Abstract
B cells, αβ T cells and γδ T cells are conserved lymphocyte subtypes encoding their antigen receptors from somatically rearranged genes. αβ T cells undergo positive selection in the thymus by engagement of their T cell receptors (TCRs) with self-peptides presented by major histocompatibility complex molecules1. The molecules that select γδ T cells are unknown2–4. Vγ5+Vδ1+ cells comprise 90% of mouse epidermal cd T cells4. By mapping and genetic complementation using a strain showing loss of Vγ5+Vδ1+ cells due to a failure of thymic selection, we show that this defect is caused by mutation in Skint1, a newly identified gene expressed in thymus and skin that encodes a protein with immunoglobulin-like and transmembrane domains. Skint1 is the prototypic member of a rapidly evolving family of at least 11 genes in mouse, with greatest similarity to the butyrophilin genes. These findings define a new family of proteins mediating key epithelial-immune interactions.
In recent years, T cells expressing γδ TCRs have emerged as an important component of the immune repertoire. γδ T cells are predominant in various murine epithelia, including those of the skin, intestine, lung and reproductive tract2,3. These epithelia constitute the primary barrier to diverse environmental insults. In mouse, epithelial compartments contain γδ T cells with specific TCR V gene segments2,3. For example, in epidermis, 95% of T cells are γδ+, and of these, 90% express the Vγ5Vδ1 TCR4. These γδ T cells are generated by positive selection in the fetal thymus, after which they migrate to the skin1–4. Such stereotypical TCRs are proposed to respond to common antigens and provide signals of infection or other physiologic perturbation2–4. The role of γδ T cells is demonstrated by mice genetically engineered to lack all such cells. These mice are susceptible to bacterial, protozoal and viral infection and morbidity5–11, cutaneous carcinogenesis12 and autoimmune and allergic inflammation13–15, and they show defects in wound repair16 and development of immune memory17.
In contrast to the well-defined mechanisms of positive selection in αβ T cell development, the mechanisms selecting any γδ T cell repertoire are unknown. Although it has been hypothesized that ligands might be expressed in fetal thymic and target organ epithelia, serving in both positive selection and tissue localization and maintenance, no such molecules have been identified2–4.
Recently, the FVB/N Mus musculus mouse strain from Taconic Laboratories (FVBTac) has been found to have a selective deficiency for epidermal Vγ5+Vδ1+ T cells; this defect is not observed in other strains, including the FVB/N strain from Jackson Laboratories (FVBJax). This deficiency is attributable to loss of thymic positive selection of Vγ5+Vδ1+ T cells and is complemented in culture by wild-type thymic stromal cells4. The trait demonstrated autosomal recessive transmission in an FVBTac x FVBJax cross, suggesting use of positional cloning to identify this gene and gain insight into γδ T cell development.
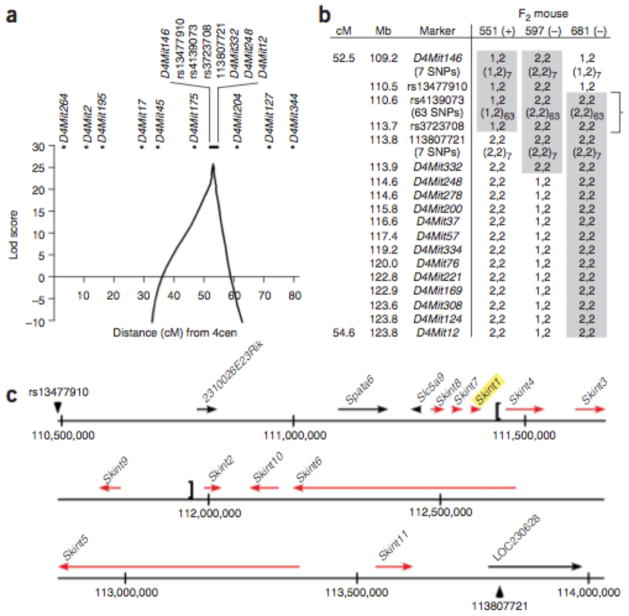
In an F2 cross between FVBTac and C57BL/6J (B6), proportions of epidermal Vγ5+Vδ1+ T cells fell cleanly into high and low modes in a proportion closely approximating 3:1 (74:28), supporting simple autosomal recessive transmission with complete penetrance (Fig. 1). We genotyped 143 informative genetic markers distributed across the genome in the F2 mice. Multipoint analysis yielded a maximum lod score of 25.3 (odds in favor of linkage >1025:1) to a 2-cM segment of chromosome 4 bounded by D4Mit146 and D4Mit12 (Fig. 2a). Lod scores were strongly negative (below −2) across all other chromosomes.
