Abstract
The diameters of several frozen-hydrated, spherical viruses were determined using polyoma virus as either an external or an internal calibration standard. The methods described provide a reproducible and accurate way to calibrate microscope magnification. The measured diameters are in excellent agreement with respective measurements previously reported for aqueous samples at room temperature using X-ray diffraction methods. These results indicate that the native morphology and dimensions of biological macromolecules are better preserved in frozen-hydrated samples when compared with more conventional electron microscopy techniques such as negative-staining, metal shadowing or thin-sectioning.
1. Introduction
Accurate determination of particle dimensions in electron micrographs depends both on the calibration of the microscope magnification value and on the specimen preparation and examination procedures. Under typical working conditions, the nominal magnification value displayed on most microscopes only approximates the correct value to within 5–15%. The actual magnification is influenced by several factors including lens hysteresis, fluctuations of the acceleration voltage and variations in the axial position of the specimen. Distortions of the specimen due to preparation artifacts or beam damage may also affect the accuracy of dimension measurements.
Determination of operational microscope magnification values requires the use of calibration methods. Diffraction grating replicas and numerous types of periodic biological and non-biological specimens, mounted on separate grids, often serve as useful external standards. Such standards are photographed immediately before or after recording images of the unknown specimen. However, these measurements may be subject to many of the same instrumental errors mentioned above. In addition, calibrations made with replica gratings, for example, are only about 10% accurate at magnifications exceeding 30,000 × because it is not possible to record enough grating spacings in one micrograph to obtain statistically significant measurements.
More accurate results may be obtained if a suitable internal calibration standard is photographed in the same field of view as the object of interest. Polystyrene latex spheres have been extensively used in this way since 1949 [1]. They are available in sizes ranging from 0.08 to 90 µm with a variation of sphere size within given samples of about 3% [2]. Latex spheres are rather unreliable because of their sensitivity to the electron beam. The diameter of latex spheres shrinks by 2% within the first few seconds exposure to the electron beam and reduces another 14% after 15 minutes in the beam [3].
Thin catalase platelet crystals, with known unit cell dimensions [4–6], are widely used as an internal standard for calibration of negatively stained specimens photographed at magnifications between 20000 × and 60000 ×. Use of such stained crystals may lead to inaccuracies because they (and the stain) are sensitive to the electron beam, especially when the sample is suspended over holes in a carbon support film [7]: crystals tend to shrink in unsupported areas. Crystals, attached to and fully supported by the carbon film, anisotropically shrink when dried. Specimen dehydration reduces the axial crystal dimension to about 70% of the fully hydrated dimension even before the specimen is subjected to the vacuum of the microscope. Irradiation for prolonged periods further reduces the crystal thickness to about 40% of its initial value and this is often accompanied by concomitant, but smaller, non-uniform changes in the cell dimensions within the platelets [7].
The sample of interest and the magnification standard are both subject to the same artifacts of drying and radiation-induced changes. The reported diameters of spherical viruses embedded in various thicknesses of negative stain are often much lower than measurements made of hydrated samples using X-ray diffraction techniques presumably due to masking of surface features in thick layers of stain and/or isotropic shrinkage of particles in the dried layer of stain (table 1). This is especially prevalent when virus particles are suspended over holes in the carbon support film. Particles attached to the support film may be larger than expected as a consequence of flattening induced by the surface tension forces present during dehydration. Some samples apparently resist such flattening by becoming “wrapped” in the support film [25]. This effect may possibly account for observed variations in the amount of stain retained by samples. Negative stains are also known to be sensitive to electron irradiation since they tend to migrate over the accessible surface of the specimen during the initial stages of exposure [26,27]. This, in turn, affects the accuracy of identifying stain-excluding boundaries. The artifacts mentioned above confound attempts to accurately measure dimensions within specimens.
Table 1.
A comparison of diameter measurements (nm) obtained by electron microscopy of unstained frozen-hydrated virions and negatively stained virions, by X-ray diffraction techniques, and by other methods
| Virus | Frozen-hydrated | Negatively stained | X-ray solution scattering | Other methods | |||
|---|---|---|---|---|---|---|---|
| Polyoma | 45.3 | [8] | 49.5 | [9] a) | 40–45 | [10] b) | |
| 38 | [11]c) | ||||||
| SV40 | 49.4 ±1.8 | 45 | [12] | 48 | [13] | 44.1 ±0.6 | [14]d) |
| NβV | 39.4 ± 1.6 | 35 | [15] | 39.4 | [18] | 35.7 | [16]f) |
| 39.5 | [16] | ||||||
| 30, 50 | [17]e) | ||||||
| CPMV | 27.5 ± 0.8 | 20.0, 24.0 | [19]g) | 26.8, 28.4 | [20h) | ||
| 28.4 | [21] | ||||||
| 26.0, 26.7, 30.8 | [21]i) | ||||||
| BMV | 26.8 ± 1.0 | 26.0 | [22] | 26.0 | [22] | 29.0 | [23]j) |
| 27.1 ± 1.1 | [4] | ||||||
| FHV | 31.2 ± 1.6 | 29 | [24] | 31.2 | [18] | ||
Packing diameter in body-centered cubic crystals.
Chromium-shadowed.
Thin-sectioned specimen.
Platinum-, palladium-, gold-, silver-shadowed.
Values are diameters for virions embedded in stain over holes in a carbon substrate and for virions flattened over the substrate, respectively.
Calculated diameter based on the density difference between virus and medium, the viscosity of the medium, and the sedimentation coefficient of the virus.
Circularly averaged diameter and distance between opposite 5-fold icosahedral vertices, respectively.
Packing diameter of virions in cubic (3-fold to 3-fold) and hexagonal crystals, respectively.
Distance between outer surfaces along icosahedral 2-fold, 3-fold, and 5-fold axes, respectively.
Freeze-dried and metal-shadowed.
Cryo-microscopy of biological specimens has emerged as one technique capable of minimizing or eliminating many of the problems and artifacts associated with negative-staining and other conventional electron microscopy preparative procedures [28–32]. The crystalline structure of a variety of frozen-hydrated biological specimens is preserved to near atomic resolution. Examples include: purple membrane [33,34], catalase [35], crotoxin [36,37], rabbit immunoglobulin FC fragment [38], and T4 DNA helix destabilizing protein [39]. Preparation procedures involving vitrification of the sample also appear to protect non-crystalline biological specimens against the distorting forces of dehydration and ice crystal formation [31]. The spherical virus particle samples used in our experiments are easily frozen in a thin layer of vitrified water, supported over holes in a carbon film, and subsequent images recorded with minimal irradiation techniques [31,32].
In this report, polyoma, a spherical animal virus whose diameter is accurately known from X-ray diffraction measurements [9], has been used as both an internal and external standard for calibrating the diameter of a variety of other spherical viruses examined in the frozen-hydrated state. This allows accurate determination of the instrument magnification and has also made it possible to calibrate previously recorded micrographs of simian virus 40 (SV40)* [40]. Our results clearly demonstrate that the gross morphology of biological specimens is well preserved in the frozen-hydrated state since the dimensions of the spherical virus particles studied are, within experimental error, identical to measurements made by X-ray diffraction of aqueous samples at room temperature.
