Abstract
Single crystals of d-ribulose-1,5-diphosphate carboxylase from tobacco leaves, Nicotiana tabacum (variety Turkish Samsun), have been examined by X-ray diffraction, electron microscopy, and optical diffraction. Twelve molecules are loosely packed into a body-centered cubic unit cell, space group 14132 with cell dimension a = 383 Å. The asymmetric unit is one quarter of a molecule, and the minimum molecular symmetry is 222. This symmetry when combined with estimates of the two subunit masses and stoichiometry is compatible with a molecular structure of the composition L8S8 (L is large subunit, S is small). If all bonds between large and small subunits are equivalent, the true molecular symmetry is 422; this symmetry is consistent with molecular images in micrographs.
1. Introduction
d-Ribulose-1,5-diphosphate carboxylase (3-phospho-D-glycerate carboxy-lyase (dimerizing), EC4.l.l.39) is an oligomeric enzyme which plays a central role in the metabolism of higher plants and certain autotrophic micro-organisms. It catalyzes the carboxylation and cleavage into triose phosphates of D-ribulose-1,5-diphosphate in the initial “dark reaction” in photosynthesis,
This enzyme comprises approximately 40% of the total soluble protein in the mobile phase of higher plant chloroplasts (Kawashima & Wildman, 1970a) and can be purified in large quantities from tobacco and spinach leaves.
Studies, reviewed by Kawashima & Wildman (1970a) and Siegel et al. (1972), have defined several physical properties of the enzyme. The sedimentation coefficient, of RuDPCase† from various plant sources has an average value of 18·5 S. The molecular weight, determined from sedimentation velocity and diffusion experiments, and from sedimentation equilibrium, has values ranging from 480,000 to 590,000. The most common value reported is 560,000. The frictional-coefficient ratio (f/f0 = 1·11; Paulson & Lane, 1966) and molecular images in electron micrographs indicate that the molecule can be approximated by a hydrated sphere 100 to 120 Å in diameter.
Evidence for two types of subunits having distinct amino acid compositions was presented by several workers (Rutner & Lane, 1967; Sugiyama & Akazawa, 1967, 1970; Kawashima, 1969; Moon & Thompson, 1969; Sugiyama et al., 1971). The large subunit has a molecular weight of 50,000 to 60,000 and the smaller subunit has a molecular weight between 12,000 and 20,000. The large subunit is catalytically active (Nishimura & Akazawa, 1973). Murai & Akazawa (1972) found that CO2 is a homo-tropic effector in the regulation of RuDPCase. The function of the small subunit may be regulatory, but it is uncertain (Nishimura & Akazawa, 1973).
Three models for the quaternary structure of the molecule have been proposed (Haselkorn et al., 1965; Steer et al., 1968; Kawashima & Wildman, 1970b). The first two were based on molecules with 24 identical subunits and are incompatible with the more recent discovery of two different subunits. The third model proposes that the molecule has an L8S8 structure, in which eight large subunits lie on the corners of a cube with a dimeric subunit (composed of two identical small polypeptide chains) at each face of the cube in contact with four large subunits. This seems an unlikely arrangement to us because it implies two kinds of binding between the large and small subunits. This is so because polypeptide chains are themselves asymmetric objects. The highest symmetry that may be possessed by an oligomer formed from eight chains is a single 4-fold axis with four 2-fold axes perpendicular to it (point group D4). Thus the oligomer cannot display the three 4-fold axes of a cube, and the six points at the juncture of four chains are not all identical. Instead, four are of one type, and two of another type. The L8S8 structure would not have equivalent bonding between all LS pairs, and a simple self-assembly mechanism is not evident.
Kawashima & Wildman's (1971) discovery of conditions for growing large single crystals enabled us to study the subunit structure by X-ray diffraction and electron microscopy. Our initial results prove that the molecule possesses a minimum of three perpendicular 2-fold axes (222), and therefore has a stoichiometry of the form L4nS4m (n and m are integers). It is likely, moreover, that the molecule has stoichiometry L8S8, and symmetry 422 (D4).
2. Materials and Methods
(a) Purification and crystal growth
RuDPCase is isolated from Nicotiana tabacum leaves by a method previously reported (Chan et al., 1972), except that the incubation step with NaHC03 and MgCl2 is omitted prior to crystallization. The protein is purified by three recrystallizations which are performed by dialyzing the salt out of the protein solution. The protein is dissolved in the following buffer: 0·025 M-Tris·HCl (pH 7·4), 0·20 M-NaCl, 5× 10−4 M-EDTA. The crystallization solution is identical but without NaCl. Protein solution (10 to 20 mg/ml) is introduced into 1-mm Li-glass X-ray capillaries sealed at one end, and salt-free buffer is carefully layered over the protein solution. The capillaries are placed in test tubes with a 10-ml reservoir of the salt-free buffer. Single crystals grow on the inside walls of the capillaries at room temperature after 4 to 8 weeks as the salt in the protein solution diffuses into the reservoir (Kwok, 1972). All crystals grown under these conditions are colorless rhombic dodecahedrons, 0·3 to 1·0 rom on an edge.