Figure 1.
Recessive transmission of Vγ5+Vδ1+ T cell deficiency in a B6 x FVBTac F2 cross. Epidermal cells from mice were stained with antibodies to TCRγδ and the Vγ5Vδ1 idiotype and analyzed by flow cytometry. (a) Representative results from F2 mice harboring high or low proportions of Vγ5+Vδ1+ T cells. Numbers in upper quadrants indicate the percentage of total epidermal cells within the quadrant; numbers in parentheses indicate percentage of γδ+ T cells that are Vγ5+Vδ1+. (b) Percentages of epidermal γδ T cells that are Vγ5+Vδ1+ in individual mice of the indicated groups. Brackets indicate values considered high and low for analysis of linkage
Figure 2.
Linkage of Vγ5 Vδ1 T cell deficiency to chromosome 4. (a) Multipoint lod scores for linkage of Vγ5+Vδ1+ T cell deficiency across chromosome 4. 4cen, centromere of chromosome 4. The maximum lod score is 26.1, with a lod −3 interval of 3.3 Mb. (b) Haplotypes of mice with recombination in the interval between D4Mit146 and D4Mit12. The genetic (cM) and physical (Mb) map positions of markers are indicated. Mouse 551 had high (+) whereas mice 597 and 681 had low (−) Vg5+Vd1+ T cell levels. The B6 and FVBTac alleles are denoted 1 and 2, respectively. The genotypes showing co- segregation with the Vγ5+Vδ1+ level of each mouse are shown in gray boxes, and the minimum linked interval is indicated by a bracket. (c) Genes within the chromosome 4 linkage interval in B6. The 3.3-Mb interval between locus rs13477910 and nucleotide 113807721 is shown as a solid line, and the transcriptional orientations of genes are indicated by arrows. Skint gene depictions (red) are based on structural characterization described herein; prior aliases for predicted genes are shown in Supplementary Figure 1. The region absent in some strains is indicated with brackets.
Three mice showed recombination of the trait locus with either D4Mit146 or D4Mit12. Genotyping of 93 additional polymorphisms in the interval between them identified 65 consecutive SNPs showing complete linkage to the trait (lod score 26.1), localizing the gene responsible for Vγ5+Vδ1+ T cell deficiency to a lod −3 interval of 3.3 Mb, delimited proximally by rs13477910 and distally by a newly identified C-to-T transition at bp 113,807,721 (Fig. 2a, b).
Genotyping of 136 microsatellite markers in FVBJax and FVBTac confirmed that these two strains are completely inbred and isogenic, indicating that the Vγ5+Vδ1+ T cell deficiency mutation in FVBTac arose de novo in the ~20 years since these strains last shared a common ancestor. We consequently sequenced known and putative exons in the lod −3 interval in these strains, expecting to find a single sequence change representing the causative mutation.
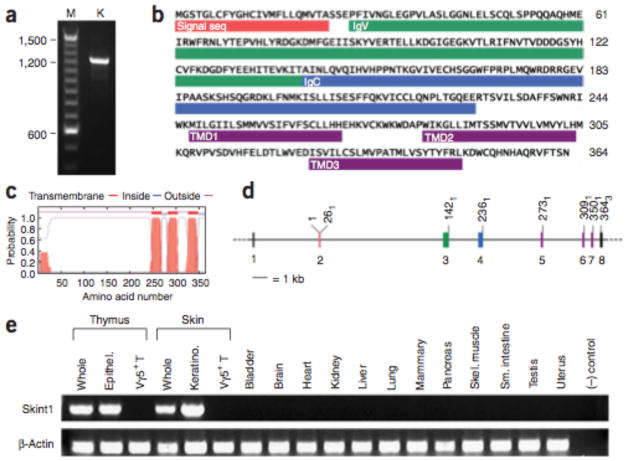
Evaluation of the linked interval identified 15 known or predicted genes (Fig. 2c and Supplementary Fig. 1 online). Sequencing of their exons and splice sites, encompassing ~30,000 base pairs, revealed a single substitution between FVBJax and FVBTac. This mutation occurred in LOC384040, a partially predicted gene that we have now fully characterized and named Skint1 (for selection and upkeep of intraepithelial T cells 1) (Fig. 3a and Supplementary Fig. 2 online). Skint1 encodes a protein of 364 amino acids predicted to include a signal sequence, sequential immunoglobulin-like domains of the IgV and IgC types, three transmembrane domains (TMDs) and a short cytoplasmic C terminus (Fig. 3b, c and Supplementary Fig. 3 online). The eight exons of Skint1 are modular, with seven protein-coding exons each encoding a distinct domain (Fig. 3b, d). Reverse transcription PCR (RT-PCR) of RNA from diverse tissues (Fig. 3e and Supplementary Fig. 4 online) revealed robust expression of Skint1 only in thymus and skin—specifically, in thymic epithelial cells and keratinocytes—but not in Vγ5+Vδ1+ T cells. Skint1 is expressed at embryonic day 15 (E15) and continuing into adulthood.