2. Materials and methods
2.1. Sample preparation and electron microscopy
Purified virus samples, stored in various buffer solutions at concentrations between 2 and 10 mg/ml, were kindly provided by colleagues (see acknowledgements). Virus isolates were mixed with the polyoma sample, and when necessary, diluted with distilled water to provide a final concentration of about 0.5–1.0 mg/ml. Small aliquots (2–5 µl) of sample were applied by adhesion to copper microscope grids covered with perforated carbon films made hydrophilic by glow discharge. Most of the sample was removed by pressing a piece of filter paper directly against the entire grid surface, wicking it nearly dry, before rapidly plunging it into liquid ethane kept slightly above its freezing point (> −183°C) [32]. The grids were then quickly transferred into liquid nitrogen where they could be stored or subsequently transferred into a specimen holder (Gatan Inc., Warrendale, PA) which maintains the specimen temperature at about −160 ° C in the microscope. Areas with frozen sample suspended over holes in the carbon substrate, and in which the thickness of the vitrified water layer was judged to be sufficient to support a monodisperse distribution of virus particles, were located by scanning grids at very low magnification (1000–2000 ×) with an electron dose rate below 10 e−/nm2 s. Micrographs of the frozen-hydrated samples were recorded at nominal magnifications of 36,000 × and 49,000 × on Kodak SO-163 film using minimal dose techniques (900–1200 e−/nm2), on a Philips EM420 electron microscope operated at 80 kV. Lens currents were fully excited and returned to preset levels before each exposure by the use of the lens normalization operation on the microscope. Exposed micrographs were developed for 12 min in full-strength Kodak D-19 developer at 20°C to maximize the sensitivity of the photographic emulsion.
2.2. Micrograph selection and digitization
The selection of micrographs for digital processing was based on several criteria. Images judged to be free of the effects of residual objective lens astigmatism and specimen drift and recorded at an optimum defocus level (1–2 µm underfocus) were identified. Regions in those images in which the sample consisted of a fairly uniform, monodisperse distribution of virus particles were digitized at 25 µm intervals. The raster step size of the rotating drum microdensitometer (Optronics C-4100, Optronics Inc., Chelmsford, MA) was carefully determined to minimize systematic errors in the calibration measurements. A contact photograph of a microscope eyepiece reticule, calibrated with a microcomparator (Nikon Profile Projector model 6C, Nippon Kogaku, K.K., Japan), was digitized and its Fourier transform was computed. The distance between prominent peaks in the Fourier transform, resulting from the periodically spaced lines in the reticule grating image, enabled an accurate measure of the raster step size to be calculated.
The emulsion side of the selected micrograph was securely taped to an opening ( ~ 3.2 × 2.6 cm) cut in a thin sheet-metal mask, curved to fit the cylindrical microdensitometer drum. The micrograph was carefully mounted to the mask to insure that the entire scanned area of the emulsion coincided with the focal surface of the scanner optics, thereby avoiding additional sources of error in the magnification calibration. Digitized images were stored on magnetic tape and transferred to disk storage on a VAX/VMS 11/750 or 8550 minicomputer (Digital Equipment Corporation, Maynard, MA) for further analysis using interactive FORTRAN routines with various data displayed on a 1280 by 1024, 8-bit per pixel, color raster graphics device (Lexidata 3400, Adage Inc., Billerica, MA).
2.3. Virion particle image selection and diameter determination
Individual virion images were selected for analysis if the particle appeared to be intact and not distorted in any obvious fashion, and also not in contact with or superimposed upon the edge of a carbon hole or any other virions. Each virion image was boxed and “floated” [41] by selecting a circular area, centered about the particle and just large enough to encompass the entire particle without including extraneous sample (e.g. surrounding vitrified water and neighboring particles). To float each “boxed” particle image, the average intensity value at the periphery of the circular area (corresponding to the projected density of the surrounding vitrified water) was subtracted from every pixel within the circular boundary. The processing system is designed to detect and ignore density values from neighboring particles that occasionally happen to overlap the chosen circular boundary from which the average float value is obtained.
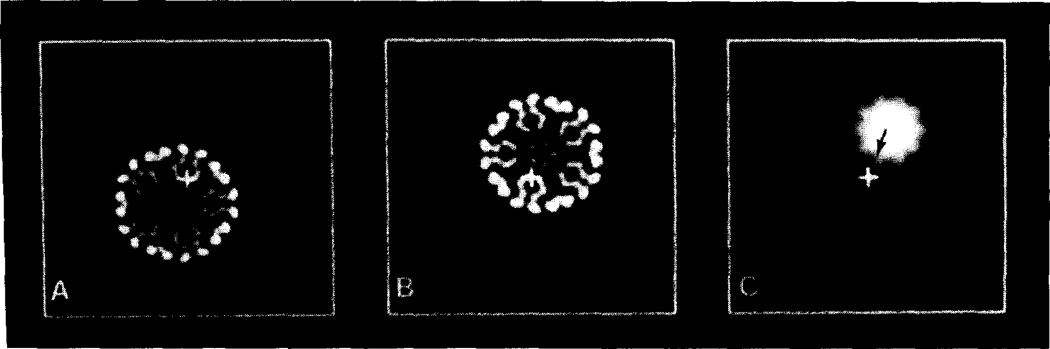
The position corresponding to the center of each particle was determined using a cross-correlation procedure (appendix A). In some instances a different cross-correlation procedure was used to select and average particle images (appendix B). Individual particle images or particle averages were circularly averaged about their respective centers (figs. 1A and 1B) and mean particle diameters were measured by locating the “edges” of the circularly averaged particles (fig. 1C). For the spherical viruses studied, location of the mean particle boundary was usually straightforward since the boundary between particle and surrounding vitrified water manifests an abrupt change in density (hence contrast) which is relatively easy to identify. All images of a particular virus were analyzed in a similar fashion and the results averaged to determine a mean and standard deviation (in pixels) for the diameter. These measurements were then calibrated against similar measurements made on the polyoma standard, using the 49.5 nm diameter determined by X-ray diffraction measurements [9]. An average data set consisted of 28 particle images (range: 18–44).
Fig. 1.
Determination of virion image radius. (A) Digital image of an unfiltered, floated, circularly boxed polyoma virion. The virion image is displayed with enhanced positive contrast. The higher density of the virus compared to the surrounding vitrified water makes it appear darker. The center and edge of the particle are identified respectively by the base and tip of the arrow (same for B). (B) Image shown in (A) circularly averaged about the particle center. The particle edge is easier to identify compared to (A) due to the abrupt change in the averaged density. (C) Radial density plot corresponding to the intensities along the arrow in (B). The arrow identifies the location of the particle boundary. Since the position of the abscissa axis is established by the floating procedure, there is some uncertainty in identifying the exact position of the particle boundary, although the uncertainty is small since the abrupt change in contrast is confined to a narrow range of radii.
2.4. Calibration of the polyoma standard
Electron diffraction patterns of frozen-hydrated samples of tobacco mosaic virus (TMV) were recorded to test the assumption that the diameter of the polyoma standard in vitrified water is identical to the X-ray measured value. TMV was chosen because it is a universally accepted biological standard which produces a strong, characteristic fiber diffraction pattern [42]. Furthermore, the TMV diffraction pattern can be easily calibrated by simultaneously recording the diffraction “powder pattern” from cubic ice crystals allowed to condense on the frozen-hydrated sample.
