Abstract
The three-dimensional structure of intact human rhinovirus 14 (HRV-14) complexed with Fab fragments (Fab17-IA) from a strongly neutralizing antibody that binds bivalently to the virion1,2 has been determined to 4.0 Å resolution by a combination of X-ray crystallography and cryo-electron microscopy. In contradiction to the most commonly held model of antibody-mediated neutralization, Fab17-IA does not induce a conformational change in the HRV-14 capsid. Instead, the paratope of the antibody undergoes a large conformational change to accommodate the epitope. Unlike any previously described antibody–antigen structure, the conserved framework region of the antibody makes extensive contact with the viral surface. Fab17-IA penetrates deep within the canyon in which the cellular receptor for HRV-14 binds3,4. Hence, it is unlikely that viral quaternary structure evolves merely to evade immune recognition. Instead, the shape and position of the receptor-binding region on a virus probably dictates receptor binding and subsequent uncoating events and has little or no influence on concealing the virus from the immune system.
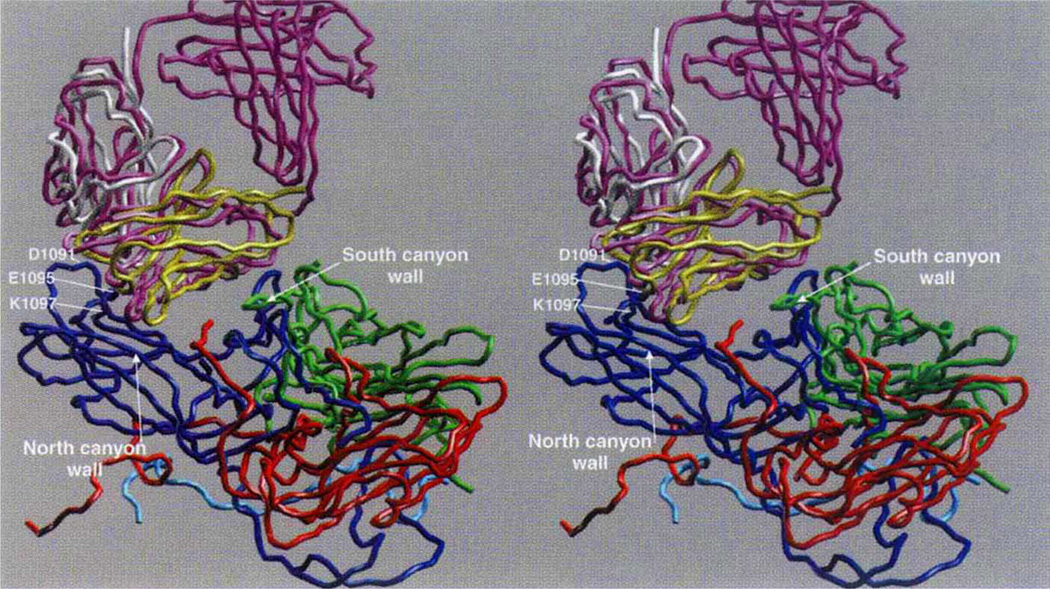
The capsid of HRV-14 is composed of 60 copies of four viral proteins, VP1–VP4. Each of the first three proteins has a relative molecular mass of ~30,000 (Mr ~ 30K) and a similar ²-barrel structure. The smaller VP4 (Mr ~ 7K) lies at the capsid-RNA interface with an extended structure3. The monoclonal antibody 17-IA binds to the NIm-IA site which was defined by natural escape mutations at residues D1091 and E1095 on the B–C loop of VP1 (the first digit designates the viral protein and the remaining three designate the residue number). The NIm-IA epitope lies at the ‘north rim’ of a depression (canyon) on the viral surface that encircles each of the five-fold icosahedral axes of the virus3 and is where the receptor, ICAM-1, binds4.
