doi: 10.1016/0022-2836(84)90245-6. [DOI] [PubMed] [Google Scholar]
- Frank J, Verschoor A, Boublik M. Science. 1981;214:1353–1355. doi: 10.1126/science.7313694. [DOI] [PubMed] [Google Scholar]
- Frank J, Verschoor A, Wagenkn-Echt T. In: New Methodologies in Studies of Protein Confirmation. Wu TT, editor. New York: Van Norstrand-Rhinehold; 1985. pp. 36–89. [Google Scholar]
- Gonias SL, Figler NL. J. Biol. Chem. 1989;264:9565–9570. [PubMed] [Google Scholar]
- Gonzalez RC, Wintz P. Digital Image Processing. 2nd ed. London: Addison-Wesley; 1987. pp. 170–171. [Google Scholar]
- Ikai A, Nishigai M, Osada T, Arakawa H, Kikuchi M. J. Protein Chem. 1987;6:81–93. [Google Scholar]
- Ikai A, Osada T, Nishigai M. J. Biochem. 1988;103:218–224. doi: 10.1093/oxfordjournals.jbchem.a122251. [DOI] [PubMed] [Google Scholar]
- Imber M, Pizzo S. J. Biol. Chem. 1981;256:8134–8139. [PubMed] [Google Scholar]
- Jensen PEH, Sottrup-Jensen L. J. Bill. Chem. 1986;261:15,863–15,869. [Google Scholar]
- Jones JM, Creeth JM, Kekwick RA. Biochem J. 1972;127:187–197. doi: 10.1042/bj1270187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson L-J, Neuenschwander DE, Strickland DK. Biochemistry. 1989;28:7636–7643. doi: 10.1021/bi00445a020. [DOI] [PubMed] [Google Scholar]
- Nishigai M, Osada T, Ikai A. Biochim. Biophys. Acta. 1985;831:236–241. doi: 10.1016/0167-4838(85)90040-8. [DOI] [PubMed] [Google Scholar]
- Oliver RM. In: Methods in Enzymology. Hirs CHW, editor. Vol. 27. San Diego: Academic Press; 1973. pp. 616–672. [DOI] [PubMed] [Google Scholar]
- Osada T, Masaaki N, Ikai A. J. Ultrastruct. Mol. Struct. Res. 1986;96:136–145. doi: 10.1016/0889-1605(86)90014-5. [DOI] [PubMed] [Google Scholar]
- Saxton WO, Baumeister W. J. Microsc. 1982;127:127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Schramm HJ, Schramm W. Hoppe-Seyler’s Z. Physiol. Chem. 1982;363:803–812. doi: 10.1515/bchm2.1982.363.2.803. [DOI] [PubMed] [Google Scholar]
- Sottrup-Jensen L. J. Biol. Chem. 1989;264:11,539–11,542. [Google Scholar]
- Starkey PM, Barrett AJ. In: Protainases in Mammalian Cells and Tissues. Barreit AJ, editor. Amsterdam: Elsevier/Worth-Holland; 1977. pp. 663–696. [Google Scholar]
- Steiner JP, Bhattacharya P, Strickland D. Biochemistry. 1985;24:2993–3001. doi: 10.1021/bi00333a028. [DOI] [PubMed] [Google Scholar]
- Stoops JK, Bretaudiere J-P, Strickland DK. Biochem. Biophys. Res. Commun. 1989;161:216–220. doi: 10.1016/0006-291x(89)91583-0. [DOI] [PubMed] [Google Scholar]
- Stoops JK, Momany C, Ernst SR, Oliver RM, Schroeter JP, Bretaudiere J-P, Hackert ML. J. Electron Micros. Tech. 1991;18:157–166. doi: 10.1002/jemt.1060180210. [DOI] [PubMed] [Google Scholar]
- Strickland DK, Bhattacharya P. Biochemistry. 1984;23:3115–3123. doi: 10.1021/bi00309a002. [DOI] [PubMed] [Google Scholar]
- Strickland DK, Steiner JP, Migliorini M, Battery FD. Biochemistry. 1988;27:1458–1466. doi: 10.1021/bi00405a010. [DOI] [PubMed] [Google Scholar]
- Tapon-Bretaudjere J, Bros A, Couture-Tosi E, De-Lan E. EMBO J. 1985;4:86–69. doi: 10.1002/j.1460-2075.1985.tb02321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unser M, Trus BL, Steven AC. Ultramicroscopy. 1987;23:39–52. doi: 10.1016/0304-3991(87)90225-7. [DOI] [PubMed] [Google Scholar]
- Unser M, Trus BL, Frank J, Steven AC. Ultramicroscopy. 1989;30:429–439. doi: 10.1016/0304-3991(89)90074-0. [DOI] [PubMed] [Google Scholar]
- Van Heel M, Frank J. Ultmmicroscopy. 1981;6:187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]
- Van Heel M, Stoffler-Meilicke M. EMBO J. 1985;4:2389–2395. doi: 10.1002/j.1460-2075.1985.tb03944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JH., Jr Am. Stat. Assoc. J. 1963;58:236–244. [Google Scholar]