TMV samples (15 mg/ml) were prepared for cryo-microscopy as described in section 2.1. The high particle concentration favors the alignment of the particles into near-parallel arrays. Once in the microscope, the anticontamination shield on the specimen cold holder was retracted and the specimen left exposed to the microscope vacuum for a period of time (~ 1 h) to allow water molecules in the column to condense on the specimen in the form of cubic ice [29]. Electron diffraction patterns were obtained with the first condenser lens fully excited, with a 12.5 µm second condenser lens aperture in place, and with the objective lens aperture removed. The grid was scanned for suitable specimen areas in defocussed diffraction mode and focussed diffraction patterns were recorded on Kodak SO-163 film with a 90 s exposure. The film was developed in Kodak D-19 (1: 2) for 4 min.
Diffraction pattern films were digitized as described in section 2.2 at two raster settings (25 µm and 100 µm) to allow accurate measurement of both the strong 1.15 nm 6th layer line of TMV and the [111] 0.366 nm diffraction ring of cubic ice [43].
Mixtures of TMV and polyoma were imaged (section 2.1) to calibrate the diameter of polyoma against the TMV standard. Fourier transforms of individual, boxed TMV particles were computed, and the 2.3 nm 3rd layer line reflection was used to calibrate the image pixel size. The average diameter of the polyoma particle images (in pixels) was determined (section 2.3) and scaled to the TMV measurements.
3. Results
3.1. Virion morphology
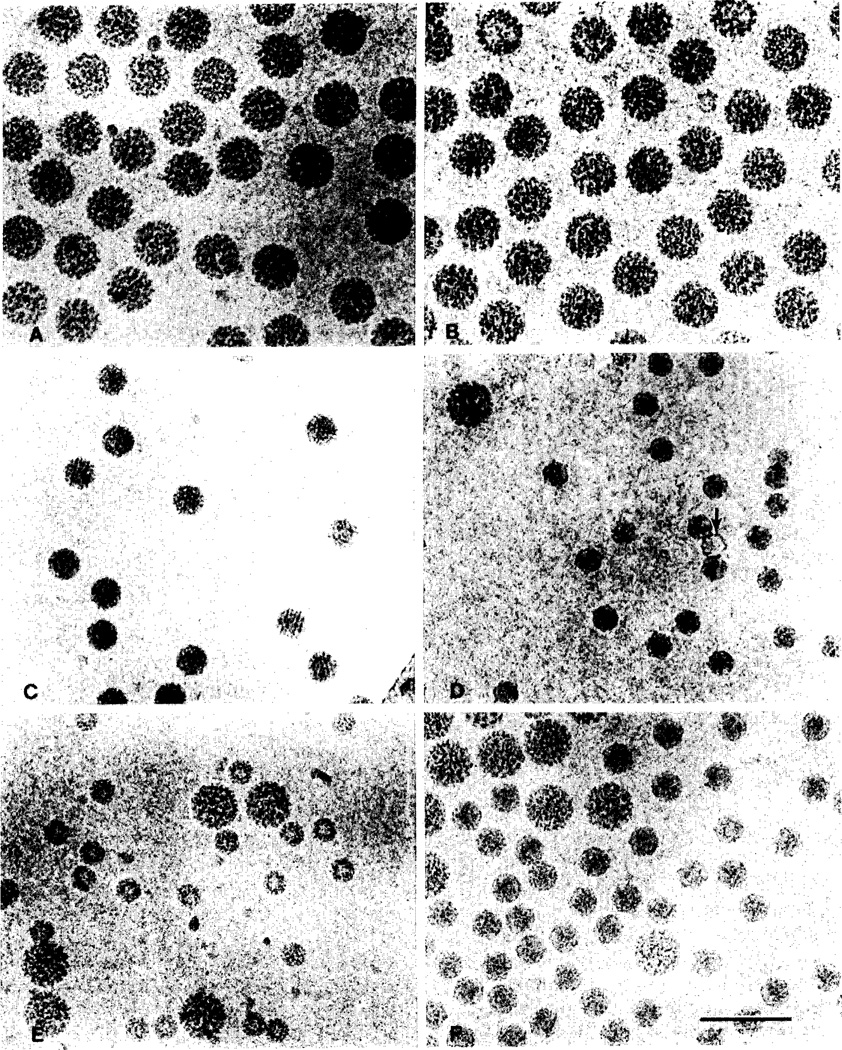
The overall morphology of unstained virus particles, which are frozen and embedded in vitrified water, appears to be well-preserved (fig. 2). Virions are imaged with positive contrast; that is, particles appear darker than the background due to the higher mass density of the protein/nucleic acid complex compared to the supporting matrix. Images of viruses with a nearly spherical surface morphology, such as polyoma and SV40 (figs. 2A and 2B), display circular profiles: particle distortion due to flattening is not visibly detectable. The profiles of polyoma particles tilted up to 30° in the microscope (data not shown) are also circular, thus indicating there is no apparent compression or expansion of the particles in a direction normal to the plane of the vitrified water layer. Small icosahedral viruses such as the T=1 cowpea mosaic virus (CPMV; fig. 2D) and the T = 3 Flock House virus (FHV; fig. 2F) often appear spherical and nearly featureless when contrasted with negative stain. Images of the same viruses, unstained and frozen in vitrified water, frequently exhibit prominent hexagonal profiles implying that their surface morphology is markedly non-spherical. Capsids (particles devoid of nucleic acid) appear as hollow shells and are easily distinguished from complete virions (fig. 2D, arrow).
Fig. 2.
Unstained, frozen-hydrated spherical viruses, suspended over holes in a carbon substrate. Magnification is identical for all images (bar = 100 nm). (A) Polyoma. (B) SV40. (C) NβV. (D) CPMV/polyoma mixture; the arrow identifies a CPMV empty capsid. (E) BMV/polyoma mixture. (F) FHV/polyoma mixture.
3.2. Diameter measurements
Results of the diameter measurements for several vitrified spherical viruses are listed in table 1. The measurement for each unknown virus was normalized with the 49.5 nm diameter of polyoma virus [9]. Previous measurements of the diameters of these viruses, examined in the electron microscope by negative-staining techniques or examined by various X-ray techniques, are included in table 1 for comparison with the frozen-hydrated results. The last column lists published measurements obtained from a variety of other physical-chemical methods.
The results clearly show that the diameter of the negatively-stained specimens listed in table 1 is typically lower than corresponding measurements obtained by X-ray techniques. Differences between the corresponding values reported in table 1 vary from 6% for SV40 to almost 30% for circularly averaged CPMV. The opposite trend has been observed for some reported values of bromegrass mosaic virus (BMV) and Nudaurelia capensis β virus (NβV); namely, that the stained particles had larger diameters than those obtained by X-ray measurements. Variations between X-ray values and those obtained from a variety of other electron microscopy and hydrodynamic measurements have also been observed. Measurements from frozen-hydrated specimens are generally more reproducible than those obtained from negatively-stained specimens and are consistent with measurements made using X-ray techniques: the X-ray diffraction and cryo-microscopy measurements generally vary by no more than 3% and the results reported for NβV and FHV are nearly identical.