(b) X-ray diffraction
The capillaries in which crystals are formed are sealed with the least possible manipulation of the crystals, and used directly for the diffraction experiments. Crystals are aligned by cone-axis oscillation (Climenson & Eisenberg, 1974) and reciprocal lattice sections are recorded, at a precession angle of 3°, perpendicular to the (100), (110), and (Ill) crystallo-graphic directions. All photographs are taken on an Elliott rotating anode with a 100 μm focal cup, operating at 40 kV and 17 mA with the X-rays brought to fine focus by a Franks focusing device (Harrison, 1968; Franks, 1955). The specimen to film distance is 60 mm.
(c) Electron microscopy
Micrographs are recorded in a Siemens 1A electron microscope operating at 80,000 V and at magnifications of 30,000 × and 40,000 ×. Views of single, isolated RuDPCase molecules are obtained by applying a dilute solution of the protein (~0·1 mg/ml) to a carbon-covered grid, permitting adhesion to occur, and negatively staining the molecules with a filtered 2% solution of uranyl acetate in water. Microcrystalline fragments, formed by crushing single crystals, are similarly prepared for microscopy.
Single crystals are fixed for thin sectioning by addition of glutaraldehyde to the crystallization solution until a final concentration of 0·5% is attained, and left for 24 h. After washing, the crystals are dehydrated by transferring them for 1-h periods into acetone/ water solutions of 30%, 50%, 75%, 90% and four changes of 100% acetone. Infiltration of the plastic embedding medium, Vestopal-W (Martin-Jaeger, Geneva, Switzerland), is accompanied by transferring crystals into Vestopal/acetone solutions of increasing Vestopal concentration (v/v): 8 h in 25% Vestopal; 8 h in 50%; 1 day in 75%; 2 days in 100%; and 1 day in a second change of 100% Vestopal. Single crystals are oriented in drops of partially polymerized, highly viscous Vestopal to which initiator and activator had been added 2 or 3 days before. The plastic is then polymerized by heating the drops for 1 week in an oven kept at 60°C. The hardened samples are attached with epoxy to gelatin capsules filled with polymerized Vestopal, and the entire capsule is mounted in a goniometer holder which is fitted to the cutting arm of the LKB U1tratome. Thin sections are cut with freshly prepared glass knives, collected on carbon stabilized nets on grids, and positively stained with 2% potassium permanganate and lead citrate (Reedy, 1965).
(d) Optical diffraction
Optical diffraction patterns are recorded using a helium-neon laser and a diffractometer constructed by M. Reedy and F. Eiserling. The diffractometer constant is 1·441 (1-mm periodic spacings in the micrograph give rise to 1·441-mm diffraction spot spacings in the transform). The electron micrograph images, which were recorded on film, are contact printed onto glass plates which are immersed in oil between two plano-convex lenses. Circular apertures are used as masks to select desired regions of the micrograph for optical transformation and to avoid the characteristic “cross-like” diffraction pattern formed by rectangular masks.
3. Results
(a) Electron microscopy of single molecules
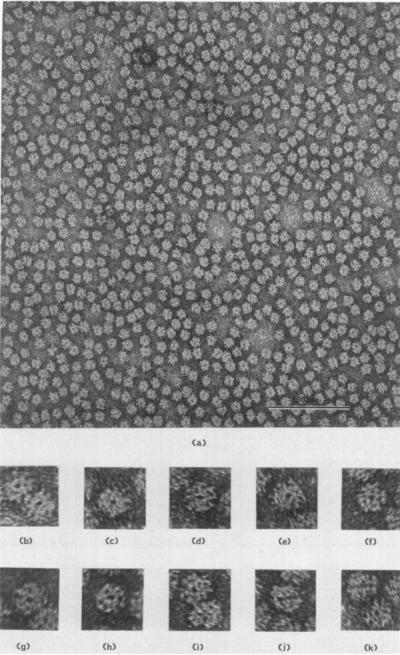
Electron microscopy of dissolved RuDPCase that has been purified to the extent of three recrystallizations, reveals a homogeneous population of circle-shaped molecules all 100 to 120 Å in diameter (Plate I). A common feature of a large portion of the molecular images in all preparations is a central dark-staining region, 20 to 25 Å in diameter. This could arise either from a hole extending through the molecule or a depression in the surface. RuDPCase isolated from other sources has a similar appearance in electron micrographs (Branton & Park, 1967; Gunning et al., 1968; Akazawa et al., 1972; Shively et al., 1973). This feature may be associated with a view along a 4-fold axis of a molecule with 422 symmetry (see Discussion). Enlarged images of molecules in this orientation are presented in sections (b) to (i) of Plate I. Often views of the molecule will show a line of stain through the center of the molecule (Plate I, (j) and (k)), indicating that there is a single hole extending through the entire molecule. This shows that the molecule possesses a unique axis of symmetry, and this orientation could be a view of the molecule perpendicular to the possible 4-fold axis.