Figure 3.
Skint1 gene and protein structure. (a) Amplification of the full-length coding region of Skint1 from keratinocyte cDNA. The DNA sequence reveals a consensus Kozak sequence for translation initiation and a continuous open reading frame of 364 amino acids (Supplementary Fig. 2). (b) Deduced amino acid sequence of Skint1. The locations of the signal sequence, IgV, IgC and transmembrane domains (TMDs) are indicated. (c) Hydropathy plot from the TMHMM program predicts three TMDs with high probability, yielding an extracellular N terminus and cytoplasmic C terminus. (d) Alignment of cDNA with genomic DNA reveals that Skint1 is encoded in modular exons. Exon colors correspond to those of the encoded domains shown in b. The codon position of the last base of each exon is indicated. (e) Tissue distribution of Skint1 expression. Products of RT-PCR with Skint1-specific primers visualized on an agarose ge (Skint1); Actb amplification is shown as a control (b-Actin). Thymic epithelium and Vγ5+ T cell RNAs were obtained at E15–16; RNAs from all other tissues were obtained at adulthood. Skint1 expression is detected in whole thymus and thymic epithelium and in whole skin and keratinocyte.
The mutation that distinguishes FVBTac from FVBJax changes codon 324 of Skint1 from GAA (glutamate) to TAA (stop) (Fig. 4a), truncating the encoded protein before the putative third TMD (Fig. 4b). The Glu324Stop mutation is absent in B6 and 21 other strains, and it precisely co-segregates with Vγ5+Vδ1+ T cell deficiency in the F2 cross as well as in the previously described4 isogenic FVBJax x FVBTac backcross (lod score 29.5). Because the de novo mutation rate in mouse is estimated to be ~4.5 ×10−9 substitutions per site per year18, we would anticipate finding such a substitution between FVBJax and FVBTac in <0.5% of random 30-kb segments.
Figure 4.
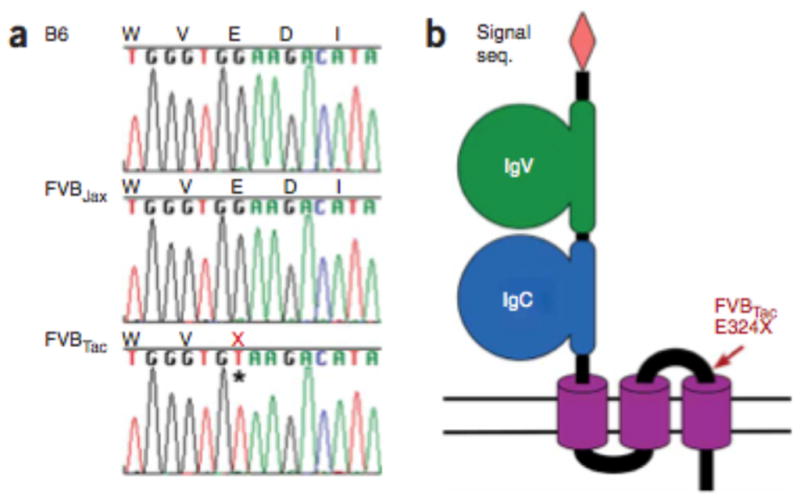
Premature termination codon in Skint1 in FVBTac. (a) Genomic DNA sequence of Skint1 codons 322–326 from B6, FVBJax and FVBTac mice reveals a premature termination mutation in FVBTac (asterisk). (b) Schematic representation of the inferred protein structure of wild-type Skint1. Location of the FVBTac E324X mutation is indicated by ‘X’ and an arrow. The structure is inferred from domain predictions for Skint1 and its paralogs.
