Summary
We present the first three-dimensional reconstruction of a prolate, tailed phage, and its empty prohead precursor by cryo-electron microscopy. The head-tail connector, the central component of the DNA packaging machine, is visualized for the first time in situ within the Bacillus subtilis dsDNA phage ϕ29. The connector, with 12- or 13-fold symmetry, appears to fit loosely into a pentameric vertex of the head, a symmetry mismatch that may be required to rotate the connector to package DNA. The prolate head of ϕ29 has 10 hexameric units in its cylindrical equatorial region, and 11 pentameric and 20 hexameric units comprise icosahedral end-caps with T=3 quasi-symmetry. Reconstruction of an emptied phage particle shows that the connector and neck/tail assembly undergo significant conformational changes upon ejection of DNA.
Introduction
Most viruses, whether animal, plant, or bacterial, utilize a complex assembly cascade, but little is known about genome packaging or its release during host cell infection (Rossmann, 1994). Genetic and physiological studies of dsDNA bacteriophages have established their assembly pathways in some detail, yet the precise mechanism of DNA packaging remains obscure. These phages package their genomes into precursor capsids (pro-heads) at a specific vertex that contains the head–tail connector, the crux of the DNA packaging machine. Among the well-studied dsDNA phages, ϕ29, which infects Bacillus subtilis, is unique in that in vitro assembly is well characterized and highly efficient because every ϕ29 prohead correctly packages its genome (Anderson and Reilly, 1993). In addition, capsid conformational changes in ϕ29 occur during DNA packaging without protein cleavage or shell expansion. Studies of ϕ29 can help pinpoint the capsid structural changes that accompany DNA packaging and that, thus far, have only been inferred by comparison of procapsids and mature particles in other viral systems. These include the ssDNA bacteriophage ϕX174 (Ilag et al., 1995; Dokland et al., 1997); the dsDNA bacteriophages P22 (Prasad et al., 1993), λ (Dokland and Murialdo, 1993), and HK97 (Conway et al., 1995); and also the mammalian herpes simplex virus (Trus et al., 1996; Zhou et al., 1998).
Proheads, the first particles produced in the ϕ29 assembly pathway, consist of a head–tail connector (gp10), scaffolding protein (gp7), major capsid protein (gp8), head fibers (gp8.5), and a 174-base RNA (pRNA) (Figure 1; Table 1). The connector is a preformed oligomer with 12- or 13-fold symmetry (Carazo et al., 1986; Dube et al., 1993; Tsuprun et al., 1994; Guasch et al., 1998) that interacts with scaffolding protein to direct prohead formation and to determine head length (Guo et al., 1991). A cyclic hexamer of pRNA, formed by intermolecular base pairing of identical subunits, binds to the connector to assemble a structure that might function as “gears” in the proposed rotary motion required for DNA packaging (Guo et al., 1998; Hendrix, 1998; Zhang et al., 1998).
Figure 1. Simplified Bacteriophage f29 Assembly Pathway.
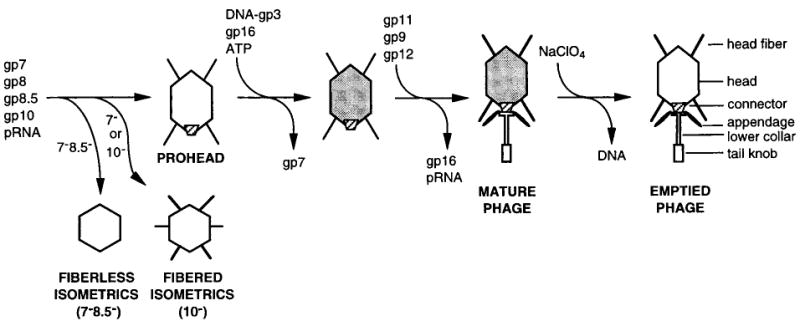
Table 1. Components of Bacteriophage ϕ29.
| Component | Function | Prohead/Virion | Na | Mass (kDa) | |
|---|---|---|---|---|---|
| gp7 | scaffolding protein | P | ∼180 | 11.3 | |
| gp8 | major capsid protein | P | V | 235 | 49.8 |
| gp8.5 | head fiber protein | P | V | 55 × 2 | 29.5 |
| gp9 | tail knob protein | V | ∼10 | 67.6 | |
| gp10 | head–tail connector | P | V | 12 or 13 | 35.9 |
| gp11 | lower collar | V | ∼12 | 33.8 | |
| gp12* | appendage | V | ∼12 × 5 | ∼80 | |
| gpRNA | prohead RNA (pRNA) | P | 6 | 57.6 | |
| gp3 | DNA terminal protein | V | 2 | 29.5 | |
| dsDNA | genome | V | 1 | 12,800 | |
is cleavage product of gp12 (92.1 kDa).