3.3. Calibration of the polyoma standard
Calibrations of particle dimensions and the micrograph magnification require that the microdensitometer raster step size be known and also that the diameter of the frozen-hydrated polyoma virions remain unchanged compared with virions at room temperature in crystals or in solution. Measurements of the microdensitometer step size, in the x (spindle) and y (rotation) directions, were found to lie within 0.25% of the expected value so no systematic error is believed to have been introduced into the measurements from digitization of film data.
Electron diffraction patterns of TMV samples embedded in vitrified water and contaminated with a layer of cubic ice were recorded to determine if the change of water from a liquid to a solid phase produces dimensional changes in biological specimens. The spacing of layer lines in the helical diffraction pattern of TMV was calibrated with the powder diffraction rings of the 0.366 nm [111] Bragg reflection of cubic ice [43]. The average pitch of the 1-start helix of TMV (obtained from three different diffraction patterns) was determined to be 2.28 nm. This is equivalent, within the errors of measurement, to the universally accepted standard value of 2.3 nm [44]. If the electron diffraction calibration of TMV is correct, and the measured polyoma diameters are scaled accordingly, the diameters of the other viruses listed in table 1 would systematically drop by slightly less than 2%. Some particle diameters (e.g. CPMV) would deviate marginally from the X-ray values, whereas other particles (e.g. BMV) would have dimensions which more closely match the X-ray results. Since our measurement of TMV differs insignificantly from the established standard, we assume that the TMV dimensions are not detectably changed in vitrified water. Thus, in this study, we accepted the X-ray value [9] for the diameter of polyoma as the standard for calibrations of the other virus particles.
3.4. Reproducibility of diameter measurement
The reproducibility of measurements using frozen-hydrated polyoma virus as a calibration standard was tested in several ways. Diameter measurements were obtained from images of five distinct icosahedral viruses (from plant, insect and animal hosts) suspended in different aqueous buffers. The microscopy was performed with a range of different operating conditions.
3.4.1. Internal versus external standard calibrations
Reproducibility of the measurements was excellent when polyoma was used as either an internal or external standard (table 1). Diameters of SV40 (fig. 2B) and Nβ3V (fig. 2C) were determined using polyoma as an external standard. The similarities in size and morphology between polyoma and SV40 precluded mixing them together (fig. 2A and 2B). Polyoma was used as an internal standard to measure the diameters of BMV (fig. 2E) and FHV (fig. 2F). CPMV virions were measured both with (fig. 2D) and without (data not shown) polyoma virus in the sample. Statistical comparisons of the two types of CPMV samples, recorded on consecutive days, showed no significant differences in the diameter measurements at the 95% level using the Student's t test. The value listed for CPMV (table 1) is an average of the external and internal standard measurements.
3.4.2. Effect of specimen location
NβV diameter measurements were made on specimens imaged in different areas of the same grid and also on different grids; the differences observed were statistically insignificant. Thus, reproducibility of the measurements is not noticeably affected by location of the sample on the grid or placement of the grid in a given specimen holder under reasonably controlled experimental conditions (i.e. the specimen height in the microscope is reproducibly maintained using flat, securely held grids).
3.4.3. Effect of defocus
A focal series of SV40 was recorded (data not shown) to determine if the accuracy of the diameter measurements could be influenced by strong Fresnel fringes that appear at the periphery of the virus particle images due to phase contrast produced at different levels of defocus. Micrographs at 1.0, 1.5, and 2.0 µm underfocus were recorded in quick succession from the same specimen area with a total electron dose of < 2000 e−/nm2. The diameters of identical sets of SV40 particle images varied by less than 1% indicating that the exact focal setting was not a significant factor in the diameter measurements reported in table 1.
3.4.4. Particle concentration
Ideally, the frozen-hydrated sample consists of a monodisperse distribution of virus particles such that the concentration of particles is high enough to allow a statistically significant number of measurements to be made from a single micrograph. In such micrographs (e.g., figs. 2A–2E), individual particle images were circularly averaged to improve the accuracy of the diameter measurements (section 2). Mean diameters were then computed from each set of particle measurements. In micrographs where the concentration of particles was too high to allow accurate boxing of individual particles (e.g., figs. 2F and 5A), an alternate procedure, in which particle averages are obtained by cross-correlation methods, was used to measure diameters (appendix B). A micrograph of FHV was analyzed by both procedures to test the self-consistency of the two methods. The results varied by less than 2% which is within the range of variation observed for each of the separate methods. The diameter for FHV reported in table 1 is the average of the two measurements obtained from the same set of particle images and is not significantly different from the X-ray solution scattering measurements.
Fig. 5.
Cross-correlation procedure to select and average virion images. (A) Image of a frozen-hydrated mixture of polyoma (larger particles) and FHV. The increase in average background density from left to right is due to an increase in vitrified water thickness over the hole in the carbon substrate. (B) Fourier-space “ prefiltered” image of (A) obtained by Fourier reconstruction methods [66] which remove low frequency fluctuations ( > ~ 100 nm) mainly contributed by the variation in the thickness of the vitrified water. The circled FHV (left) and the polyoma (right) images were used a reference images for the cross-correlation procedure. Both reference particles were boxed with identical size circular boundaries to minimize ambiguities in discriminating correlation peaks for the two different virions (see footnote in appendix B). (C) Schematic representation of (B), identifying the approximate location of the FHV and polyoma particles. (D) Cross-correlation pattern between the polyoma reference (circled in (B)) and image (A). Areas of high correlation are represented by brighter colors. The arrow identifies the strongest correlation peak, locating the polyoma reference. (E) Cross-correlation pattern between the polyoma reference circled in (B)) with image (B). The removal of low-frequency features enhances the ability to clearly identify all the polyoma positions. Virion images in (B), identified by the peaks, were averaged, and this average was used as the reference in a subsequent cycle of cross-correlation averaging. (F) Cross-correlation pattern between the reference average image and (B). The peaks identifying the positions of the polyoma particles are much sharper and clearer than those in (E). The inset shows the averaged polyoma image obtained by combining those particles identified by the peaks in the last correlation pattern computed. (G) Same as (D) with the exception that the FHV image circled in (B) was used as the reference. The arrow locates the position of the FHV reference. (H) Same as (E) using the FHV reference (circled in (B)). (I) Same as (F) using an averaged FHV image. The inset shows the averaged FHV image obtained after the second cycle of correlation averaging.
4. Discussion
4.1. Preservation of biological samples in the frozen-hydrated state
The use of frozen-hydrated spherical viruses as a magnification standard is especially suitable for two primary reasons: (i) quick freezing of particulate biological samples in a thin layer of vitrified water provides an excellent way to preserve the samples without introducing many of the artifacts associated with other preparative procedures and (ii) measurements obtained using a frozen-hydrated calibration standard are more reproducible than those obtained using other available standards.