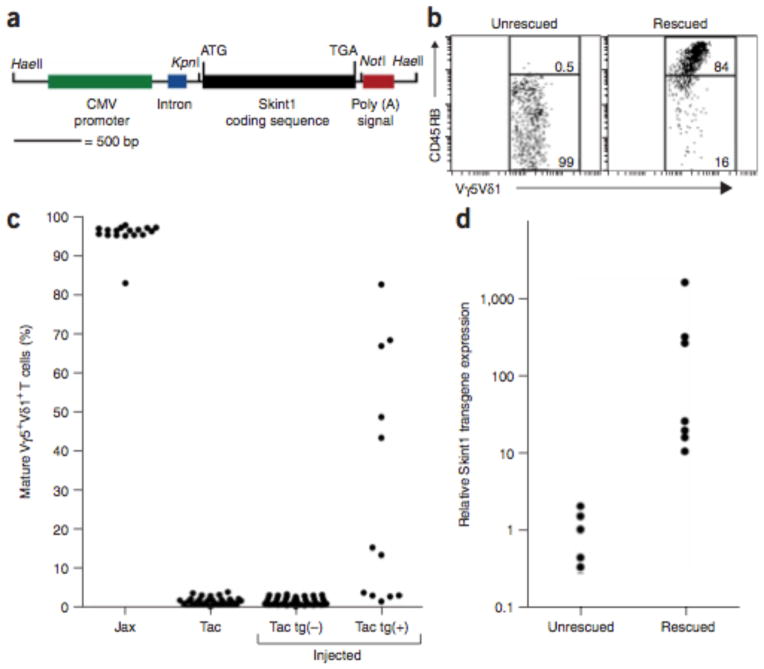
Vγ5+Vδ1+ deficiency in FVBTac is detectable at E17 as deficiency of mature Vγ5+Vδ1+ T cells in the thymus (defined as Vγ5+Vδ1+ T cells expressing high levels of CD45RB)4. We attempted genetic rescue of this defect by injection of single-cell FVBTac embryos with a construct containing wild-type FVBJax Skint1 (Fig. 5a); injected embryos were implanted into foster mothers and killed at E17. Twelve of 52 injected embryos harbored the transgene. Of these, 7 showed phenotypic rescue, with 13–83% mature Vγ5+Vδ1+ cells (Fig. 5b, c); these levels are a minimum of 10 s.d. removed from the mean of the uninjected population (mean 1.5% ± 0.9% mature cells, range 0.0–3.6%). None of the 40 transgene-negative embryos showed more than 3.1% mature cells (mean 1.3% ± 0.9%; P = 6 ×10−6 for association of rescue with presence of transgene). Transgenic embryos with phenotypic rescue had median Skint1 expression that was 25-fold higher than that of embryos that were not rescued (P = 0.0025; Fig. 5d). The finding of a truncating mutation in Skint1 that distinguishes isogenic strains and is completely linked to Vγ5+Vδ1+ T cell deficiency, along with phenotypic rescue by wild-type Skint1, constitutes formal proof that mutation of Skint1 is the cause of Vγ5+Vδ1+ T cell deficiency in FVBTac mice.
Figure 5.
Rescue of Vγ5+Vδ1+ T cell deficiency by a wild-type Skint1 transgene. (a) Structure of the linearized Skint1 transgene. (b) E17 thymocytes prepared from individual transgene-injected mice were stained with antibodies to TCRγδ, the Vγ5Vδ1 idiotype and CD45RB and were analyzed by flow cytometry. Representative results from a transgene-negative embryo (left) and a transgenepositive, rescued embryo (right) are shown. In the unrescued embryo, 0.5% of Vγ5+Vδ1+ T cells are mature (that is, express high levels of CD45RB); in the rescued embryo, 84% of Vγ5+Vδ1+ T cells are mature. (c) The percentage of mature Vγ5+Vδ1+ T cells from mice of indicated groups. ‘Jax’ and ‘Tac’ represent the FVBJax and FVBTac lines; ‘tg(−)’ and ‘tg(+)’ represent transgene-negative and transgene-positive injected embryos. Phenotypic rescue is seen only among transgene positive embryos. (d) Expression of the Skint1 transgene in thymus of unrescued and rescued transgene-positive embryos shown in log scale. Rescued mice have high Skint1 transgene expression.