N = copy number. Data for gp9, 11, and 12* are from D. L. A., unpublished.
The in vitro ϕ29 DNA packaging system is relatively simple. It requires the prohead with pRNA, the supercoiled DNA substrate with the covalently bound terminal protein (DNA-gp3), the packaging ATPase (gp16), and ATP (Guo et al., 1986; Grimes and Anderson, 1997). The head becomes more angular (Bjornsti et al., 1983), and the surface charge decreases during packaging of DNA-gp3 and expulsion of gp7 (D. L. A., unpublished). Furthermore, packaging of ϕ29 DNA in vitro proceeds with distinct pauses that may accompany capsid structural transitions (Bjornsti et al., 1983). Virions mature in vivo or in vitro when gp16 and pRNA are released as the lower collar (gp11), tail knob (gp9), and appendages (gp12*) are assembled sequentially onto a stable, DNA-filled head (Figure 1). Mutant infections or in vitro assembly can be used to halt the process at discrete steps, making the morphogenesis pathway of ϕ29 (Figure 1) accessible to analysis by cryo-electron microscopy.
New methods based on reconstruction of isometric ϕ29 particles were devised to determine the structures of the 420 × 540 Å prolate prohead, the mature virion, and the DNA-emptied particle. These isometric particles, which lack connector and pRNA, are produced by suppressor-sensitive mutants, defective for the connector (“10−,” missing gp10) (Hagen et al., 1976), or mutants with a temperature-sensitive scaffolding protein (“7−”) (D. L. A., unpublished) (Figure 1). The present studies reveal the structure of the head–tail connector in pro-heads, virions, and DNA-emptied particles. The connector protrudes into and appears to make loose association with a vertex of the head shell. Such weak interactions may be necessary to allow the connector to rotate and package DNA as proposed (Hendrix, 1978; Grimes and Anderson, 1997). Release of DNA from the mature virion is accompanied by conformational changes in the connector, lower collar, and tail knob.
Results and Discussion
Isometric Particles
Cryo-electron microscopy (cryoEM) and 3D image reconstruction defined the shell subunit lattice of defective ϕ29 isometric particles, the prohead intermediate, and the mature and emptied virions (Figure 2; Table 2). Reconstructions were first determined from images of fiberless and fibered isometric particles (Figures 2A and 2B) produced in infections with mutants defective for head fibers and scaffolding protein (7−/8.5−) or scaffolding protein (7−) alone, respectively. The structures of these particles showed how the major capsid (gp8) and head fiber (gp8.5) proteins are organized (Figures 2F and 2G) and also served as initial models and controls for computing 3D reconstructions of the proheads and mature virions. Both types of isometric particle appear empty and exhibit thin, circular profiles in cryo-electron micrographs (Figures 2A and 2B), suggesting that the particles have icosahedral (532) symmetry. Indeed, the subsequent 3D reconstructions showed clear T=3 quasi-symmetry with 180 copies of gp8 organized as 20 hexameric structures at each of the 3-fold axes and 12 pentameric structures at each of the 5-fold axes (Figures 2F and 2G). The T=3 arrangement (Caspar and Klug, 1962) of gp8 subunits forms a spherical particle that is 425 Å in diameter along each 2-fold axis.
Figure 2. Cryo-Electron Microscopy and Three-Dimensional Image Reconstructions of Bacteriophage ϕ29 Particles.
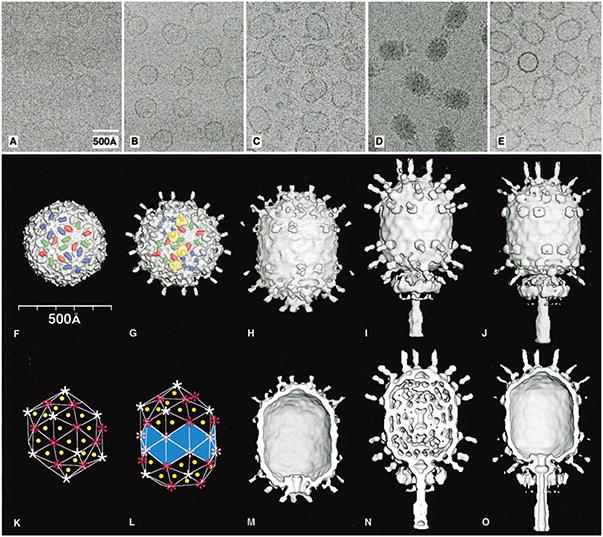
Micrographs of ϕ29 particles embedded in vitrified ice (A–E; scaled to the same magnification), and surface-shaded views of the corresponding 3D reconstructions (F–J,M–O). (A and F) Fiberless, isometric particles. (B and G) Fibered, isometric particles. (C and H) Proheads. (D and I) Mature phage. (E and J) Emptied phage. Prolate particles generally orient with the long axis nearly parallel to the vitreous ice surface (C–E). Fiberless (F) and fibered (G) isometric particles show protruding domains of gp8 colored red, green, and blue to highlight the three quasi-equivalent subunits in the T=3 capsid, and four fibers are highlighted in yellow. (K) T=3 triangulation net consisting of 60 triangles that define the lattice organization of the isometric structures in (F) and (G). Yellow dots mark the location of several of the 60 quasi-3-fold axes and hence the location of the fibers. The sites of pentamers and hexamers are identified by white and red symbols, respectively. (L) Q=5 triangulation net with the bottom vertex missing showing the lattice organization of the prolate structures in (H–J). Fifty-five triangles contain quasi-3-fold axes (yellow dots), whereas the remaining 20 (light blue), at the equator, have local 3-fold symmetry. Pink symbols mark the sites of equatorial hexamers. (M–O) Same as (H–J), with the front halves removed to reveal internal features such as the thin capsid wall, the dsDNA, and the structures of the connector and tail assemblies. Table 2 lists details about the microscopy and data analysis.