PLATE I.
(a) A field of RuDPCase molecules negatively stained with uranyl acetate. Magnification 250,000 ×. (b) to (i) Enlarged images of molecules displaying a central hole. This orientation may correspond to a view parallel to a unique 4-fold axis of molecular symmetry. Magnification 750,000 ×. (j) and (k) Enlarged images displaying a central line of stain through the molecules.
(b) Crystals
RuDPCase crystallizes into large, non-birefringent, rhombic dodecahedrons with well-defined faces. Nevertheless, the crystals are extremely sensitive to mechanical manipulations; the slightest shock induces decay to a thin film of protein lying at the bottom of a large drop of solvent. This property forces us to grow crystals inside X-ray capillary tubes. The liquid of crystallization surrounding the crystal is carefully removed, leaving the crystal in a small pool of mother liquor. This pool is necessary to preserve crystalline structure for more than ten hours in the X-ray beam. With it crystals diffract up to 100 hours.
The density of the crystals (ρ = 1·058±0·005 g/cm3) has been measured (Kwok, 1972) and the weight fraction of protein in the crystals (Xp = 0·21 ±0·02) has been estimated knowing the partial specific volume of the protein in dilute solution (v̄p = 0·73±0·01 cm3/g; Kwok, 1972) and the partial specific volume of the pure solvent (v̄s = 1·001±0·002 cm3/g). The unusually low values for the density and weight fraction of protein in the crystals, along with the extreme sensitivity of the crystals, implies that there is loose packing of the molecules in the unit cell. These observations are consistent with the model of the crystal structure to be presented.
(c) X-ray diffraction
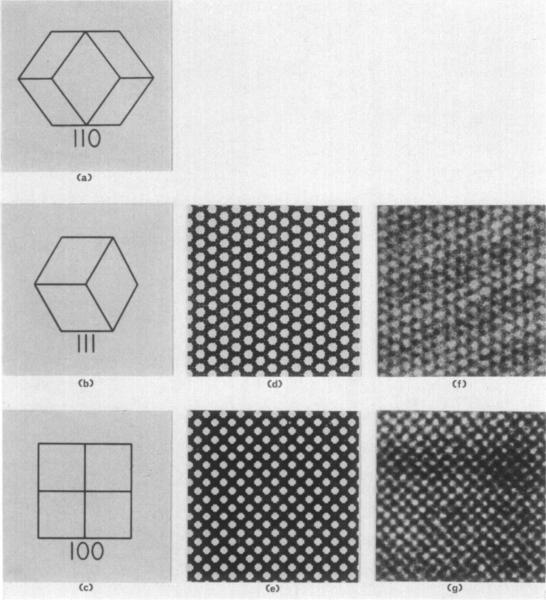
Three zero-level precession photographs (Plate III (1), (m), (n)) were obtained with the 2, 3, and 4-fold crystallographic axes (Plate III( a), (b), (c)) precessing 3° about the X-ray beam. These orientations enable the (hhl), (h, k, h + k), and (hkO) sections of the reciprocal lattice to be recorded. The symmetries of these three sections are, respectively, 2mm, 6mm, and 4mm. These symmetries are more apparent on the original films. The Laue symmetry is m3m and the structure is in the cubic crystal class with point symmetry 432. The lattice is body-centred with a= 383 A. This restricts the possible space groups to 1432 and 14132.
PLATE III.
(a) to (c) Drawings of a rhombic dodecaherdral crystal viewed parallel to the 2, 3, and 4-fold axes, respectively. The crystal depicted in (a) is rotated 90° about the 2-fold axis with respect to the diffraction pattern to its right. The orientation of the crystal drawn in (b) is identical with the orientation of the sections and diffraction pattern3 in (d), (f), (h), (j), and (m). The crystal in (c) is rotated 45° with respect to the (e), (g), (i), (k), and (n).
(d) and (e) Projection drawings representing magnified (112,000 ×) images of the molecular packing in space group 14132 viewed perpendicular to the 111 and 100 crystal planes. Molecules are colored black to correspond with electron microgmphs of positively stained thin sections of embedded crystals appearing in (f) and (g). The drawings depict projections of the structure thicker than one unit cell.
(f) and (g) Electron micrographs of positively stained thin sections of fixed, dehydrated, and embedded form I RuDPCase crystals at a magnification of 112,000 × . Sections are perpendicular to the 3 and 4-fold axes of the crystal and are therefore views of the III and 100 crystallographic planes.