The variable domains (VH · VL) of Fab17-IA make extensive contact with both the north and south walls of the canyon region. At the north wall, the antigen-binding region (paratope) contacts ~550 Å2 of the viral surface around NIm-IA site (Fig. 1). The heavy-chain hypervariable region dominates the contact surface: 23 residues from the heavy chain but only 9 residues from the light chain. As was proposed on the basis of cryo-electron microscopy on this HRV/Fab complex5, coulombic interactions dominate the paratope–epitope interface with roughly two-thirds of the total buried viral surface being contributed by charged side chains. Fab17-IA contacts ~49, 12 and 0.3 Å2 of the K1097, K1085 and K1236 side chains, respectively. These contact areas correlate with a decrease in Fab affinity by 44-, 9.3- and 0.8-fold, respectively, when residues K1097, K1085 and K1236 are separately mutated to glutamate5. Additional charge interactions include Arg 91L (Fabl7 numbering corresponds to Kabat6: the last letter designates the chain type) which forms salt bridges with the two natural escape mutation sites (Glu 1095, Asp 1091), and Asp 54H and Asp 56H interdigitate between Arg 1094 and Lys 1097. Four residues, His 1078, Val 1079, Thr 1080 and Asp 1101, which are all considered to lie at the bottom of the north side of the canyon, contact the bound Fab17. Thus, these interactions provide strong evidence that antibodies are capable of binding deep into viral crevices.
Figure 1.
Cα backbone of the initial and final models. VP1 is blue, VP2 green, VP3 red, initial Fab purple, final VL domain white, and final VH domain yellow. The RNA interior of the virus is towards the bottom of the figure, and some key contact residues around the HRV14 Nlm-IA site are labelled.
The south wall of the canyon (~300 Å2) is also contacted by Fab17-IA. This region is nearly identical to that recognized by the HRV-14 receptor ICAM-1 (ref. 4) and includes a fairly even distribution of charged, polar and hydrophobic residues. Approximately one-third of this area involves contacts with heavy-chain CDR3 residues (where CDR is complement-determining region) that mostly interact with the carboxy terminus of VP3 near the base of the canyon. The remaining ~200 Å2 of the surface contacted by the VH domain comprises residues from the conserved framework regions FR1 and FR3. The importance of these framework contacts is suggested by results on another Fab (Fab12-IA) that binds to HRV-14 in the same orientation as Fab17-IA and only differs from Fab17-IA by 5 amino acids (Z. C. Che, T. J. S., N.H.O. and T.S.B., manuscript in preparation). Both monoclonal antibodies probably came from the same mother cell but differ as a result of somatic mutations. Two of the five amino-acid differences between antibodies 12-IA and 17-IA lie within the CDR1 loop (32H and 34H); the remaining three are in the FR3 region (68H, 82aH and 76H). The first two of these FR3 residues (68H and 82aH) contact the south wall of the canyon. Although somatic mutations occur throughout the variable domain7, it is unlikely that mutations in these framework sites that contact the viral surface are merely coincidental.