Table 2. Data Collection and Processing.
| Fiberless Isometrics | Fibered Isometrics | Prohead | Mature Phage | Emptied Phage | |
|---|---|---|---|---|---|
| Underfocus (μm)a | 2.1 | 1.7 | 2.3 | 2.8 | 2.6 |
| Number of particlesb | 203 | 204 | 1081 | 290 | 579 |
| 122 (203) | 128 (204) | 1037 (2081) | 190 (290) | 432 (579) | |
| Correlation coefficientc | 0.355 (0.033) | 0.312 (0.046) | 0.333 (0.047) | 0.378 (0.052) | 0.309 (0.041) |
| Resolution (Å) | 21 | 21 | 27 | 36 | 33 |
| Symmetry imposed | 532 | 532 | 5 | 5 | 5 |
| Origin and orientation refinement methodd | PFT | CL/PFT | PFT | PFT | PFT |
Determined from the contrast transfer function of the microscope.
The number of particles included in each 3D map. The total number of boxed particles is given in parentheses.
Averaged real space correlation coefficient and standard deviation (in parentheses) over all radii for all particles.
CL, common lines method (Fuller et al., 1996). PFT, model-based polar-Fourier-transform method (Baker and Cheng, 1996).
Close inspection of the reconstructions of the isometric particles (Figures 2F and 2G) shows that each gp8 subunit consists of two domains, one that forms a sausage-like, protruding feature (44 × 24 × 25 Å3), and one that forms a thin, contiguous shell structure (11 to 19 Å thick). The internal gp7 scaffolding protein, proposed to be associated with hexameric capsomers (Anderson and Reilly, 1993), is missing in the reconstructions because it was mostly lost during sample preparation, as shown by SDS gels (data not shown). Comparison of the fibered and fiberless particles shows that the head fibers attach to the gp8 subunits at quasi-3-fold axes that relate a pair of hexamers and one pentamer (Figure 2G). Each head fiber consists of a disk-like platform at its base from which a rigid fiber extends radially outward by about 250 Å. The platform interacts with the protruding domains of three gp8 subunits at the quasi-3-fold positions. The amino terminus of gp8.5 (residues 1 to 60) has a sequence consistent with a β secondary structure, whereas the remaining sequence (61–280) is predicted to be mostly α helical (Experimental Procedures). The carboxy-terminal 35 residues consist of heptad repeats typical of coiled coils. Thus, fibers probably attach to the head by means of their amino-terminal, bulky base and extend radially outward as a coiled coil. Although no function has been ascribed for these fibers (Tosi and Anderson, 1973; Reilly et al., 1977), their location suggests a possible role in stabilizing the capsid.
The Prolate Head
CryoEM images of three types of prolate particles, including the prohead, mature virus, and emptied virus (Figures 2C–2E), were used to compute 3D reconstructions (Figures 2H–2J). Each prolate head consists of two icosahedral (half T=3) caps separated by an equatorial band of 10 hexamers for a total of 30 hexamers compared to 20 in the isometric particles (Figures 2K and 2L). These prolate particles are described by the elongation number Q=5, from which the number of subunits (240) may be calculated by the relation 30(T+Q) (Aebi et al., 1974). However, because the connector replaces five subunits at the portal vertex, the predicted number of gp8 subunits is 235. Consistent with this Q=5 model for ϕ29, and in contrast to a previous, elongated T=1 model (Carrascosa et al., 1981), recent biochemical analysis indicates ∼233 copies of gp8 in each prolate head (D. L. A., unpublished). Thus, prolate phage heads, like those of ϕ29 and T4, appear to incorporate extra hexamers in the equatorial region during assembly. Also, unlike the procapsids of the T=7 bacteriophages λ (Dokland and Murialdo, 1993), HK97 (Conway et al., 1995), and P22 (Thuman-Commike et al., 1996), whose hexamers are skewed, the ϕ29 prohead hexamers appear regular and display local 6-fold symmetry.