Viruses, as well as a variety of other biological macromolecules, have been shown to be well preserved in vitrified water. Diameter measurements for adenovirus, λ bacteriophage, and the isometric head of T4 bacteriophage [45] all show close agreement with respective low-angle X-ray diffraction measurements [31]. Three-dimensional reconstructions have been calculated from images of frozen-hydrated virions of Semliki Forest [46], Sinbis [47], SV40 [40], Rotavirus [48] and NβV [49]. The overall morphology of all of these viruses is well preserved in vitrified water, suggesting that particle flattening and other gross deformations do not occur. Calculation of scattering curves from the three-dimensional maps of Sinbis virus [47] and NβV [50] agree quite closely with X-ray solution scattering experiments. In addition, even though the virus reconstructions have been computed with 10-fold averaging (522 symmetry), they still possess near perfect 532 symmetry as would be expected for particles with icosahedral symmetry. Negatively stained La Crosse virus [51] and Influenza virus [52] often exhibit a pleomorphic appearance or are otherwise flattened, whereas frozen-hydrated preparations of both these viruses show spherical particles which exhibit many features not apparent in negative stain. Virions in the present study maintained a symmetrical profile when in the frozen-hydrated state. No detectable flattening of polyoma virions was observed even in tilted samples.
Preservation of biological structure in vitrified water has also been reported for a variety of non-virus samples. Measurement of the lattice constants of frozen-hydrated catalase platelet crystals were within 1% of those obtained by X-ray diffraction [31], and electron diffraction patterns have been recorded which demonstrate structural preservation of features as small as 3.4 A in catalase crystals [53]. Unstained, frozen-hydrated microtubules [54] have a uniform mean diameter which corresponds closely to X-ray fiber diffraction data. Diffraction intensities along F-actin filament [55] and gap junction membrane [56] layer lines also closely agree with those from X-ray diffraction. The remarkable agreement between dimensions of aqueous samples measured by X-ray diffraction and frozen-hydrated specimens measured by electron microscopy is somewhat surprising since water expands 2–3% upon freezing to the vitreous state. One might expect such expansion to create forces which would act to compress the biological sample [43]. Apparent resistance of the sample to structural change caused by vitrification could indicate that water redistributes during freezing or that differences exist between the density of vitreous water in the free solvent compared with the hydration shell surrounding the biological molecule [29].
4.2. Reproducibility of measurements
The reproducibility of measurements made with vitrified samples indicates that these samples make more attractive calibration standards compared to conventional ones. Replica gratings and polystyrene spheres are generally unsuitable at the magnifications typically required in virus imaging. Replica gratings are also difficult to use as internal standards. The smallest commercially available polystyrene spheres are about ten times larger than many viruses. Also, the hydrophobic nature of polystyrene spheres tends to force them out of solution onto the surface of the water-hydrophylic support film interface as the aqueous sample evaporates and thins just before freezing [31]. Many viruses have hydrophilic surfaces and thus tend to diffuse into thicker regions of the aqueous sample as it dries. Catalase platelet crystals are more suitable than the standards mentioned above. However, they are somewhat impractical to use as an internal standard due to the difficulty of recording, in the same micrograph, sufficient numbers of virus particles together with a crystal.
Polyoma was chosen as a calibration standard because it has a predominantly spherical external morphology and a diameter accurately known from X-ray crystallography [9]. Furthermore, polyoma maintains its spherical shape and is randomly oriented when quickly frozen. The average standard deviation in measurements of the polyoma diameter is about 2% of the mean. The diameters of other viruses standardized against polyoma closely match the corresponding values obtained by X-ray diffraction methods (table 1). This is true despite the variety of conditions employed while making measurements: use of polyoma as an internal or external standard, placement of the grid in the microscope holder, the level of defocus of the image, and the choice of method used to measure the particle diameters (appendices A and B).
4.3. Limitations affecting accuracy and reliability of measurements
Vitrification of virus particles does not appear to cause significant changes or distortions of the particle dimensions. Electron diffraction measurements of the TMV helix pitch (6.84 nm) closely agree both with original measurements (6.78 nm [57,58]) and subsequent remeasurements (6.9 nm [44]) of fibers examined at room temperature by X-ray diffraction. If accurate, our measurements of TMV dimensions indicate that the helical particle distorts less than 1% in the axial direction (changes in particle diameter were not measured). Since this difference is statistically within the level of error in the measurements, more extensive, controlled experiments would be necessary to detect if compression or expansion of TMV occurs. In the absence of these measurements, we assume there is no detectable distortion of the TMV particles.
When calibrated against TMV, the mean diameter of polyoma is about 48.6 nm. This implies that polyoma is compressed by less than 2%. Discrepancy between this value and that determined by X-ray diffraction might arise from systematic errors such as those which would occur if measurements were made from TMV particles whose axes are tilted away from a direction perpendicular to the electron beam (i.e. the long axis of the TMV particles might be slightly tilted within the plane of supporting layer or the region of the EM grid examined may not be perpendicular to the electron beam). Other systematic errors might result from differential distortion in differently shaped particles or particles with anisometric shapes (e.g. from differences in the packing density of subunits in axial and equatorial directions).
The reliability of the particle measurements might also be influenced by the choice of averaging method. These methods produce a measure of mean particle diameters which must represent an under-estimate of the maximum particle diameter. In this study, the mean diameter for polyoma is taken as the interparticle packing distance in crystals studied by X-ray crystallography [9]. This distance is equal to the contact distance between particles, measured along the body diagonal of the 57.2 nm cubic unit cell, which coincides with the 3-fold icosahedral particle axis. The particle diameter is slightly larger when measured between 5-fold vertices since the pentavalent capsomeres extend radially 0.2–0.3 nm further than the hexavalent capsomeres [9]. The mean diameter measurement is also affected by the presence of deep, solvent-filled channels in the capsid surface, resulting from the large gaps between neighboring, radially projecting capsomeres. The presence of vitrified water in these gaps would act to lower the average electron density in spherically averaged data, thus causing the mean diameter to lie somewhere between high and low extremes of the capsid surface topography (i.e. the tips of the pentavalent capsomeres to the surface connecting the base of neighboring capsomeres). The measured diameter of the markedly non-spherical CPMV is another example of a virus whose mean value lies somewhere between the minimum and maximum radial limits of the surface.
Clearly, the choice of standard and methods of measurement are critical factors which may introduce systematic errors into the calibration measurements. Nevertheless, the techniques presented here provide strong support for the notion that values obtained from frozen-hydrated specimens are more consistent with values obtained from X-ray diffraction compared to those obtained by conventional microscopy techniques.
4.4. Advantages of cryo-microscopy over other preparative techniques
Quick freezing of biological samples in a thin layer of vitrified water circumvents several artifacts encountered with negative staining and other commonly used preparative techniques. Dehydration-induced distortion (e.g. particle flattening) is one of the major problems associated with electron microscopy of biological specimens. This phenomenon significantly affects the reliability of diameter measurements made from viruses prepared with conventional techniques. Earnshaw et al. [59] compared the diameter values of a number of viruses prepared by a variety of electron microscopy techniques with corresponding values obtained from X-ray diffraction. Values obtained from positively stained or thin-sectioned virions varied as much as 31% from the X-ray-derived values. Measurements of negatively stained virions showed the closest agreement with the X-ray measurements but still deviated from the latter values by as much as 10% in some instances. The diameter of negatively stained polyoma, 45.3 nm [8], although smaller than the correct value, is still significantly larger than that determined from particles in thin-sectioned material (38 nm [11]). An extreme example of preparation-induced artifact was obtained for chromium-shadowed polyoma virus where particles with elliptical outlines of dimensions 24 × 59 nm were observed [10].