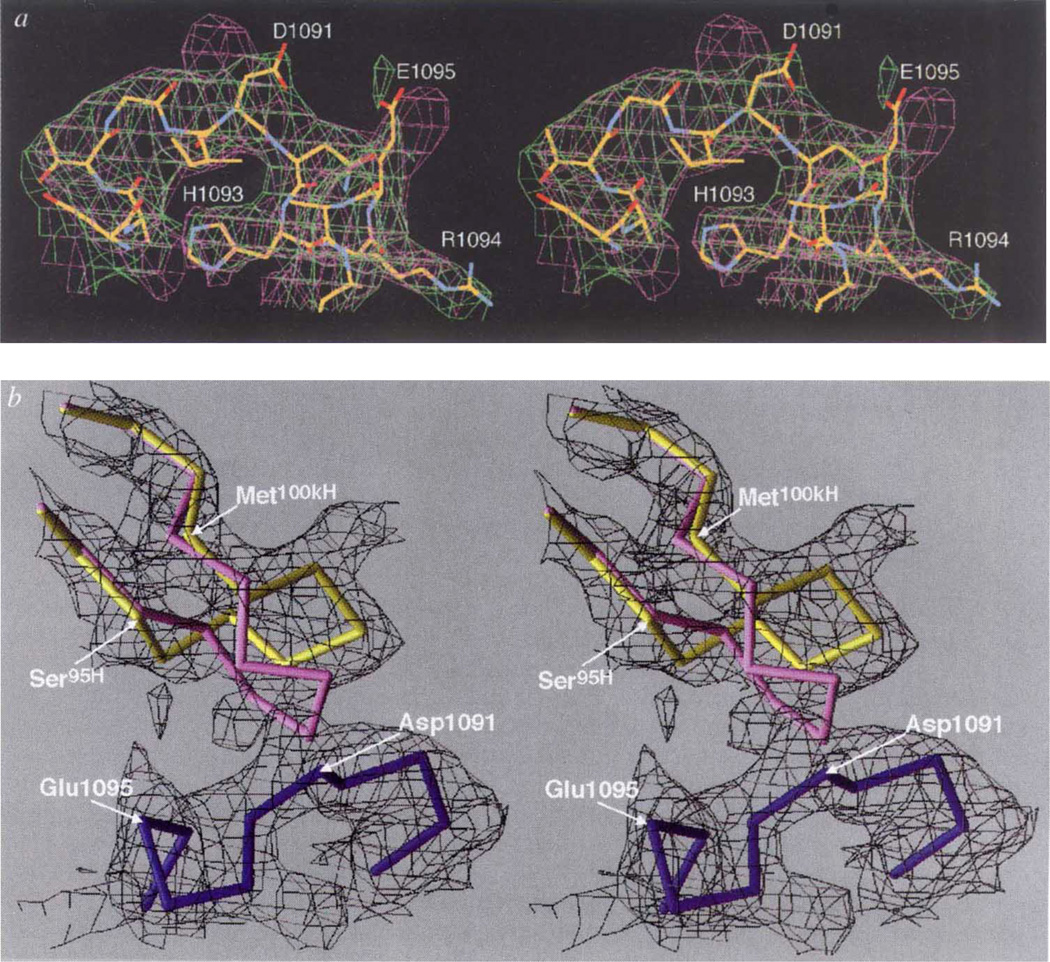
Several large changes in the Fab17-IA structure accompany binding to HRV-14, although the virus capsid remains essentially rigid (Fig. 2a). A large conformational change occurs in the heavy-chain CDR3 loop (Fig. 2b, c, d) where the Cα atom of Tyr102H moves up to 5.6 Å from its position in the unliganded Fab structure (Fig. 2b). This movement opens the cleft between the heavy- and light-chain hypervariable regions (Fig. 2c, d) in a manner quite similar to that in the heavy-chain CDR3 loop of other Fab/antigen complexes8 and generates additional space for Arg 91L to move further into the cleft, where it forms salt bridges with Asp 1091 and Glu 1095. The only changes in the HRV-14 capsid around the epitope are confined to the side chains on the NIm-IA loop that contact the Fab.
Figure 2.
Conformational changes at the paratope/epitope interface. a, Comparison between Nlm-IA loop region in the unliganded HRV-14 (mauve) and the Fab/virus complex (green) electron density maps. The atomic coordinates of the native HRV-14 structure were used to calculate the unliganded HRV-14 electron density map, whereas the electron density map of the virus–Fab complex was calculated using phases from 20-fold real-space averaging. The structure of the liganded structure is coloured according to atom type (carbon, yellow; oxygen, red; nitrogen, blue). b, Initial (unliganded Fab structure) and final structures of the heavy-chain CDR3 are purple and yellow, respectively. The Nlm-IA site is shown in dark blue. The black cage is the 4Å electron density map calculated using the experimentally determined phases from 20-fold real-space averaging and phase extension. For this diagram, the VH domains of the initial and final models were aligned to each other. c, d, Stereo diagram of electrostatic potential and structural changes in the hypervariable region before (c) and after (d) binding to the Nlm-IA site. The view is towards the hypervariable region, with the VL domain towards the top of the diagram and the VH domain towards the bottom. Surfaces of positive and negative potential are depicted blue and red, respectively. The largest conformational changes occur in the heavy-chain CDR3 loop and in the side chain of Arg91L. White crosses provide points of reference in the stereo views.