The ϕ29 head fibers are not as distinct in either the micrographs or the reconstruction of the prohead compared to the mature phage. This probably reflects lower occupancy of the fiber-binding sites in the prohead. The fibers, which are dispensable for prohead assembly and DNA packaging (Reilly et al., 1977), only extend from the quasi-3-fold axes at the two caps and are missing at the equatorial band where there are no pentamers of gp8 (Figures 2H–2J; yellow dots in Figure 2L). This feature is consistent with the location of fibers at the 60 quasi-3-folds in the isometric particle (Figures 2G and 2K). However, prolate heads have but 55 fibers because the replacement of five gp8 subunits by the connector precludes the binding of five fibers. In fact, the base of each fiber makes extensive contact with a gp8 subunit in a pentamer at each quasi-3-fold position and makes little or no contact with the hexamer subunits. Therefore, perhaps the pentamers must differ in structure from hexamers at the quasi-3-fold axis. This explains why fibers are missing from the equatorial region and is also consistent with the putative dimeric nature of the fibers (D. L. A., unpublished).
There is some evidence for the presence of scaffolding proteins in the form of weak density associated with each equatorial hexamer on the internal surface of the prohead reconstruction (data not shown). Although the amount of scaffold appears to be substoichiometric, these proheads are fully active in DNA packaging in vitro.
The nucleic acid within the mature phage head is mostly separated by a gap from the inner wall of the protein capsid (Figure 2N), which is probably the reason why little if any of the symmetry of the assembled protein is imposed on the packaged dsDNA. The DNA does appear to be loosely organized into layers or shells, but these are not as distinct or as compact as in the genomes of some dsDNA bacteriophages (Lepault et al., 1987; Cerritelli et al., 1997) or in the dsRNA viruses (Dryden et al., 1993; Caston et al., 1997; Grimes et al., 1998) and the dsDNA herpes simplex virus (Booy et al., 1991).
The Neck/Tail Assembly
The neck/tail complex in full and empty virions is ∼440 Å long and consists of four proteins (gp10, 11, 12, and 9) that form, respectively, the head-tail connector, lower collar, appendages, and distal tail knob (Figures 2I, 2J, 2N, 2O, and 3). The reconstructions of the three prolate particles (Figures 2M–2O) show the putative connector density clearest in proheads (Figure 2M). At a high-density contour level, a clear boundary delineates the connector and the capsid shell, with the broad end of the connector slightly larger than a gp8 pentamer (132 Å diameter). The connector is ∼80 Å long, has a conical shape (140 to 80 Å external diameter), and an axial channel (60 to 35 Å diameter). Its broad and narrow ends protrude on the inside and outside of the capsid shell, respectively (Figure 2M). Slight variations in the connector structure in different prolate particles (Figures 2M–2O) presumably reflect influences imposed by the genome or by different conformations of gp8. These reconstructed views of the connector are consistent with electron microscopy studies of stained samples of isolated connectors and two-dimensional crystals that showed the connector to consist of two cylindrical regions with an axial channel (Carazo et al., 1986). Low electron density at the connector periphery suggests the presence of weak interactions where the symmetry mismatch between connector (12- or 13-fold) and shell (5-fold) occurs. Indeed, weak interactions would facilitate the hypothesized rotation of the connector that utilizes ATP hydrolysis to drive DNA translocation (Hendrix, 1978; Grimes and Anderson, 1997).
Figure 3. Schematic Diagram of an Emptied ϕ29 Phage.
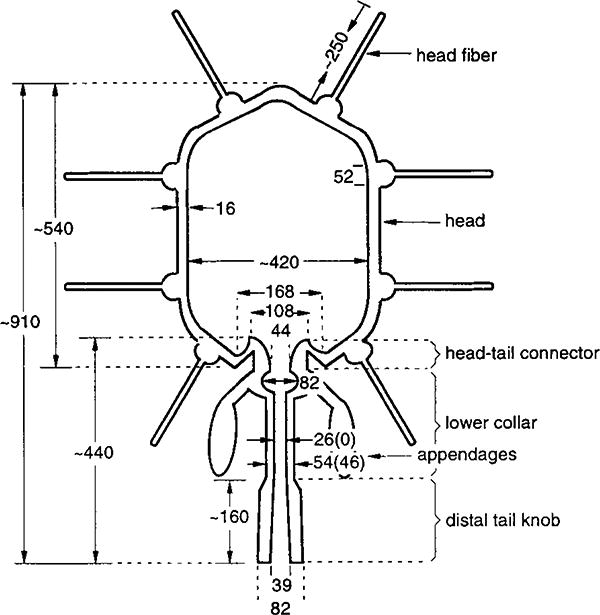
Distances are given in Å. Numbers in parentheses correspond to the mature phage. The 420 Å dimension of the prolate head refers to the external diameter.