NβV diameter measurements vary over a wide range (30–50 nm), presumably due to artifacts induced by preparation conditions which cannot be carefully controlled. For example, negatively stained particles imaged in a single micrograph were quite small (30 nm) where the particles were embedded in stain suspended over holes in the carbon substrate and quite large (50 nm) in areas where the particles were in contact with the support film [17]. These differences are likely to reflect the presence of differential compression forces caused by contraction of stain in the holes [26] and flattening caused by surface tension forces present during drying of particles attached to the carbon substrate. In addition, the masking of surface features by dense layers of stain could also contribute to wide variation in diameter measurements of negatively stained particles. The diameter measurements of Juckes [16] seem somewhat surprising since his negative stain value (39.4 nm) is consistent with the X-ray solution scattering value (39.4 nm [18]) whereas hydrodynamic measurements [16] indicated a smaller diameter (35.7 nm).
The average diameter of CPMV determined by X-ray solution scattering is 28.4 nm [20,21]. The outer dimensions of CPMV vary significantly since the surface morphology is markedly non-spherical. The smallest dimension, between opposite surfaces along icosahedral 2-fold axes, is 26.0 nm. The distance between opposed 5-fold vertices (30.8 nm) [21] represents the maximum diameter of CPMV. Measurements from a three-dimensional reconstruction of negatively stained CPMV particles indicated a spherical average and maximum diameter of 20.0 and 24.0 nm respectively [19]. The mean diameter of frozen-hydrated virions (27.5 nm) is in excellent agreement with the X-ray solution scattering value (table 1).
In almost all the examples reported in table 1, with the exception of some NβV and BMV values, the reported diameters of negatively stained virions were significantly smaller than the corresponding values determined by X-ray diffraction or cryo-microscopy. These differences may reflect a general tendency for stains to mask surface features or compress particles when they are suspended over holes in the carbon substrate.
5. Conclusion
Our results add further support to a growing body of evidence which shows that the preparation of vitreous, hydrated biological samples helps preserve structural dimensions and features in a more faithful manner than other classical electron microscopy techniques. The use of biological specimens such as TMV (helical), polyoma (spherical) and catalase (crystalline sheet), coupled with non-distorting, frozen-hydrated preparation techniques, provides a convenient and accurate means to calibrate measurements of unknown, similarly-shaped particles.
Acknowledgments
We gratefully acknowledge the following individuals for providing purified virus samples: W. Murakami (Brandeis University) for polyoma; M. Bina (Purdue University) for SV40; D. Hendry (University of South Africa) for NβV; J. Johnson (Purdue University) for CPMV, BMV and FHV; and M. Cahoon (Brandeis University) for TMV. We thank E. Jacqueline Drak for assistance in developing the technique described in appendix A. and B. Fuller, H. Hinkel and M. McDonough for technical assistance. Insightful comments from reviewers J. Dubochet and S. Fuller are also gratefully acknowledged. Research supported by National Institutes of Health grant GM33050 (to T.S.B.).
Appendix A
Initial location of spherical particle centers using a rotational cross-correlation procedure
A necessary requirement for the successful determination of the three-dimensional structure of spherical viruses is that several unique views (images) of the virus particle be combined in such a way that the relative view orientations are properly maintained and a common phase origin (particle center) is identified [60]. Determination of the view orientation for particles with icosahedral symmetry relies on accurate identification of the particle center, since this is the point of intersection of the 2-, 3-, and 5-fold axes of icosahedral symmetry. To reconstruct the three-dimensional structure of SV40 [40,61], a cross-correlation procedure was devised to aid in initially locating the center of each particle.
A projected image of the top half of a computer-generated, T = 3 icosahedral model, viewed along a five-fold axis (fig. 3A), illustrates the procedure. The model image was generated so the center of the boxed area (“ + ” symbol) and the center of the particle did not coincide. A second image (fig. 3B) was produced by rotating the first image (fig. 3A) 180° about an axis perpendicular to and coincident with the center of the box. The cross-correlation function [62] was computed between the original and rotated images (fig. 3C). The position of the highest peak in the correlation pattern (base of arrow) relative to the origin of the pattern (“ + ”) identifies the vector difference between the centers of the particles in the original and rotated images. The center of the unrotated particle relative to the center of the box it lies within (fig. 3A) is one-half of this vector distance, measured from the box center in the same direction as the arrow in fig. 3C. The position of the correlation peak (fig. 3C) is accurately identified using interactive graphics software as depicted in fig. 4.
Fig. 3.
Cross-correlation method for initial location of the center of images of “spherical” particles. (A) Incorrectly “boxed” image of a model T = 3 icosahedral particle, viewed down a five-fold axis. The center of the model is slightly below and to the left of the box center (“ + ”). (B) Same as (A) after 180° rotation. (C) Cross-correlation pattern between (A) and (B). The vector difference between the highest peak in the cross-correlation pattern (base of arrow) and the origin of the pattern (“ + ”), identifies the direction and magnitude of the difference between the centers of the unrotated (A) and rotated (B) model images. The center of the particle in (A) is one-half the vector distance identified in (C) from the center of the “boxed” area in (A).
Fig. 4.
Pattern produced by cross-correlating a polyoma virion image with the same image rotated by 180°. The inset at the upper right is a magnified contour display of the center of the pattern. This facilitates the location of the peak position in the cross-correlation pattern using an interactive graphics cursor (“×”).
As noted by Frank [62], the procedure described above locates the centrosymmetric origin (2-fold axis) of the boxed image. Thus, one possible pitfall of this technique is that the particle center and “best” two-fold axis may not coincide, as might be true for some noisy, low-irradiation micrographs of unstained biological specimens. To reduce the possibility of mis-identifying the true particle center, the cross-correlation procedure is normally performed on images prefiltered to remove high resolution features (usually finer 2.5 nm resolution). Despite these precautions, the location of a few particle centers still remain equivocal. For example, multiple peaks sometimes appear in the cross-correlation pattern which make it difficult to objectively choose the “correct” center.
Another method (not illustrated), which overcomes some of the artifacts possible in the above procedure, is to circularly average an average image using the technique described in appendix B and then compute a cross-correlation function between each individual image and the circular average. The peak in each of these correlation patterns then identifies the vector difference between the image pairs and fixes the center of each individual particle. The advantage of this procedure is that the circular average has a well-defined center so the corresponding correlation patterns have clearer and sharper peaks compared to patterns calculated according to the first method described.
Appendix B
A cross-correlation procedure for selection and averaging of spherical virus images
An alternative approach, independently developed but following similar strategies to those described by Frank and Wagenknecht [63], is sometimes useful for measuring particle diameters, especially in samples with mixtures of virions of distinguishable size or shape or when the distribution of virions in the frozen-hydrated sample is unfavorable (e.g. highly concentrated). In this method, selection and averaging of individual particle images are automated with cross-correlation procedures [64,65].