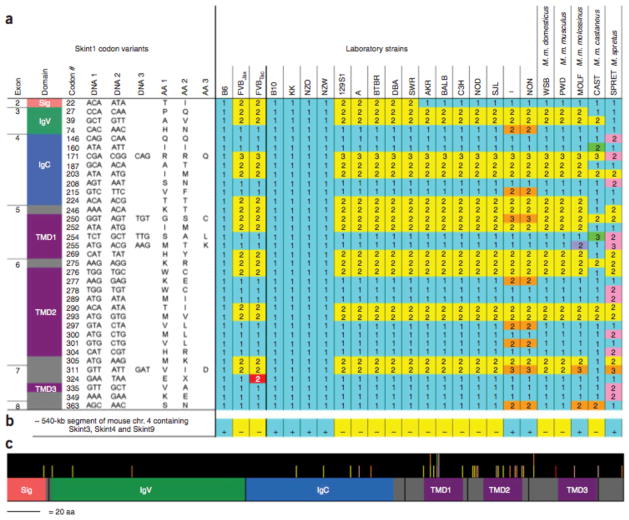
There is a markedly high prevalence of missense versus synonymous substitutions in Skint1 among strains, a hallmark of positive genetic selection (Fig. 6). For example, there are 16 missense versus 2 synonymous substitutions between B6 and FVB, and the Ka/Ks value (the adjusted ratio of missense to synonymous substitutions) of 3.5 is 32-fold higher than the genome-wide Ka/Ks of 0.11 for mouse versus rat19. Among 16 other laboratory strains, there are 4 diverse Skint1 haplotypes, each fixed multiple times in distantly related strains, consistent with all having existed among their common ancestors. In 5 additional strains derived from wild mice, there is further variation; most notably, Mus spretus shows 21 missense substitutions versus 2 synonymous substitutions compared with B6. These substitutions are distributed throughout the protein but are concentrated within the first two putative TMDs and the intervening loop.
Figure 6.
Skint1 haplotypes in diverse mouse strains. (a) Variants at 40 Skint1 codons, resulting in 35 missense substitutions, 4 silent substitutions and 1 premature termination (found only in FVBTac), are listed next to a schematic of the protein structure, with domains colored as in Figure 3, showing the domain location of the substitutions. The sequences present in B6, FVBJax and FVBTac, as well as 16 other laboratory strains and 5 strains derived from wild mice, are indicated with numbers corresponding to the codons listed at the left. Blue shading within the table highlights B6-like sequence (the reference sequence); Skint1 variation in other strains is shaded yellow (FVB-like), orange (found in I and NON but not FVB), lavender (found only in MOLF), green (found only in CAST) and pink (found only in SPRET). The stop mutation unique to FVBTac is shaded red. Four Skint1 haplotypes are observed among the 16 laboratory strains studied. (b) Presence (+) or absence (−) of an ~540 kb segment of mouse chromosome 4 containing Skint3, Skint4 and Skint9. The segment is present in B6 and 8 other distantly related mouse strains, but absent in FVB and 13 other strains. (c) Skint1 protein schematic, drawn to scale, indicating where missense substitutions occur relative to the protein structure; bar color denotes the strains harboring the substitutions, as shown in a. Missense substitutions are concentrated within the first two TMDs and the intervening loop.
Skint1 orthologs in other species were identified by alignments of syntenic segments (Supplementary Figs. 5 and 6 online). Rat and cow have Skint1 orthologs with identical organization and inferred protein topology and similar sequence. In contrast, Skint1 is absent in dog, and has multiple in-frame premature termination codons in both human and chimpanzee. We infer that Skint1 was present in a common mammalian ancestor but has been lost at least once in the mammalian lineage. It is noteworthy that the presence of Skint1 is correlated with presence of a restricted cutaneous T cell population. Rat, like mouse, has a prevalent Vγ5+Vδ1+ T cell population in epidermis20, and cow skin harbors high levels of γδ T cells, with predominant Vγ3 and Vγ7 chains21,22. In contrast, human skin does not possess comparably high levels of γδ T cells, nor do human T cells have a monomorphic TCR23–25.
Mouse Skint1 paralogs were identified and characterized (Supplementary Figs. 3 and 5–7 online). There are ten intact paralogs, all clustered with Skint1 on chromosome 4 (Skint2 – Skint11, Fig. 2c). These paralogs all show expression in thymus and/or skin; several also show expression in other tissues (Supplementary Fig. 8 online). All have predicted motifs and topologies like those of Skint1, including multiple TMDs. Most have multiple isoforms arising from alternative splicing, including isoforms with no predicted TMDs. Skint5 and Skint6 have an additional 49 and 40 exons, respectively, each composed of 60–63 base pairs encoding related amino acid sequences. These paralogs also show rapid evolution. Skint2 shows evidence of positive selection between B6 and FVB (Ka/Ks = 3.6). In addition, an B540 kb segment that contains Skint3, Skint4 and Skint9 (Fig. 2c) is present in B6 but entirely absent in FVB. This segment is present in 8 distantly related strains and absent in 13 others (Fig. 6). Only three Skint1 paralogs in mouse (Skint2, Skint7 and Skint10) are present in the rat syntenic region, and only one (Skint7) in cow. Rat has three additional paralogs not present in mouse, and cow has one; none of these paralogs are present in human, chimp or dog. It is clear that the Skint family is undergoing rapid evolution.