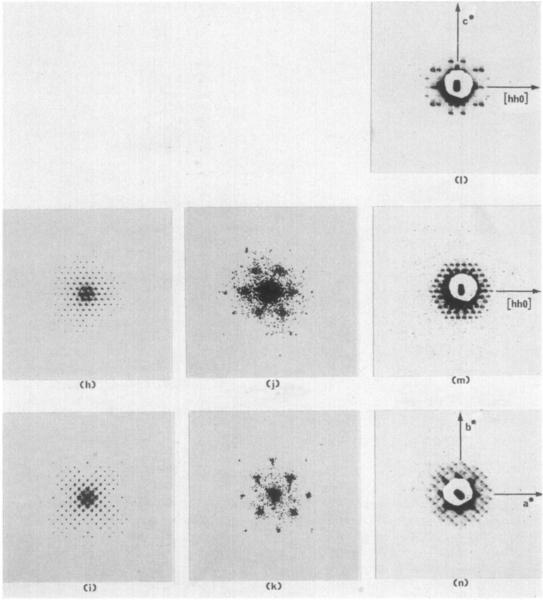
(h) and (i) Optica1 diffraction patterns of the drawings in (d) and (o) reproduced at the same scale as the X-ray precession photographs in (m) and (n).
(j) and (k) Optical diffraction patterns of the electron micrographs in (f) and (g). The patterns are enlarged four times larger than the scale of the diffraction patterns in (h) and (i) and (m) and (n).
(I) to (n) Three-degree zero-level precession photographs taken with the precession axis parallel to Lhe 2, 3, and 4-fold crystal axes. The central portion of each pattern is hidden hy the shadow of the lead beam stop . An image of the focused X-ray beam appears in the center of each pattern and defines t he position of the 0,0,0 reflection. The diffraction patterns have been magnified 4·5 times from the original photographs.
The absence of the 6,0,0 reflection favors space group 14132 since it is extinguished in this space group. Though it is always possible to observe an accidental absence, we present evidence below that 14132 is the correct space group.
When we calculate n, the number of molecules per unit cell, from the relation
| (1) |
where N is Avagadro's number,
ρ is crystal density,
Xp is weight fraction of protein in the crystal,
V is volume of the unit cell and
M is molecular weight,
we find n = 13·5±1·8. The symmetry of the two possible space groups permits only some multiple of eight or 12 molecules per unit cell. This indicates that 12 is the probable value for this crystal form. Given the molecular weight, partial specific volume of the protein, and the partial specific volume of the solvent, one can calculate expected densities for unit cells with n = 12, 16, or 24,
| (2) |
With 12, 16, or 24 molecules per unit cell, ρ is calculated to be 1·053, 1·071, and 1·107 g/cm3, respectively (assuming M = 560,000). Thus, the measured value for the crystal density (ρ = 1·058±0·005 g/cm3; Kwok, 1972) is consistent with a unit cell containing 12 molecules. With 12 molecules per cell, the molecule must sit on special positions; there are two possible packing arrangements for n = 12 in each of the two space groups (Henry & Lonsdale, 1969).
From the unit cell volume (5·62 × 10−17 cm3) and the number of molecules per unit cell, we calculated the solvent-content parameter, VM = 8·36 Å 3/dalton (Matthews, 1968). This high value (see Discussion), and the fact that the diffraction patterns extend only to very low resolution (~50 Å), are further evidence that the crystal structure is very open and susceptible to distortions.
(d) Microcrystals
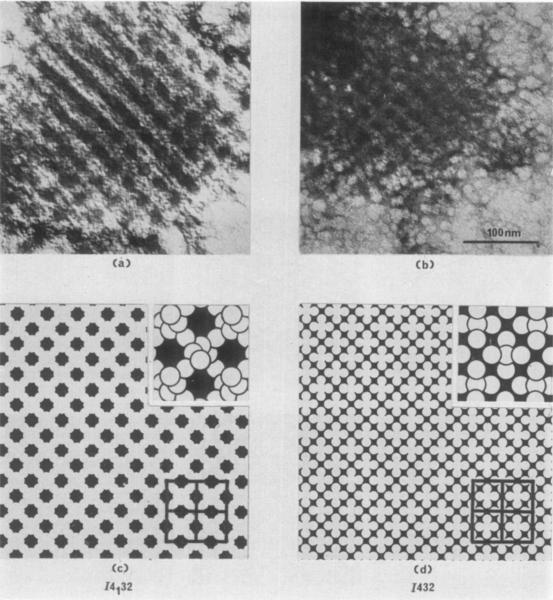
The images of two fragments of microcrystals negatively stained with uranyl acetate are reproduced in Plate II(a) and (b). Below these images are two projection drawings (Plate II(c) and (d)) of a view perpendicular to the 100 crystallographic planes representing the possible molecular packing for unit cells of space groups 1432 and 14132 with 12 molecules per unit cell. In the drawings, the molecules are depicted as white circles (surrounded by black stain) 120 Å in diameter placed at the 12 special positions (International Tables for X-ray Crystallography, vol. 1) in a cubic unit cell with a = 383 Å. It is apparent that the closer approximation to the crystal structure of the small fragments is given by the 14132 space group drawing. A second packing with 12 molecules exists for both space groups, but in each the projected structure is identical to the drawings of Plate II(c) and (d).
PLATE II.
(a) and (b) Fragments of form I RuDPCase crystals, negatively stained in uranyl acetate. Magnification 200,000 ×.