A single particle image representing each type of virion appearing in the micrograph is selected and boxed from the scanned micrograph (figs. 5A and 5B) *. Each reference image is cross-correlated with the entire scanned area (main image) and peaks in the resultant pattern (figs. 5E and 5H) identify the locations of regions in the main image most similar to the reference. The peaks, sorted according to their magnitude, rank-order the corresponding set of individual particle images: high values correspond to particles most similar to the reference and low values correspond to particles most dissimilar to the reference. Those particles which correlate most strongly with the reference are averaged by summing the individual particle images located in the original micrograph (figs. 5A and 5B) at positions identified by the peaks in the correlation pattern (e.g., figs. 5E and 5H). This average is used as a reference image in a second cycle of cross-correlation with the main image (figs. 5F and 5I). Any bias introduced by the particular choice of the initial reference used in the first cycle is reduced during this refinement cycle. The diameter of the averaged particle image (insets to figs. 5F and 5I) is measured as described in the text, (section 2) for all virus types.
* In mixed samples where the different virions can be distinguished by size, the different reference particles (fig. 5B, circled virions) are boxed within circular boundaries of identical size to help minimize the generation of “false” correlation peaks. That is, the surrounding background density (vitrified water) in the boxed image of the smaller of the two reference particles serves as an additional constraint in guiding the correlation procedure to discriminate between different size particles (i.e those which do versus those which do not fill the entire circular boundary).
The two major disadvantages to this procedure are (i) it is very difficult to distinguish different particles that are similar in size and morphology (e.g. polyoma and SV40) and (ii) the procedure is extremely sensitive to long-range (low resolution) variations such as differences in the thickness of vitrified water across a hole in the carbon substrate (figs. 5A, 5D and 5G). The second problem is minimized by prefiltering (in Fourier space) the entire scanned image to remove low-frequency fluctuations (generally coarser than 100 nm resolution) and compute the cross-correlation averages with a main image with a uniform background (figs. 5B, 5E and 5H). This procedure does not completely eliminate variations arising, for example, from approximately linear changes in sample thickness across the field of view. Linear gradients can be removed by established procedures [67] after which subsequent processing can be applied.
Footnotes
Abbreviations used: simian virus 40, SV40; Nudaurelia capensis beta virus, NβV; cowpea mosaic virus, CPMV, bromegrass mosaic virus, BMV; Flock House virus, FHV; tobacco mosaic virus, TMV.
References
- 1.Backus RC, Williams RC. J. Appl. Phys. 1949;20:224. [Google Scholar]
- 2.Sjostrand FS. Electron Microscopy of Cells and Tissues. Vol. 1. New York: Academic Press; 1967. p. 365. [Google Scholar]
- 3.Cermola M, Schreil W-H. J. Electron Microsc. Tech. 1987;5:171. [Google Scholar]
- 4.Luftig R. J. Ultrastruct. Res. 1967;20:91. doi: 10.1016/s0022-5320(67)80038-8. [DOI] [PubMed] [Google Scholar]
- 5.Wrigley NG. J. Ultrastruct. Res. 1968;24:454. doi: 10.1016/s0022-5320(68)80048-6. [DOI] [PubMed] [Google Scholar]
- 6.Unwin PNT. J. Mol. Biol. 1975;98:235. doi: 10.1016/s0022-2836(75)80111-2. [DOI] [PubMed] [Google Scholar]
- 7.Berriman J, Leonard KR. Ultramicroscopy. 1986;19:349. doi: 10.1016/0304-3991(86)90095-1. [DOI] [PubMed] [Google Scholar]
- 8.Wildy P, Stoker MGP, Macpherson IA, Home RW. Virology. 1960;11:444. [Google Scholar]
- 9.Rayment I, Baker TS, Caspar DLD, Murakami WT. Nature. Vol. 295. London: 1982. p. 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahler H, Rowe WP, Lloyd BJ, Hartley JW. J. Natl. Cancer Inst. 1959;22:647. [PubMed] [Google Scholar]
- 11.Howatson AF, Almeida JD. J. Biophys. Biochem. Cytol. 1960;8:828. doi: 10.1083/jcb.8.3.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayor HD, Jamison RM, Jordan LE. Virology. 1963;19:359. doi: 10.1016/0042-6822(63)90075-8. [DOI] [PubMed] [Google Scholar]
- 13.Lattman EE. Science. 1980;208:1048. doi: 10.1126/science.6246579. [DOI] [PubMed] [Google Scholar]
- 14.Koch MA, Eggers HJ, Anderer FA, Schlumberger HD, Frank H. Virology. 1967;32:503. doi: 10.1016/0042-6822(67)90302-9. [DOI] [PubMed] [Google Scholar]
- 15.Hendry DA, Bekker MF, van Regenmortel MHV. South African Med. J. 1968;42:117. [PubMed] [Google Scholar]
- 16.Juckes IRM. J. Gen. Virol. 1979;42:89. [Google Scholar]
- 17.Finch JT, Crowther RA, Hendry DA, Struthers JK. J. Gen. Virol. 1974;24:191. doi: 10.1099/0022-1317-24-1-191. [DOI] [PubMed] [Google Scholar]
- 18.Johnson JE. personal communication. [Google Scholar]
- 19.Crowther RA, Geelen JLMC, Mellema JE. Virology. 1974;57:20. doi: 10.1016/0042-6822(74)90104-4. [DOI] [PubMed] [Google Scholar]
- 20.White JM, Johnson JE. Virology. 1980;101:319. doi: 10.1016/0042-6822(80)90513-9. [DOI] [PubMed] [Google Scholar]
- 21.Stauffacher CV, Usha R, Harrington M, Schmidt T, Hosur MV, Johnson JE. In: Crystallography in Molecular Biology. Moras D, Drenth J, Strand-berg B, Suck D, Wilson K, editors. New York: Plenum; 1987. p. 293. [Google Scholar]
- 22.Anderegg JW, Wright M, Kaesberg P. Biophys. J. 1963;3:175. doi: 10.1016/s0006-3495(63)86813-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaesberg P. In: Roberts RB, editor. Cambridge MA. Proc. 1st Natl. Biophysics Conf; New Haven, CT: Yale University Press; 1957. p. 244. 1959. [Google Scholar]