Removed from the Fab-virus contacts (>12Å away), unassigned electron density was observed inside the HRV-14 drug-binding cavity and this distorts the canyon floor just as WIN compounds do9. We attribute this density to the cryoprotectant PEG400 which was present in high concentration (40%) for these crystal diffraction experiments; this density was not observed in the native HRV-14 electron density maps. Alternatively, Fab–virus interactions might facilitate the binding of this small compound but, because conformational changes do not occur at the contact interfaces upon Fab binding, it is unlikely that the canyon floor would be affected by Fab binding. Nevertheless, the appearance of this density raises the possibility that previously observed ‘pocket factors’10, which bind to the same site on other serotypes of HRV and give a nearly identical appearance, may have come from the crystallization mother liquor rather than from the host cell.
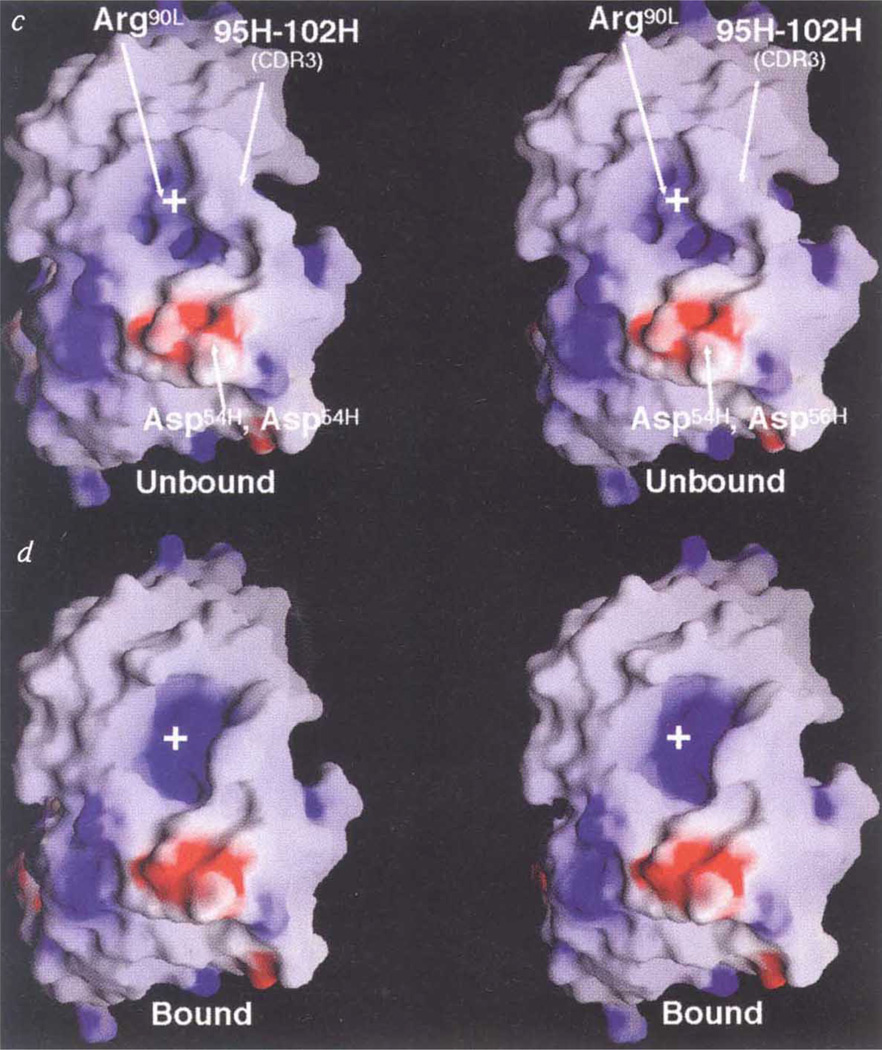
The canyon hypothesis3,11 proposes that conserved residues in the floor of the HRV-14 canyon bind to the cell receptor (ICAM1), but that the small dimensions of the canyon should limit accessibiity to antibodies (Fig. 3). Also, HRV-14 morphology should allow residues at the top of the canyon to mutate freely without affecting virus–receptor interactions. Although a number of natural escape mutation sites lie at the upper north rim of the canyon, our crystal structure analysis shows that Fab17-IA contacts extensive regions inside the canyon, including most of those residues involved in receptor recognition. Thus, the lack of mutations elsewhere in the Fab contact region is more likely to be due to the deleterious effects of such mutations, rather than to antibody accessibility. Indeed, mutation of Lys 1097, which lies halfway down the north canyon wall and interacts with Asp 5411 and Asp 56H, to Gln or Glu reduces virus viability in addition to blocking antibody binding.