The protrusion of the connector into the capsid shell gives proheads (Figures 2C and 2M) and empty heads (Figures 2E and 2O) their characteristic “bullet” shapes, with opposite ends pointed and flattened. The connector protrudes less into full heads (Figures 2N and 2O), though most of the fine details of its structure are obscured by density that also appears to plug the channel at the broad end of the connector. The connector attaches to the lower collar, which consists of a bulge region and a thin tube that extends ∼160 Å to the tail knob (Figures 2N, 2O, and 3). The bulge is ascribed to the lower collar because no such feature has been reported for connectors (Carazo et al., 1986; Tsuprun et al., 1994). In mature phage (Figure 2N), the distinction between the connector and lower collar is less apparent than in the emptied phage (Figure 2O). Also, the absence of an axial channel in the mature collar tube is consistent with expansion of the tube from 46 to 54 Å in its outer diameter, without change of length, upon DNA release (Figure 3).
Early results demonstrated that 12 appendages are attached to the lower collar and function as adsorption organelles in the mature phage (Anderson et al., 1966; Tosi and Anderson, 1973; Carrascosa et al., 1982). However, the 5-fold averaging inherent in the reconstruction process (Experimental Procedures) produces 10, rather than 12, large density features in a “miniskirt” attached to the bulge of the lower collar (Figures 2I, 2J, 2N, and 2O). Thus, the incorrect symmetry may have been enforced in the neck/tail portion of the reconstructed density maps. Nevertheless, the appearance of discrete, rather than completely cylindrically symmetric features, demonstrates that the tail assembly in different particles must generally be oriented in the same way relative to the head. This argues against the hypothesis that the tail assembly should be randomly oriented with respect to the head (Carazo et al., 1986).
The distal portion of the neck/tail assembly consists of the tail knob, which has an outside diameter larger than that of the lower collar tube (Figures 2N, 2O, and 3). In the emptied virion, an axial channel of varying diameter traverses the connector, lower collar, and tail knob (Figure 3). Hence, the tail structure can adapt itself to permit a dsDNA of ∼20 Å diameter to pass through it while being injected into the host cell. This process must be triggered at the time of cell recognition by the appendages or penetration of the tail through the cell wall. In mature phage, the axial channel is filled with electron-dense material except at the bulge in the lower collar and at a point just inside the distal end of the tail (Figure 2N). It appears as if some or all of this material is lost from the tail knob during DNA ejection.
DNA Packaging and Release
The connector is believed to be the origin from which prohead assembly is initiated (Guo et al., 1991; Anderson and Reilly, 1993). This correlates with the notion that the pentameric opening in the phage head occupied by the connector is too small to accommodate insertion of a rigid connector after prohead shell assembly occurs. The symmetry mismatch between the 12- or 13-meric connector and the pentameric hole may facilitate movements necessary for DNA packaging (Hendrix, 1978) in which a loosely fitting connector may act like the commutator of an electric motor. Upon completion of DNA packaging into the head, addition of the tail components appears to provide a gating structure that retains DNA in the head until interaction with the host triggers its release. Adsorption of ϕ29 appendages to the B. subtilis cell wall may trigger conformational changes that are transmitted to other parts of the tail via the bulge in the lower collar. These changes, for example, result in expansion of the lower collar tube (without change in length) upon ejection of the DNA in vitro.
The reconstructions presented here provide a stimulus to expand and extend structural analyses of intermediates as a means to visualize the dynamic processes of nucleic acid packaging and subsequent ejection. This will be possible because ϕ29 mutant infections yield well-characterized assembly intermediates and because in vitro assembly of ϕ29 rivals in vivo assembly in efficiency (Anderson and Reilly, 1993). More generally, the union of 3D reconstruction with physiological studies of mutants and the extension of structural analysis to higher resolutions (Baker and Johnson, 1996) should yield a better understanding of viral morphogenesis and infection.
Experimental Procedures
Production of ϕ29 Particles
Fibered and fiberless isometric particles were produced by infection of Bacillus subtilis RD2 (sup−) with the mutants ts 7(224)-sus 16(300)-sus 14(1241) or ts 7(224)-sus 8.5(900)-sus 16(300)-sus 14(1241), respectively, as described (Wichitwechkarn et al., 1989), at 45°C. Genes 7, 8.5, 16, and 14 encode scaffolding protein, head fibers, packaging ATPase, and a cell lysis protein, respectively. Particles were purified by centrifugation in a 10%–46% OptiPrep densitygradient containing 25 mM Tris-HCl (pH 7.8), 5 mM MgCl2, 50 mM NaCl, and 2 mM sodium azide (0.5× TMSA buffer). Isometric particles composed of the major capsid protein (gp8), with or without head fibers (gp8.5), and without connectors (gp10) and scaffolding protein (gp7), were the predominant particles isolated. Proheads were produced in B. subtilis SpoOA12 (sup−) cells infected with the mutant sus 16(300)-sus 14(1241) and purified by successive centrifugation in a 10%–60% OptiPrep density gradient and an isopycnic CsCl gradient (ρ = 1.22 g/cm3), both in 0.5 × TMSA. ϕ29 was produced as described (Anderson et al., 1966). DNA was released from ϕ29 at 2 × 1012 particles/ml by incubation in 1 M sodium perchlorate in 50 mM Tris-HCl (pH 7.8), 10 mM MgCl2, and 100 mM NaCl (TMS buffer) for 16 hr at 37°C. ϕ29 and ghosts were purified by isopycnic centrifugation in CsCl (ρ = 1.33) in TMS.