- 24.Dealing SC, Scotti PD, Wigley PJ, Dhana SD. New Zealand J. Zool. 1980;7:267. [Google Scholar]
- 25.Kellenberger E, Häner M, Wurtz M. Ultramicroscopy. 1982;9:139. doi: 10.1016/0304-3991(82)90236-4. [DOI] [PubMed] [Google Scholar]
- 26.Unwin PNT. J. Mol. Biol. 1974;87:657. doi: 10.1016/0022-2836(74)90076-x. [DOI] [PubMed] [Google Scholar]
- 27.Baker TS. In: Sturgess JM, editor. Toronto. Proc. 9th Intern. Congr. on Electron Microscopy; Toronto: Microscopical Society of Canada; 1978. p. 2. 1978. [Google Scholar]
- 28.Chiu W. Ann. Rev. Biophys. Chem. 1986;15:237. doi: 10.1146/annurev.bb.15.060186.001321. [DOI] [PubMed] [Google Scholar]
- 29.Dubochet J, Adrian M, Chang J-J, Homo J-C, Lepault J, McDowall AW, Schultz P. Quart. Rev. Biophys. 1988;21:129. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 30.Stewart M, Vigers G. Nature. Vol. 319. London: 1986. p. 631. [DOI] [PubMed] [Google Scholar]
- 31.Adrian M, Dubochet J, Lepault J, McDowall AW. Nature. Vol. 308. London: 1984. p. 32. [DOI] [PubMed] [Google Scholar]
- 32.Milligan RA, Brisson A, Unwin PNT. Ultramicroscopy. 1984;13:1. doi: 10.1016/0304-3991(84)90051-2. [DOI] [PubMed] [Google Scholar]
- 33.Henderson R, Baldwin JM, Downing KH, Lepault J, Zemlin F. Ultramicroscopy. 1986;19:147. [Google Scholar]
- 34.Jaffe JS, Glaeser RM. Ultramicroscopy. 1987;23:17. doi: 10.1016/0304-3991(87)90223-3. [DOI] [PubMed] [Google Scholar]
- 35.Taylor KA, Glaeser RM. J. Ultrastruct. Res. 1976;55:448. doi: 10.1016/s0022-5320(76)80099-8. [DOI] [PubMed] [Google Scholar]
- 36.Jeng TW, Chiu W. In: Bailey GW, editor. Phoenix, AZ. Proc. 41st Annual EMSA Meeting; San Francisco, CA: San Francisco Press; 1983. p. 430. 1983. [Google Scholar]
- 37.Deng LL. In: Bailey GW, editor. Baltimore, MD. Proc. 45th Annual EMSA Meeting; San Francisco, CA: San Francisco Press; 1987. p. 646. 1987. [Google Scholar]
- 38.Grant RA, Degn LL, Chiu W, Robinson J. In: Bailey GW, editor. Phoenix, AZ. Proc. 41st Annual EMSA Meeting; San Francisco, CA: San Francisco Press; 1983. p. 730. 1983. [Google Scholar]
- 39.Grant RA, Schmid MF, Chiu W, Deatherage JF, Hosoda J. Biophys. J. 1986;49:251. doi: 10.1016/S0006-3495(86)83638-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baker TS, Drak J, Bina M. Proc. Natl. Acad. Sci. USA. 1988;85:422. doi: 10.1073/pnas.85.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeRosier DJ, Moore PB. J. Mol. Biol. 1970;52:355. doi: 10.1016/0022-2836(70)90036-7. [DOI] [PubMed] [Google Scholar]
- 42.Holmes KC. In: Jurnak FA, McPherson A, editors. Virus Structures; Biological Macromolecules and Assemblies; New York: Wiley; 1984. p. 121. [Google Scholar]
- 43.Dubochet J, Lepault J, Freeman R, Berriman JA, Homo J-C. J. Microscopy. 1982;128:219. [Google Scholar]
- 44.Caspar DLD. personal communication. [Google Scholar]
- 45.Baschong W, Aebi U, Baschong-Prescianotto C, Dubochet J, Landmann L, Kellenberger E, Wurtz M. J. Ultrastruct. Mol. Struct. Res. 1988;99:189. doi: 10.1016/0889-1605(88)90063-8. [DOI] [PubMed] [Google Scholar]
- 46.Vogel RH, Provencher SW, von Bonsdorff M, Adrian C-H, Dubochet J. Nature. Vol. 320. London: 1986. p. 533. [DOI] [PubMed] [Google Scholar]
- 47.Fuller SD. Cell. 1987;48:923. doi: 10.1016/0092-8674(87)90701-x. [DOI] [PubMed] [Google Scholar]
- 48.Prasad BVV, Wang GJ, Clerx JPM, Chiu W. J. Mol. Biol. 1988;199:269. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- 49.Olson NH, Baker TS, Bomu W, Johnson JE, Hendry DA. In: Bailey GW, editor. Baltimore, MD. Proc. 45th Annual EMSA Meeting; San Francisco, CA: San Francisco Press; 1987. p. 650. 1987. [Google Scholar]
- 50.Olson NH, Baker TS, Bomu W, Johnson JE. manuscript in preparation. [Google Scholar]
- 51.Talmon Y, Prasad BVV, Clerx JPM, Wang G-J, Chiu W, Hewlett MJ. J. Virol. 1987;61:2319. doi: 10.1128/jvi.61.7.2319-2321.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Booy FP, Ruigrok RWH, van Bruggen EFJ. J. Mol. Biol. 1985;184:667. doi: 10.1016/0022-2836(85)90312-2. [DOI] [PubMed] [Google Scholar]
- 53.Taylor KA, Glaeser RM. Science. 1974;186:1036. doi: 10.1126/science.186.4168.1036. [DOI] [PubMed] [Google Scholar]
- 54.Mandelkow E-M, Rapp R, Mandelkow E. J. Microscopy. 1986;141:361. doi: 10.1111/j.1365-2818.1986.tb02729.x. [DOI] [PubMed] [Google Scholar]
- 55.Trinick J, Cooper J, Seymour J, Egelman EH. J. Microscopy. 1986;141:349. doi: 10.1111/j.1365-2818.1986.tb02728.x. [DOI] [PubMed] [Google Scholar]
- 56.Unwin PNT, Ennis PD. Nature. Vol. 307. London: 1984. p. 609. [DOI] [PubMed] [Google Scholar]
- 57.Bernal JD, Fankuchen I. J. Gen. Physiol. 1941;25:111. doi: 10.1085/jgp.25.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watson JD. Biochim. Biophys. Acta. 1954;13:10. doi: 10.1016/0006-3002(54)90265-6. [DOI] [PubMed] [Google Scholar]
- 59.Earnshaw WC, King J, Eiserling FA. J. Mol. Biol. 1978;122:247. doi: 10.1016/0022-2836(78)90040-2. [DOI] [PubMed] [Google Scholar]
- 60.Crowther RA. Phil. Trans. Roy. Soc. B261. London: 1971. p. 221. [DOI] [PubMed] [Google Scholar]
- 61.Baker TS, Drak J, Bina M. Biophys. J. 1989;55:243. doi: 10.1016/S0006-3495(89)82799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frank J. Topics Current Phys. 1980;13:187. [Google Scholar]
- 63.Frank J, Wagenknecht T. Ultramicroscopy. 1984;12:169. [Google Scholar]
- 64.Saxton WO. In: Electron Microscopy in Molecular Dimensions. Baumeister W, Vogell W, editors. Berlin: Springer; 1980. p. 245. [Google Scholar]
- 65.Frank J, Goldfarb W. In: Electron Microscopy in Molecular Dimensions. Baumeister W, Vogell W, editors. Berlin: Springer; 1980. p. 261. [Google Scholar]
- 66.Baker TS. Image processing of biological specimens: A bibliography. In: Griffith JD, editor. Electron Microscopy in Biology. Vol. 1. New York: Wiley; 1981. p. 189. [Google Scholar]
- 67.Aebi U. PhD Thesis. University of Basel; 1977. [Google Scholar]