We have identified Skint1 as a previously unknown gene required for positive selection of Vγ5+Vδ1+ T cells in the embryonic thymus and normal levels of these T cells in skin. Skint1 represents the prototype of a family of proteins expressed in thymus and skin; these proteins are unusual among immunoglobulin superfamily members for having multiple transmembrane domains. Numerous proteins with immunoglobulin-like domains play a role in direct cell-cell signaling in the immune system. Our findings suggest that Skint1 may act by engaging a cell surface molecule on immature Vγ5+Vδ1+ T cells in the embryonic thymus. This target could be the Vγ5Vδ1 TCR itself; however, this is not necessarily so. There are many alternative possible mechanisms for Skint1 action; discrimination among these will require further experimentation. The expression of Skint1 in keratinocytes suggests a role in epidermis, possibly in the selective localization and maintenance of Vγ5+Vδ1+ T cells in skin. Indeed, FVBTac mice transgenic for wild-type Skint1 also show rescue of Vγ5+Vδ1+ T cell deficiency in the skin (data not shown). Nonetheless, this result does not establish whether rescue in skin requires epidermal expression of the transgene, or whether expression in thymus is sufficient.
Because other immunoglobulin-like molecules involved in heterotypic immune cell-cell interactions commonly act as heterodimers, it will be pertinent to establish whether Skint1 has a heterodimeric partner; genes (such as Skint7) that show coevolution with Skint1 in species with restricted epidermal γδ T cell populations are obvious candidates. The observation that Skint1 expressed in diverse mammalian cell lines does not reach the cell surface is consistent with the notion that a heterodimeric partner is needed (data not shown).
The Skint family is most similar to butyrophilin (Btn) and the Btn-like (Btnl) proteins, a distinct family of largely unknown function26,27 (Supplementary Figs. 5 and 6). The similarity resides in the immunoglobulin-like domains; Btn proteins differ by possession of only one TMD and the presence of C-terminal PRY/SPRY domains. Like the Skint family in mouse, the BTN family has expanded in the primate lineage, and it is noteworthy that many BTN genes are expressed in thymus, B cells and/or T cells. The recent finding that variants in BTNL2 are associated with sarcoidosis, a disease characterized by exaggerated macrophage and helper T cell activity, motivates further investigation of the Btn family in immune function28.
The rapid evolution and positive selection of Skint family members raises the questions of what has driven these events and whether the marked variation at this locus contributes to trait variation among inbred strains. For example, susceptibility to chemically induced skin tumors, a trait associated with γδ T cell deficiency12, has recently been shown to be linked to a segment of mouse chromosome 4 that includes the Skint cluster29. It will be of particular interest to determine whether Skint proteins are generally involved in the selection and maintenance of γδ T cell populations and/or in other aspects of immune function.
Methods
Mice. C57BL/6J (B6) and FVB/NJ (FVBJax) mice were purchased from Jackson Laboratories and FVB/NTac (FVBTac) mice were purchased from Taconic Farms. Mice were bred and maintained in accordance with approved protocols and procedures of the Yale University Institutional Animal Care and Use Committee.
Intercross and phenotyping. B6 and FVBTac mice were crossed to produce F1 B6/FVBTac mice, which were then intercrossed to produce F2 mice. Epidermal cell suspensions from 15 B6, 24 FVBTac, 14 F1 and 102 F2 mice at 3–4 weeks of age were prepared and stained as previously described4. Vγ5+Vδ1+ T cells were detected by flow cytometry using antibodies to TCRd and Vg5 (PE-GL3 and FITC-536, BD Pharmingen), antibody to Vd1 in conjunction with Vg5 (17D1, previously described)4,30 and Cy5-conjugated goat anti–rat IgM secondary to 17D1 (Jackson ImmunoResearch Laboratories). Data were collected on a FACSCalibur (BD), with electronic gates set on live cells as assessed by light scatter and propidium iodide exclusion, and analyzed with CellQuest software (BD).
Genetic and physical maps. Genetic and physical map positions of all markers and genes are relative to NCBI Mus musculus reference assembly build 35.1, which correspond to those on the University of California Santa Cruz (UCSC) Mouse August 2005 (mm7) assembly18. DNA sequences were aligned to this assembly via BLAT search.