(c) Drawing illustrating the packing of 12 molecules per unit cell in space group 14132. The drawing represents a projection of the magnified (200,000 ×) crystal structure viewed along the crystallographic axis (perpendicular to the 100 crystallographic planes). Molecules are depicted as white circles, 120 Å in diameter, to correspond with images of negatively stained individual molecules. The black areas represent the presence of heavy met.al contrasting agent (uranyl acetate). Four unit cells are outlined in the lower right-hand corner. An enlarged view of the molecular packing at a magnification of 400,000 × is drawn in tho upper right-hand corner ; individual molecules are outlined in black to reveal the spatial arrangement of molecules in the special positions within the crystal.
(d) A drawing similar to (c), representing a view perpendicular to the 100 crystallographic planes of space group 1432 with 12 molecules per unit cell.
Note the similarity of (a) and (b) to (c), and dissimilarity to (d).
(e) Thin sections
By preparing thin sections from embedded RuDPCase crystals we can directly observe the crystal structure and verify that the packing arrangement is compatible with space group 14132. Thin sections were cut parallel to the 100 and 111 crystallo-graphic planes. The sections were stained positively, thus protein appears black. Electron micrographs of selected sections are reproduced in Plate III (f) and (g) opposite the corresponding projection drawings for space group 14132 (Plate III (d) and (e)). Notice in the drawings that the molecules are depicted as black circles to signify positive staining of the crystal section. The white regions represent solvent in the crystal. The correspondence between the micrographs and drawings is clear, indicating that the crystal structure is well represented at this resolution by the model. The large proportion of unstained regions in the micrographs verifies the experimental evidence that indicates a large fraction of solvent in the crystal.
In micrographs showing larger areas of the crystal, it is possible, by viewing the micrograph at a glancing angle, to notice distortions in the crystal lattice. The unit cells generally extend in a straight line for no more than 20 repeats before a noticeable curvature can be recognized. These distortions in the crystal sections could represent distortions in the crystal as grown, and therefore could explain the low resolution attained in the X-ray diffraction experiments, or they could be artifacts of treatment of the crystals during preparation for microscopy. In either case, the micrographs do show that the crystal structure has been preserved to some extent after fixation, dehydration, embedding and sectioning. The unit-cell dimensions of the sectioned crystals agree with the dimensions from the X-ray photographs. In all cases the dimensions of the sectioned crystals are approximately 5 to 10% lower than those for the hydrated crystals. This is expected since most specimens decrease in size as the embedding medium hardens during polymerization.
Two problems arise during sectioning of crystals. The first is proper orientation of the crystal during the cutting of sections. It is necessary to section the embedded crystal as nearly parallel as possible to the desired crystallographic planes. The appearance of the molecular packing and the cell dimensions change with orientation. Off-axis cutting gives rise to widely spaced “beat patterns” as the section passes through different layers of unit cells. By measuring the orientation and period of the beat, corrections of the cutting angle can be made in order to produce correctly oriented sections.
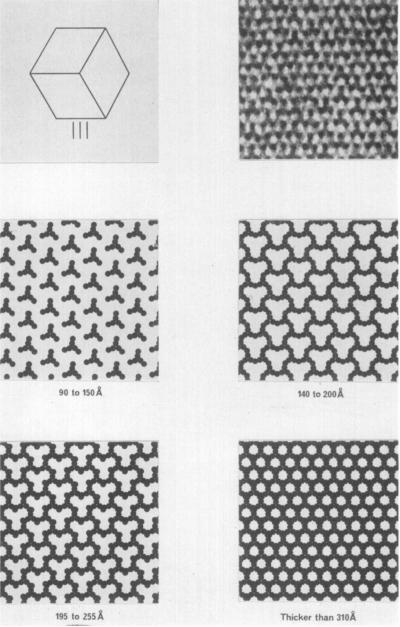
The second problem is that the appearance of the crystal structure viewed in a particular direction depends on how thick the section is, if the thickness of the section is less than one unit cell. This is illustrated in Plate IV for the view perpendicular to the III planes in the crystal. The drawings, again based on circular molecules, show how the appearance of the positively stained crystal structure changes as sections of varying thickness are cut. A crystal with unit-cell dimensions as large as 383 Å makes it possible to obtain sections that are less than one unit cell thick. Plate V(a) and (b) shows examples of sections whose thicknesses are probably less than 300 Å. These micrographs illustrate sections with folds as a result of the procedure of picking up sections onto the electron microscope grid. The thickness of the fold gives a maximum value for twice the thickness of the unfolded section (Small, 1968). The appearance of a thin section could also arise from incomplete but uniform penetration of stain, but the fold thicknesses suggest this is not the case here. The micrographs in Plate III(f) and (g) illustrate views of the crystal that contain copies of all 12 of the symmetry-related molecules per unit cell.
PLATE IV.
Projection drawings of sections of various thicknesses from the unit cell of a form I crystal. All views are perpendicular to the 111 crystal planes, and are at a magnification of 150,000 ×. Molecules are shown black, to represent positive staining; solvent is white. In the upper left-hand corner, a rhombic dodecahedral crystal is shown with a view parallel to its 3-fold axis. In the upper right-hand corner is shown an electron micrograph of a positively stained section of thickness less than one unit cell. Note the similarities to the drawings for thicknesses of 90 to 255 Å.