Electron Microscopy
Aliquots (3.5 μl) of purified ϕ29 samples were prepared for cryoEM essentially as previously described (Olson and Baker, 1989). Images were recorded on film at 36,000 or 38,000× nominal magnification in Philips EM420 or CM200 FEG microscopes, respectively, and with electron dose levels about 20 e−/Å2. Some of the prolate particle samples were tilted up to 9° in the microscope (Dryden et al., 1993). Table 2 lists the defocus levels for images used in 3D reconstruction.
3D Image Reconstruction
Suitable micrographs of fields of ϕ29 particles were selected and digitized at 14 μm intervals (3.9 or 3.7 Å/pixel at the specimen) on a Zeiss PHODIS scanner. Individual particle images were boxed, floated, and preprocessed to normalize mean intensities and variances and remove linear background gradients as described (Belnap et al., 1996).
Common-lines procedures (Fuller et al., 1996) were used to determine an initial set of orientations for the fibered isometric particles. The model-based, polar-Fourier-transform method (PFT) (Baker and Cheng, 1996) was then used to refine particle orientations and centers. Both orientation search procedures and the computation of the 3D reconstruction assumed the isometric particles obeyed icosahedral, point-group symmetry. The reconstruction of the fibered isometric particle was then used as a model to determine the orientations of the fiberless ϕ29 particles.
For the processing of the prolate particle images, an initial model was generated from the 3D density map of the fiberless isometric particle assuming the proposed Q=5 symmetry. The first reconstruction was computed with Fourier-Bessel methods (Fuller et al., 1996), modified to assume 52 instead of 532 point-group symmetry. A few cycles of refinement of particle orientation and origin parameters led to a reconstruction that displayed structural features with clear local 5-fold symmetry that was not enforced in the map calculation. The 2-fold ambiguity in particle orientations, created because the assumed 52 symmetry forced opposite ends of the prolate heads to be treated as the same and hence averaged together, was corrected by “eye,” since the two ends are easily distinguished in images (Figures 2C–2E). A new map was computed with only 5-fold axial symmetry imposed, and this became the “starting” model for the subsequent refinement of orientation and origin parameters and calculation of 3D maps. The presence of the head fibers at the quasi-3-fold axes, which were absent from the initial model, was a compelling validation of the final map of the prolate head.
Orientation refinement was monitored by means of correlation coefficients computed with real and reciprocal space data (Dryden et al., 1993). All maps were computed with Fourier-Bessel procedures, and eigenvalue spectra were used to monitor the conditioning of the linear, least-square equations (Fuller et al., 1996). The resolution of each map was estimated by splitting the image data into two sets and comparing structure factors obtained in the separate reconstructions. The threshold for the surface-shaded rendering was generally set about one standard deviation above the average noise level. The effect of the phase contrast transfer function (CTF) was not compensated in the reconstruction process, and data were rejected beyond the first node of the CTF.
Secondary Structure Prediction
The PredictProtein automatic network server was used for secondary structure prediction of the head fiber protein, using the profile-based neural network prediction algorithm PHDsec (Rost and Sander, 1994). The program COILS (Lupas et al., 1991) and visual inspection were also used for the prediction of coiled-coil structure.
Acknowledgments
S. Casjens and R. Hendrix predicted a T=3 extended icosahedron for ϕ29 from provisional biochemical data. We are grateful to W. Grohulski for initial image analysis studies; R. Ashmore for help with programs; G. Anand and R. Chen for production of ϕ29 particles; and W.Zhang and P. Chipman for discussions. Work was supported by NIH grants GM33050 and AI35212 to T. S. B., DE03606 and GM39931 to D. L. A., and NSF grant MCB9603571 to M. G. R. The facilities shared by the Structural Biology group at Purdue University, which have been developed and supported by grants from NIH, NSF, the Lucille P. Markey Foundation, and reinvestment funds from the office of the University Executive Vice President for Academic Affairs, have benefited T. S. B. and M. G. R.