Genotyping and analysis of linkage. Genomic DNA was prepared from mouse livers with a DNeasy kit (Qiagen). F2 mice were genotyped for 143 informative microsatellite markers distributed across the mouse genome by PCR amplification using fluorescently labeled primers, with detection on an ABI 3700 DNA analyzer and genotype calling with Genotyper software (Applied Biosystems); allele calls were verified by visual inspection. Mice with recombination within the initial linkage interval were genotyped for 12 additional informative microsatellite markers as described above and for 81 informative SNPs by PCR amplification and direct DNA sequencing. Analysis of linkage was performed using the LINKAGE program, specifying Vγ5+Vδ1+ T cell deficiency as an autosomal recessive trait with 99.99% penetrance and 0.01% phenocopies.
FVBJax and FVBTac comparative genotyping. To confirm that FVBJax and FVBTac are isogenic strains, 136 microsatellite markers distributed across the mouse genome were genotyped using genomic DNA from each line, as described above. The only marker that was biallelic between the two substrains was D15Mit107. Twenty other microsatellite markers surrounding D15Mit107, including flanking markers 0.0 cM (0.5 Mb) proximal and 1.1 cM (2.2 Mb) distal, were subsequently genotyped and found to be identical between FVBJax and FVBTac.
Analysis of genes in the linkage interval. Known and putative exons in the linkage interval were PCR amplified from B6, FVBJax and FVBTac genomic DNA, purified with a QiaQuick gel extraction kit (Qiagen) and sequenced.
Characterization of full-length Skint1. The G-to-T transversion mutation in FVBTac was found in exon 4 of NCBI record LOC384040 (as annotated in January 2006), a computationally predicted gene. Primers within predicted exons from this record were used for PCR amplification and sequencing of transcripts from thymus and skin cDNAs, revealing two additional exons and excluding LOC384040 exon 1 from the transcript. The 5′ end of the transcript was obtained via 5′ RACE amplification and sequencing from Marathon-Ready mouse E15 embryo cDNA (Clontech), revealing two more previously unannotated exons. A 1,244-bp cDNA containing the complete protein-coding sequence of the gene, now referred to as Skint1, was PCR amplified and sequenced from thymus, thymic epithelial cell, skin and keratinocyte cDNAs of B6, FVBJax and FVBTac mice, using primers in exon 1 (64 bp proximal to the start codon in exon 2) and exon 8 (85 bp distal to the stop codon). Domains in the encoded protein were identified using SignalP, TMHMM, Pfam and CDD. Expression profiles were obtained by RT-PCR using RNA from indicated tissues and cell types and Skint1-specific primers within exons 2 and 4 (633 bp; Fig. 3e) or exons 4 and 8 (749 bp; Supplementary Fig. 4). RNAs used in obtaining the data in Figure 3e were from B6 mice with the exception of thymic epithelium, thymic Vg5+ T cell and keratinocyte RNAs, which were from FVBJax mice. RNAs used in obtaining the data in Supplementary Figure 4 are from B6, FVBJax and FVBTac as indicated.
Creation of FVBTac embryos transgenic for wild-type Skint1. Full-length Skint1 coding sequence was PCR amplified from FVBJax adult skin cDNA, cloned into the pcDNA 3.1D TOPO mammalian expression vector (Invitrogen) and subcloned into the pCI mammalian expression vector (Promega). The construct was verified by DNA sequencing then linearized and trimmed by digestion with HaeII, purified by agarose gel electrophoresis and resuspended in injection buffer (Specialty Media). FVBTac single-cell embryos (~280) were each injected with ~5 fg of purified construct and implanted into seven pseudopregnant foster mice. Fifty-two resulting embryos were killed at E17. The liver and one thymic lobe of each embryo were flash-frozen in liquid nitrogen for RNA and DNA extraction, and the remainder of the thymus was prepared for phenotyping.
Transgenic phenotyping. Thymocyte cell suspensions from 17 FVBJax, 27 FVBTac and 52 transgene-injected FVBTac E17 embryos were prepared and stained as previously described4. Vγ5+Vδ1+ T cells were detected by flow cytometry as described above, and maturity was assessed with antibody to CD45RB (PE-16A, BD Pharmingen).
Transgene integration and expression. Trizol reagent (Invitrogen) was used to extract RNA and DNA from the liver and thymus of the 52 transgene-injected FVBTac embryos, and RNA was DNase-digested and purified with an RNeasy kit (Qiagen). Presence of the transgene was assessed by PCR amplification from DNA using transgene-specific primers. Expression of the transgene was measured via quantitative RT-PCR, with oligo(dT) priming of cDNA followed by PCR with transgene-specific primers; amplification with Actb (which encodes b-actin) primers served as an internal control. Quantification was performed in triplicate for each sample with SYBR Green and detection on an iCycler (Bio-Rad); expression level of the transgene was determined from the standard curve and normalization to Actb. Water blank and transgene-negative DNA samples were included as negative controls.