PLATE V.
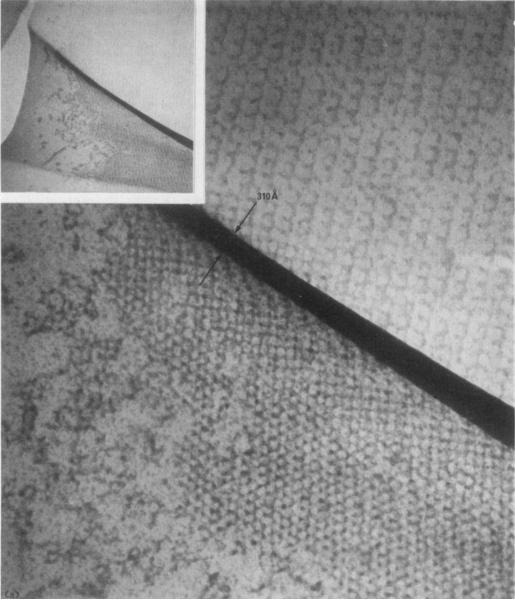
Micrograph of crystal sections showing folds produced during transfer of the sections onto electron microscope grids.
(a) A measure of the fold thickness (310 Å) enables one to estimate the section thiclmess (155 Å); (sec Small, 1968). Our interpretation, that this feature is indeed a double fold, is supported by a central line in the original negative. This proves it is possible to cut sections as thin as those illustrated in Plate IV. Magnification 180,000 ×. Inset magnification 36,000 ×.
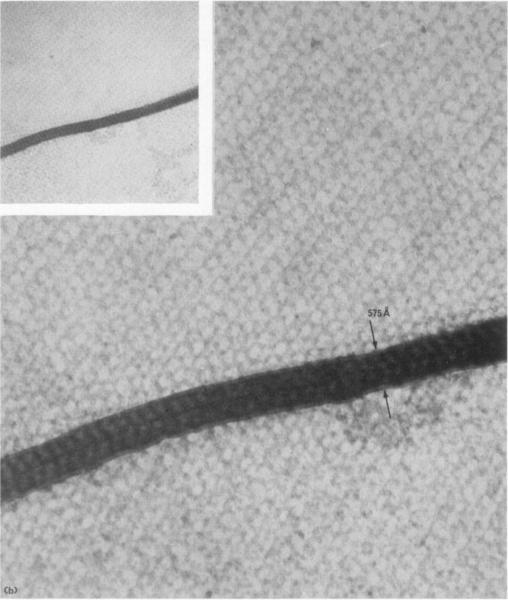
(b) A second folded section; the double thickness (575 Å) suggests the section thiclmess is 288 Å
Note the central line in the fold. Magnification 180,000 ×. Inset magnification 47,000 ×.
(f) Optical diffraction
For our proposed molecular packing structure to be correct, optical diffraction patterns from the projection drawings should correspond with the X-ray diffraction photographs. We find this to be true. The two projection drawings of Plate III(d) and (e) were used as diffraction gratings in an optical diffractometer to produce the corresponding central sections of the optical transform. These were recorded on film and are reproduced in Plate III(h) and (i) at the same relative magnification as the X-ray precession photographs. There are striking similarities in the corresponding patterns. Note, for instance, the weak (or absent) 6,0,0 reflection in the optical diffraction pattern produced from the 100 drawing (Plate III(i)). In Plate III(j) and (k) are the optical diffraction patterns from the electron micrographs of the thin sections illustrated in Plate III(f) and (g). The patterns do not extend beyond very low resolution; only the first and sometimes second-order reflections are strong enough to be recorded. Though not evident, some weak third-order reflections are present in the original negatives. The geometry of the diffraction patterns are nearly equivalent to the geometry of the reciprocal lattices of the X-ray photos and optical diffraction photos of the models. This indicates that the sections were oriented to within a few degrees of the correct cutting angle.
Where the diffracted intensities of the drawings differ significantly from those of the crystals, we attribute the differences to our crude representation of all projected matter in the drawings as black, rather than as varying shades of gray, as is obviously necessary to achieve anything near perfect agreement.
4. Discussion
Though our X-ray diffraction data are limited, when combined with micrographs, the molecular weight, and the crystal density, they suffice to determine much about the basic structure of the RuDPCase molecule. First, they reveal the minimum molecular symmetry. X-ray diffraction from the form I crystals shows that the space group of the crystals is either 1432 or, more probably, 14132, and electron microscopy (Results sections (d) and (e)) leaves no doubt that the latter space group is correct. In this space group there must be some multiple of eight or 12 molecules per unit cell. Since the crystal density and molecular weight indicate 13·5± 1·8 molecules per cell (Results, section (b)), there are clearly 12. Now, with 12 molecules per unit cell in space group 14132, each molecule must occupy a special position of 222 (D2) symmetry (Henry & Lonsdale, 1969). That is, the polypeptide chains are arranged in the molecule about three mutually-perpendicular 2-fold axes. The molecular symmetry can be higher if one or more of these molecular 2-fold axes is actually a higher even-fold axis.