Figure 3.
Fab/virus canyon interactions. a, The HRV/ICAM and HRV/Fab17-IA complexes, showing the HRV-14 canyon that is accessible to cellular receptor but inaccessible to antibody (adapted from ref. 30). b, Current view of virus/receptor and virus/antibody interactions showing that the Fab contacts a large portion of the canyon but not residues at the very bottom (the ‘floor’). Molecular surfaces of HRV-14 (c) and the Fab17-IA/HRV-14 complex(d). The view is parallel to the canyon floor with an icosahedral 5-fold axis towards the left the nearest 2-fold axis towards the right, and the RNA interior towards the bottom of the diagram.HRV-14, the variable domains of the bound Fab, and the molecular interfaces are shown in red, blue, grey, respectively. Only a small portion of the canyon floor is not contacted by the bound Fab.
The structure determination of the HRV–Fab complex clearly shows that strongly neutralizing antibodies do not have to induce large conformational changes in the virus capsid to cause neutralization. If the viral epitope maintains a fairly rigid structure about which the Fab conforms, then what is the mechanism of neutralization? Abrogation of HRV-14 cell attachment by antibody 17-IA is directly correlated with in vitro neutralization and is rationalized as a consequence of the extensive overlap between the ICAM-1 and Fab17-IA contact areas. Weakly neutralizing (strongly aggregating) antibodies and their Fabs also prevent virus binding to cell membranes12 but do not contact the proposed ICAM-binding residues (cryo-electron microscopy places Fabl ~10Å farther from the ICAM-binding region; Z. C. Che, N.H.O., T.J.S. and T.S.B., manuscript in preparation). Hence, these antibodies probably block cell attachment simply because of their large, bulky shape rather than by blocking specific interactions with the receptor. All NIm-IA antibodies stabilize HRV-14 at pH 5.0, but four other antibodies that bind to other NIm sites did not13. This secondary effect of antibody binding is unlikely to be due to bidentate binding, because antibody 1-IA, which binds monovalently5, stabilizes the virion. As Fabl does not contact the south wall, stabilization is also unlikely to be due to Fab binding in that region. The areas of contact common between Fab17 and Fab1, and therefore likely to participate in antibody-mediated stabilization, lie only about the NIm-IA site. Therefore, NIm-IA antibodies may protect HRV-14 against pH denaturation by binding to a region, near the receptor recognition site, which participates in the uncoating process. Antibodies that bind in this manner may exhibit post-absorption neutralization effects, as observed with poliovirus14.
As antibody production (B-cell stimulation) is driven by antigen binding and not by neutralization efficiency, the different antibodies generated in vivo (which may bind to very different regions of the virus) often have different neutralization efficacies and behaviour. However, in vivo this can be compensated for by the synergism that antibodies exhibit with other immune system components. Many different viruses are known in which antibodies neutralize weakly or not at all in vitro, yet still yield in vivo protection. For example, with foot-and-mouth disease virus (FMDV), antibody-mediated in vivo processes such as opsonization (antibody-mediated phagocytosis) and the reticuloendothelial system can play a dominant role in protection15.