References
- Aebi U, Bijlenga R, Broek JVD, Broek RVD, Eiserling F, Kellenberger C, Kellenberger E, Mesyanzhinov V, Muller L, Showe M, et al. The transformation of tau particles into T4 heads. II. Transformations of the surface lattice and related observations on form determination. J Supramol Struct. 1974;2:253–275. doi: 10.1002/jss.400020218. [DOI] [PubMed] [Google Scholar]
- Anderson D, Reilly B. Morphogenesis of bacteriophage ϕ29. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus Subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology, and Molecular Genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 859–867. [Google Scholar]
- Anderson DL, Hickman DD, Reilly BE. Structure of Bacillus subtilis bacteriophage ϕ29 and the length of ϕ29 deoxyribonucleic acid. J Bacteriol. 1966;91:2081–2089. doi: 10.1128/jb.91.5.2081-2089.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- Baker TS, Johnson JE. Low resolution meets high: towards a resolution continuum from cells to atoms. Curr Opin Struct Biol. 1996;6:585–594. doi: 10.1016/s0959-440x(96)80023-6. [DOI] [PubMed] [Google Scholar]
- Belnap DM, Olson NH, Cladel NM, Newcomb WW, Brown JC, Kreider JW, Christensen ND, Baker TS. Conserved features in papillomavirus and polyomavirus capsids. J Mol Biol. 1996;259:249–263. doi: 10.1006/jmbi.1996.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsti MA, Reilly BE, Anderson DL. Morphogenesis of bacteriophage phi29 of Bacillus subtilis: oriented and quantized in vitro packaging of DNA-gp3. J Virol. 1983;45:383–396. doi: 10.1128/jvi.45.1.383-396.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booy FP, Newcomb WW, Trus BL, Brown JC, Baker TS, Steven AC. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo JM, Donate LE, Herranz L, Secilla JP, Carrascosa JL. Three-dimensional reconstruction of the connector of bacteriophage ϕ29 at 1.8nm resolution. J Mol Biol. 1986;192:853–867. doi: 10.1016/0022-2836(86)90033-1. [DOI] [PubMed] [Google Scholar]
- Carrascosa JL, Mendez E, Corral J, Rubio V, Ramirez G, Salas M, Vinuela E. Structural organization of Bacillus subtilis phage ϕ29. A model Virology. 1981;111:401–413. doi: 10.1016/0042-6822(81)90343-3. [DOI] [PubMed] [Google Scholar]
- Carrascosa JL, Vinuela E, Garcia N, Santisteban A. Structure of the head-tail connector of bacteriophage ϕ29. J Mol Biol. 1982;154:311–324. doi: 10.1016/0022-2836(82)90066-3. [DOI] [PubMed] [Google Scholar]
- Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Caston JR, Trus BL, Booy FP, Wickner RB, Wall JS, Steven AC. Structure of L-A virus: a specialized compartment for the transcription and replication of double-stranded RNA. J Cell Biol. 1997;138:975–985. doi: 10.1083/jcb.138.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli ME, Cheng N, Rosenberg AH, McPherson CE, Booy FP, Steven AC. Encapsidated conformation of bacteriophage T7 DNA. Cell. 1997;91:271–280. doi: 10.1016/s0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- Conway JF, Duda RL, Cheng N, Hendrix RW, Steven AC. Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. J Mol Biol. 1995;253:86–99. doi: 10.1006/jmbi.1995.0538. [DOI] [PubMed] [Google Scholar]
- Dokland T, Murialdo H. Structural transitions during maturation of bacteriophage lambda capsids. J Mol Biol. 1993;233:682–694. doi: 10.1006/jmbi.1993.1545. [DOI] [PubMed] [Google Scholar]
- Dokland T, McKenna R, Ilag LL, Bowman BR, Incardona NL, Fane BA, Rossmann MG. Structure of a viral procapsid with molecular scaffolding. Nature. 1997;389:308–313. doi: 10.1038/38537. [DOI] [PubMed] [Google Scholar]
- Dryden KA, Wang G, Yeager M, Nibert ML, Coombs KM, Furlong DB, Fields BN, Baker TS. Early steps in reovirus infection are associated with dramatic changesinsupramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube P, Tavaras P, Lurz R, van Heel M. The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J. 1993;12:1303–1309. doi: 10.1002/j.1460-2075.1993.tb05775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller SD, Butcher SJ, Cheng RH, Baker TS. Three-dimensional reconstruction of icosahedral particles—the uncommon line. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- Grimes S, Anderson D. The bacteriophage ϕ29 packaging proteins supercoil the DNA ends. J Mol Biol. 1997;266:901–914. doi: 10.1006/jmbi.1996.0843. [DOI] [PubMed] [Google Scholar]
- Grimes JM, Burroughs JN, Gouet P, Diprose JM, Malby R, Zientara S, Mertens PPC, Stuart DI. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- Guasch A, Pous J, Parraga A, Valpuesta JM, Carrascosa JL, Coll M. Crystallographic analysis reveals the 12-fold symmetry of the bacteriophage ϕ29 connector particles. J Mol Biol. 1998;281:219–225. doi: 10.1006/jmbi.1998.1928. [DOI] [PubMed] [Google Scholar]