The 222 symmetry restricts the possible subunit stoichiometries of the RuDPCase molecule; each polypeptide chain must be present in a multiple of four copies (L4nS4m). The observed molecular weights of the subunits rule out the possibility of either four or 12 large chains, so that the structure must be L8S4m. In fact, physical and chemical studies have provided evidence for eight copies of the large subunit (Rutner, 1970; Sugiyama et al., 1971; Nishamura & Akazawa, 1973). The fraction of the total mass contributed by the small subunit is so small that molecules of stoichiometry L8S8, L8S12, and L8S16 are all compatible with our crystallographic data and the spread of reported molecular weights (480,000 to 590,000; Kawashima & Wildman, 1970a).
Even though all three stoichiometries are compatible with molecular weights reported for RuDPCase, the L8S8 structure is the most likely. Only in this structure can all binding domains between large and small subunits be equivalent. In an L8S16 structure, for example, each large subunit might bind two small subunits, but it then must have evolved two different binding sites for small subunits. While this is possible, the simpler pattern of equivalent binding is almost always found in oligomeric enzymes (Matthews & Bernhard, 1973). The reason is that equivalent binding domains permit a simple mode of self-assembly.
If the stoichiometry of the RuDPCase molecule is L8S8 and if binding between all LS pairs is equivalent, then the symmetry of the RuDPCase molecule must be 422 (D4). This is the symmetry of a two-layered structure, each layer of which consists of four LS pairs arranged about a 4-fold axis; perpendicular to the 4-fold axis are four 2-fold rotation axes. This symmetry is compatible with our X-ray diffraction results, that the molecule has at least 222 symmetry, because 222 is a subgroup of 422 symmetry. The symmetry 422 is also compatible with our electron micrographs of isolated molecules, which show a single, prominent hole. Higher rotational axes result often in a hole about the axis, simply because it is impossible to pack identical bulky objects tightly about an axis. Thus the single prominent hole is more likely to be associated with a unique 4-fold axis than with one of three 2-fold axes. In short, though our data demand only that the RuDPCase molecules have symmetry 222 and one of the stoichiometries L8S8, L8S12, or L8S16, the additional assumption of equivalent bonding requires a L8S8 molecule of 422 symmetry.
The combined X-ray diffraction and electron microscopic results also reveal the packing of molecules in the form I crystals of the present study. This packing is unusually loose and open. A measure of the openness of packing is the Matthews parameter (Matthews, 1968), V M, the ratio of the unit cell volume to the mass of protein contained within it. For the form I crystals, VM has the value 8·36 Å3/dalton, which is three times as large as the most common value for protein crystals. Yet this value is consistent with many of our other observations on the crystals; their extreme mechanical fragility, their low density, and their high solvent content as observed directly in micrographs of microcrystalline fragments and sectioned crystals. Still another indication of the looseness of the crystal packing is the poor resolution exhibited by X-ray photographs. There is some evidence that suggests that the major contacts between molecules in the form I crystals may be between the large subunits. Crystals with morphology identical to form I have been obtained from RuDPCase from several pure and hybrid species of tobacco plants (Chan et al., 1972). The amino acid compositions of the large subunits are quite constant in most species, whereas the compositions of the small subunits vary considerably (Kawashima & Wildman, 1970a; Kawashima et al., 1971). The slower evolution of the large subunit may be related to the observation that it is coded for by chloroplast DNA and synthesized on 70 S chloroplast ribosomes (Chan & Wildman, 1972), whereas the small subunit is coded for by nuclear DNA and synthesized on 80 S cytoplasmic ribosomes (Kawashima & Wildman, 1972). In any case, it may be a conserved domain on the surface of the large subunit, binding to another conserved domain on another molecule, that causes crystals of the same form to grow from RuDPCase of a variety of species.
Acknowledgments
We are grateful to DrS. G. Wildman for discussions and help in obtaining pure RuDPCase, to Monica Eiserling for preparing thin sections of crystals, and to Dr D. L. D. Caspar for suggestions that led to X-ray diffraction photographs. Two authors (T. S. B.) and (L. W.) are predoctoral trainees and another author (D. E.) is a Career Development awardee of the United States Public Health Service. Grant GM16925 to one of us (D. E.) and National Science Foundation grant GB13117 to another author (F. A. E.) have supported this research.
Footnotes
Abbreviation used: RuDPCase, dribulose-1,5-diphosphate carboxylase.