Our structure determination of the HRV–Fab complex has helped to define the relationship between virus architecture and receptor-binding sites. Despite its recessed receptor-binding region, HRV-14 exhibits similarities to viruses with exposed cell recognition sites: these include FMDV16,17, Sindbis virus18,19 poliovirus20, and the haemeagglutinin spike of influenza21. It appears that the location and shape of the cell-receptor-binding site on a virus is dictated by the nature of the specific receptor being recognized, as well as what processes occur subsequent to receptor binding. For HRV-14, the canyon does not protect the ICAM-1 binding site from antibody recognition, but does allow ICAM-1 to cause virus uncoating22. Finally, as was observed in the case of influenza virus21, the surface of the virus covered by the antibody is much larger than that covered by ICAM-1 (ref. 4). Therefore, many residues on the virus offer potential sites for mutation that can thwart antibody binding without affecting receptor binding, thus permitting conserved residues to be exposed to the immune system.
Methods
Crystallization
HRV-14 was purified as described23 and Fab fragments were generated from the monoclonal antibody, mAb17-IA, as described2. Virus and Fab sample, dialysed against 10 mM Tris, pH 7.5, were combined at a ratio of ~240 Fab molecules per virion and stored at 4 °Cfor between 12 h and 3 days. The complex was concentrated at 5–10 °C using centrifuge concentrators with a 10K molecular-weight cutoff. The low temperature, low-ionic-strength buffer, and high concentration (10–20 mg m1−1) facilitated the precipitation of the Fab virus complex and yielded larger crystals than when the complex was concentrated in the presence of high salt. The precipitate was resuspended in and dialysed against 10 mM Tris buffer, pH 7.5, 100 mM NaCI. The solution, at room temperature, was then passed through a 0.2-µ syringe filter and concentrated to ~0.9–1.0 mg ml−1 extinction coefficient (7.7 ml mg−1 cm−1) using a Centricon 10 filter and centrifuging at 4,000–5,000g and 17–20 °C. The complex was crystallized using the vapour diffusion method, with a reservoir containing 1 ml of 10 mM Tris buffer, pH 7.5, 250–300 mM NaCl, 10 mM CaCl2, and 0.75% PEG 8000. Each sitting drop contained 20 µl of the complex to which 20 pi of precipitating buffer (10 mM Tris buffer, pH 7.5, 10 mM CaCl2 and 0.75% PEG 8000) was added. Crystals with a cubic habit usually appeared in 10–14 days and grew up to 0.8 mm in width within one month.
Data collection
Crystals were extremely sensitive to X-ray radiation at room temperature. Consequently, data collection was done with crystals flash-frozen at −170 °C in a 40% (v/v) PEG 400 final concentration. As the crystals were grown in the presence of high NaCI concentrations, they had to be quickly transferred to solutions of diluted reservoir solution with successively higher concentrations of PEG 400. The final X-ray data set was collected on a single 0.25-mm crystal that had been soaked for 30 min each in solutions containing 30% of the reservoir solution and 10, 20, 30 and 40% (final concentrations) of PEG 400. Data were. collected at the Cornell High Energy Synchrotron Source (CHESS) with 0.908 Å X-rays and recorded on Fuji image plates. For each image, the crystal was oscillated 0.3° for 30–40 s at a crystal-to-film distance of 400 mm. Images (198) were processed using DENZO24 and the integrated intensities scaled together using SCALEPACK24 using I/σ cutoff of −0.5 (Table 1).
TABLE 1.