- Guo P, Grimes S, Anderson D. A defined system for in vitro packaging of DNA-gp3 of the Bacillus subtilis bacteriophage phi29. Proc Natl Acad Sci USA. 1986;83:3505–3509. doi: 10.1073/pnas.83.10.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Erickson S, Xu W, Olson N, Baker TS, Anderson D. Regulation of the phage ϕ29 prohead shape and size by the portal vertex. Virology. 1991;183:366–373. doi: 10.1016/0042-6822(91)90149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Zhang C, Chen C, Garver K, Trottier M. Inter-RNA interaction of phage ϕ29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- Hagen EW, Reilly BE, Tosi ME, Anderson DL. Analysis of gene function of bacteriophage ϕ29 of Bacillus subtilis: identification of cistrons essential for viral assembly. J Virol. 1976;19:501–517. doi: 10.1128/jvi.19.2.501-517.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW. Symmetry mismatch and DNA packaging in large bacteriophages. Proc Natl Acad Sci USA. 1978;75:4779–4783. doi: 10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophage DNA packaging: RNA gears in a DNA transport machine. Cell. 1998;94:147–150. doi: 10.1016/s0092-8674(00)81413-0. [DOI] [PubMed] [Google Scholar]
- Ilag LL, Olson NH, Dokland T, Music CL, Cheng RH, Bowen Z, McKenna R, Rossmann MG, Baker TS, Incardona NL. DNA packaging intermediates of bacteriophage ϕX174. Structure. 1995;3:353–363. doi: 10.1016/s0969-2126(01)00167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepault J, Dubochet J, Baschong W, Kellenberger E. Organization of double-stranded DNA in bacteriophages: a study by cryo-electron microscopy of vitrified samples. EMBO J. 1987;6:1507–1512. doi: 10.1002/j.1460-2075.1987.tb02393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Olson NH, Baker TS. Magnification calibration and the determination of spherical virus diameters using cryo-microscopy. Ultramicroscopy. 1989;30:281–298. doi: 10.1016/0304-3991(89)90057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad BVV, Prevelige PE, Marietta E, Chen RO, Thomas D, King J, Chiu W. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J Mol Biol. 1993;231:65–74. doi: 10.1006/jmbi.1993.1257. [DOI] [PubMed] [Google Scholar]
- Reilly BE, Nelson RA, Anderson DL. Morphogenesis of bacteriophage ϕ29 of Bacillus subtilis: mapping and functional analysis of the head fiber gene. J Virol. 1977;24:363–377. doi: 10.1128/jvi.24.1.363-377.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG. Viral cell recognition and entry. Protein Sci. 1994;3:1712–1725. doi: 10.1002/pro.5560031010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Sander C. Combining evolutionary information and neural network to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- Thuman-Commike PA, Greene B, Jakana J, Prasad BVV, King J, Prevelige PE, Jr, Chui W. Three-dimensional structure of scaffolding containing phage P22 procapsids by electron cryo-microscopy. J Mol Biol. 1996;260:85–98. doi: 10.1006/jmbi.1996.0383. [DOI] [PubMed] [Google Scholar]
- Tosi M, Anderson DL. Antigenic properties of bacteriophage ϕ29 structural proteins. J Virol. 1973;12:1548–1559. doi: 10.1128/jvi.12.6.1548-1559.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Booy FP, Newcomb WW, Brown JC, Homa FL, Thomsen DR, Steven AC. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- Tsuprun V, Anderson D, Egelman EH. The bacteriophage ϕ29 head-tail connector shows 13-fold symmetry in both hexagonally packed arrays and as single particles. Biophys J. 1994;66:2139–2150. doi: 10.1016/S0006-3495(94)81009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichitwechkarn J, Bailey S, Bodley JW, Anderson DL. Prohead RNA of bacteriophage ϕ29: size, stoichiometry, and biological activity. Nucleic Acids Res. 1989;17:3459–3468. doi: 10.1093/nar/17.9.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Lemieux S, Wu X, St-Arnaud D, McMurray TC, Major F, Anderson D. Function of hexameric RNA in packaging of bacteriophage ϕ29 DNA in vitro. Mol Cell. 1998;2:141–147. doi: 10.1016/s1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Macnab SJ, Jakana J, Scott LR, Chiu W, Rixon FJ. Identification of the sites of interaction between the scaffold and outer shell in herpes simplex virus-1 capsids by difference electron imaging. Proc Natl Acad Sci USA. 1998;95:2778–2783. doi: 10.1073/pnas.95.6.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]