REFERENCES
- Akazawa T, Kondo H, Shimazue T, Sugiyama T. Biochemistry. 1972;11:1298–1303. doi: 10.1021/bi00757a028. [DOI] [PubMed] [Google Scholar]
- Branton D, Park RB. J. Ultrastruct. Res. 1967;19:283–303. doi: 10.1016/s0022-5320(67)80222-3. [DOI] [PubMed] [Google Scholar]
- Chan PH, Wildman SG. Biochim. Biophys. Acta. 1972;277:677–680. doi: 10.1016/0005-2787(72)90115-3. [DOI] [PubMed] [Google Scholar]
- Chan PH, Sakano K, Singh S, Wildman SG. Science. 1972;176:1145–1146. doi: 10.1126/science.176.4039.1145. [DOI] [PubMed] [Google Scholar]
- Climenson CS, Eisenberg DS. J. Appl. Crystallogr. 1974;7:268–274. [Google Scholar]
- Franks A. Proc. Roy. Soc. ser. B. 1955;68:1054–1064. [Google Scholar]
- Gunning BES, Steer MW, Cochrane MP. J. Cell Sci. 1968;3:445–456. [Google Scholar]
- Harrison SC. J. Appl. Crystallogr. 1968;1:84–90. [Google Scholar]
- Haselkorn R, Fernández-Marán H, Kieras FJ, van Bruggen EFJ. Science. 1965;150:1598–1601. doi: 10.1126/science.150.3703.1598. [DOI] [PubMed] [Google Scholar]
- Henry NFM, Lonsdale K. International Tables for X-ray Crystallography. 3rd edn Vol. 1. The Kynock Press; Birmingham, England: 1969. [Google Scholar]
- Kawashima N. Plant and Cell Physiol. 1969;10:31–40. [Google Scholar]
- Kawashima N, Wildman SG. Annu. Rev. Plant Physiol. 1970a;21:325–358. [Google Scholar]
- Kawashima N, Wildman SG. Biochem. Biophys. Res. Commun. 1970b;41:1463–1468. doi: 10.1016/0006-291x(70)90551-6. [DOI] [PubMed] [Google Scholar]
- Kawashima N, Wildman SG. Biochim. Biophys. Acta. 1971;229:240–249. [PubMed] [Google Scholar]
- Kawashima N, Wildman SG. Biochim. Biophys. Acta. 1972;262:42–49. doi: 10.1016/0005-2787(72)90217-1. [DOI] [PubMed] [Google Scholar]
- Kawashima N, Kwok SY, Wildman SG. Biochim. Biophys. Acta. 1971;236:578–586. doi: 10.1016/0005-2795(71)90242-x. [DOI] [PubMed] [Google Scholar]
- Kwok SY. Ph.D. Thesis. University of California; Los Angeles: 1972. [Google Scholar]
- Matthews BW. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Matthews BW, Bernhard SA. Annu. Rev. Biophys. Bioeng. 1973;2:257–317. doi: 10.1146/annurev.bb.02.060173.001353. [DOI] [PubMed] [Google Scholar]
- Moon KE, Thompson E0P. Aust. J. Biol. Sci. 1969;22:463–470. [Google Scholar]
- Murai T, Akazawa T. Biochem. Biophys. Res. Commun. 1972;46:2121–2126. doi: 10.1016/0006-291x(72)90768-1. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Akazawa T. Biochem. Biophye. Res. Commun. 1973;54:842–848. doi: 10.1016/0006-291x(73)90770-5. [DOI] [PubMed] [Google Scholar]
- Paulson JM, Lane MD. Biochemistry. 1966;5:2350–2357. doi: 10.1021/bi00871a025. [DOI] [PubMed] [Google Scholar]
- Reedy MK. J. Cell Biol. 1965;26:309–311. doi: 10.1083/jcb.26.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutner AC. Biochem. Biophys. Res. Common. 1970;39:923–929. doi: 10.1016/0006-291x(70)90412-2. [DOI] [PubMed] [Google Scholar]
- Rutner AC, Lane MD. Biochem. Biophys. Res. Commun. 1967;28:531–537. doi: 10.1016/0006-291x(67)90346-4. [DOI] [PubMed] [Google Scholar]
- Shively JM, Ball FL, Kline BW. J. Bacteriol. 1973;116:1405–1411. doi: 10.1128/jb.116.3.1405-1411.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel MI, Wishnick M, Lane MD. In: The Enzymes. Boyer PD, editor. Vol. 6. Academic Press; New York: 1972. pp. 169–192. [Google Scholar]
- Small JV. In: Fourth European Regional Conference (Rome) on Electron Microscopy. Bocciarelli DS, editor. Vol. 1. Tipografia Poliglotta Vaticana; Roma: 1968. pp. 609–610. [Google Scholar]
- Steer MW, Gunning BES, Graham TA, Carr DJ. Planta. 1968;79:254–267. doi: 10.1007/BF00396032. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Akazawa T. J. Biochem. (Tokyo) 1967;62:474–482. doi: 10.1093/oxfordjournals.jbchem.a128691. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Akazawa T. Biochemistry. 1970;9:4499–4504. doi: 10.1021/bi00825a006. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Ito T, Akazawa T. Biochemistry. 1971;10:3406–3411. doi: 10.1021/bi00794a014. [DOI] [PubMed] [Google Scholar]