Distribution and Rsym of the HRV-14/Fab17-IA complex data
| Resolution (Å) | R-factor(%) | % Total data |
|---|---|---|
| 20.00-7.86 | 6.3 | 79.1 |
| 7.86-6.30 | 13.8 | 72.6 |
| 6.30-5.52 | 18.6 | 68.1 |
| 5.52-5.03 | 19.2 | 67.5 |
| 5.03-4.67 | 19.5 | 66.1 |
| 4.67-4.40 | 22.1 | 61.8 |
| 4.40-4.18 | 25.1 | 53.6 |
| 4.18-4.00 | 29.3 | 46.9 |
| Total | 14.9 | 64.5 |
Structure determination
The crystals of the virus-Fab complex belong to the rhombohedral space group, R3 (a = 372Å, α = 108.4 Å). Vm calculations author: define Vm indicated the presence of only one virus particle, saturated with Fab fragments, per unit cell. The orientation of the particle was defined by strong 5-fold (~25σ), 3-fold (~23σ) and 2-fold (~20σ) peaks in the self-rotation function, calculated using the program GRLF25. The known structures of Fab17-IA (ref. 26), HRV-14 (ref. 3), and the Fab17-IA/HRV14 cryo-EM image reconstruction5, were used to place an entire HRV-14-(Fab)60 particle in the rhombohedral cell at (0,0,0) in the orientation defined by the self-rotation function calculation. Structure-factor amplitudes and phases were calculated from the Ca coordinates of the model to 8 Å using the program SFALL from the CCP4 package and used to create a molecular envelope mask for 20-fold averaging using Rossmann’s suite of programs27. Phases were extended in steps of one reciprocal lattice point, and each step was followed by at least six cycles of 20-fold non-crystallographic averaging. Subsequent molecular masks were generated from the averaged electron density with an assumed protein content of 53% in the crystal. A total of eight different masks were used during the 47 steps of phase extensions and 300 cycles of averaging that were required to attain an initial density map at 4Å resolution. This 4Å map clearly showed that the initial position of the Fab model was up to 4 Å away from the correct position (Fig. 2); the conformation of the CDR3 loop of the heavy chain differed from that in the unliganded structure; the HRV-14 density was consistent with the initial model; and the electron density for the Fab constant domains was diffuse and uninterpretable. Therefore, the constant domains were removed from the model. The electron density was 20-fold-averaged for ~15 cycles during each of the three cycles of phase refinement. This model, without the. constant domains, A was then refined using X-PLOR28. An overall B value of 20 Å2 and all data were used for Powell positional refinement. The initial R factor for the model was 29.7% using all data between 10.0 and 4.0 Å resolution, and the R factor of the current model is 21.2%, with geometrical statistics typical for a structure determined to >3.5Å resolution. Buried surface areas were calculated using a 1.7 Å probe radius29. Atomic coordinates for this model have been submitted to the Brookhaven Protein Database for distribution.
ACKNOWLEDGEMENTS
This work was supported by grants form the NIH to T.J.S. and T.S.B. and by the Lucille P. Markey Charitable Trust (Purdue Structural Biology Center). We thank R. R. Rueckert and A. Mosser for the generous gift of hybridomas and for stimulating discussions; R. McKenna, M. Agbandje, J. Dai and M. G. Rossmann for advice on molecular replacement; M. Rossmann, I. Wilson, D. Stuart, J. Hogle and S. Sheriff for advice and discussion; and the support staff of CHESS for assistance in data collection.
References
- 1.Mosser AG, Leippe DM, Rueckert RR. Molecular Aspects of Picomavirus Infection and Detection. In: Semler BL, Ehrenfeld E, editors. Am. Soc. Microbiol. Washington, DC: 1989. pp. 155–167. [Google Scholar]
- 2.Smith TJ, Olson NH, Cheng RH, Chase ES, Baker TS. Proc. Natl Acad. Sci. USA. 1993;90:7015–7018. doi: 10.1073/pnas.90.15.7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossmann MG, et al. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 4.Olson NH, et al. Proc. Natl Acad. Sci. USA. 1993;90:507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith TJ, et al. J. Virol. 1993;67:1148–1158. doi: 10.1128/jvi.67.3.1148-1158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabat EA, Wu TT, Reid-Miller M, Perry HM, Gottesman KS. Bethesda, MD: US Department of Health and Human Services, Public Health Service, NIH; 1987. [Google Scholar]
- 7.Tomlinson IM, et al. J. Mol. Biol. 1996;256:813–817. doi: 10.1006/jmbi.1996.0127. [DOI] [PubMed] [Google Scholar]
- 8.Stanfield RL, Wilson IA. Trends Biotechnol. 1994;12:275–279. doi: 10.1016/0167-7799(94)90139-2. [DOI] [PubMed] [Google Scholar]
- 9.Smith TJ, et al. Science. 1986;233:1286–1293. doi: 10.1126/science.3018924. [DOI] [PubMed] [Google Scholar]
- 10.Oliveira MA, et al. Structure. 1993;1:51–68. doi: 10.1016/0969-2126(93)90008-5. [DOI] [PubMed] [Google Scholar]
- 11.Rossmann MG. J. Biol. Chem. 1989;264:14587–14590. [PubMed] [Google Scholar]
- 12.Colonno RJ, Callahan PL, Leippe DM, Rueckert RR. J. Viral. 1989;63:36–42. doi: 10.1128/jvi.63.1.36-42.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee W-M. thesis. USA: Univ. Wisconsin; 1991. [Google Scholar]
- 14.Wien MW, et al. Nature Struct. Biol. 1995;2:232–243. doi: 10.1038/nsb0395-232. [DOI] [PubMed] [Google Scholar]
- 15.McCullough KC, et al. J. Virol. 1992;66:1835–1840. doi: 10.1128/jvi.66.4.1835-1840.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acharya R, et al. Nature. 1989;327:709–716. doi: 10.1038/337709a0. [DOI] [PubMed] [Google Scholar]
- 17.Verdaguer N, et al. EMBO J. 1995;14:1690–1696. doi: 10.1002/j.1460-2075.1995.tb07158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K-S, Schmaljohn AL, Kuhn RJ, Strauss JH. Virology. 1991;181:694–702. doi: 10.1016/0042-6822(91)90903-o. [DOI] [PubMed] [Google Scholar]
- 19.Smith TJ, et al. Proc. Natl Acad. Sci. USA. 1995;92:10648–10652. doi: 10.1073/pnas.92.23.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harber J, Bernhardt G, Lu H-H, Sgro J-Y, Wimmer E. virology. 1995;214:559–570. doi: 10.1006/viro.1995.0067. [DOI] [PubMed] [Google Scholar]
- 21.Bizebard T, et al. Nature. 1995;376:92–94. doi: 10.1038/376092a0. [DOI] [PubMed] [Google Scholar]
- 22.Hoover-Litty H, Greve JM. J. Virol. 1993;67:390–397. doi: 10.1128/jvi.67.1.390-397.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnold E, et al. J. Mol. Biol. 1984;177:417–430. doi: 10.1016/0022-2836(84)90293-6. [DOI] [PubMed] [Google Scholar]
- 24.Otwinoski Z. In: Data Collection and Processing. Sawyer L, Isaacs N, Bailey S, editors. Warrington, UK: SERC Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- 25.Tong L, Rossmann MG. Acta Crystallogr. 1990;A46:783–792. doi: 10.1107/s0108767390005530. [DOI] [PubMed] [Google Scholar]
- 26.Liu H, et al. J. Mol. Biol. 1994;240:127–137. doi: 10.1006/jmbi.1994.1427. [DOI] [PubMed] [Google Scholar]
- 27.Rossmann MG, et al. J. Appl. Cryst. 1992;25:166–180. [Google Scholar]
- 28.Brünger AT. X-plor (Version 3.1) User's Guide. New Haven, CT: Yale Univ.; 1992. [Google Scholar]
- 29.Sheriff S. immunomethods. 1993;3:191–196. [Google Scholar]
- 30.Luo M, et al. Science. 1987;235:182–191. doi: 10.1126/science.3026048. [DOI] [PubMed] [Google Scholar]