Variant genotyping in diverse mouse strains. Genomic DNA from 16 additional laboratory strains (129S1/SvImJ, A/J, AKR/J, BALB/cJ, BTBR <T> tf/tf, C3H/HeJ, C57BL/10J, DBA/2J, I/LnJ, KK/H1J, NOD/LtJ, NON/LtJ, NZO/H1LtJ, NZW/LacJ, SJL/J and SWR/J) and 5 strains originating from wild mice (WSB/EiJ, Mus musculus domesticus; PWD/PhJ, M. m. musculus; MOLF/EiJ, M. m. molossinus; CAST/EiJ, M. m. castaneus; and SPRET/Ei, M. spretus) was used to PCR amplify and sequence the coding exons of Skint1 and to assess presence or absence of the large insertion/deletion polymorphism (the B540 kb segment of chromosome 4 containing Skint3, Skint4 and Skint9) via positive or negative PCR amplification of DNA intervals within and outside the boundaries of the segment.
Identification and characterization of Skint1 orthologs and paralogs. Full-length sequences of orthologous genes were obtained with Vista alignments of corresponding genomic intervals. Paralogs were identified via BLAST search using Skint1 protein sequences to query six-frame translations of genomes with the tblastn program and an expect threshold of 0.01. Genes with stop codons in the IgV and/or IgC domains were characterized as pseudogenes (seven in the mouse chromosome 4 cluster, one on chromosome 9). For the ten intact paralogous genes in the mouse chromosome 4 cluster, full-length gene sequences and structures were determined via PCR and RACE amplification and sequencing of transcripts from thymus and/or skin cDNAs. Orthologs and paralogs were queried for domain structure as described above. Genes were aligned with ClustalW. Ka/Ks scores were calculated with the PAL2NAL program. Expression profiles of paralogous genes were obtained via RT-PCR using RNA from tissues of adult B6 mice (extracted, purified and reverse transcribed as described above) and gene-specific primers within the exons encoding the signal sequence and IgC domain of each Skint gene.
URLs. Expression data for BTN genes: UCSC Human Genome Browser, Mar. 2006 assembly, UCSC Genes track, Microarray Expression Data description field (http://genome.ucsc.edu/cgibin/hgGateway?hgsid=103493475&clade=vertebrate&org=Human&db=hg18) and Stanford SOURCE GeneReports, Microarray Gene Expression Data field and Unigene & EST Expression Information field (http://source.stanford.edu/cgi-bin/source/sourceSearch). UCSC Mouse Genome Browser, Aug. 2005 assembly: http://genome.ucsc.edu/cgi-bin/hgGateway?hgsid=103493475&clade=vertebrate&org=Mouse&db=mm7.
Supplementary Material
Acknowledgments
We thank the Yale Transgenic Mouse and Gene Targeting Resource for injection of transgene constructs and E. Boyden, S. Boyden, A. Gharavi and C. Nelson- Williams for helpful discussions. Supported in part by the Howard Hughes Medical Institute, the Wellcome Trust and the US National Institutes of Health.
Footnotes
Accession codes. Skint1: EF494889, EF494890, EF494891, EU099296, EU099297, EU099298. Skint2: EU099299, EU099300, EF494892, EF494893. Skint3: EF494894, EF494895, EF494896. Skint4: EF494897, EF494898. Skint5: EU099301, EU099302, EU099303. Skint6: EU099304, EU099305. Skint7: EU099306, EF494899, EF494900, EF494901, EF494902. Skint8: EU099307, EF494903, EF494904, EU099308. Skint9: EF494907. Skint10: EF494905, EF494906. Skint11: EU099309. Accession codes are also listed in Supplementary Figure 3.
References
- 1.Janeway CA, Travers P, Walport M, Schlomchick M. Immunobiology. 5. Garland Publishing; New York: 2001. [Google Scholar]
- 2.Allison JP, Havran WL. The immunobiology of T cells with invariant γδ antigen receptors. Annu Rev Immunol. 1991;9:679–705. doi: 10.1146/annurev.iy.09.040191.003335. [DOI] [PubMed] [Google Scholar]
- 3.Hayday AC. γδ cells: a right time and a right place for a conserved third way of protection. Annu Rev Immunol. 2000;18:975–1026. doi: 10.1146/annurev.immunol.18.1.975. [DOI] [PubMed] [Google Scholar]
- 4.Lewis JM, et al. Selection of the cutaneous intraepithelial γδ+ T cell repertoire by a thymic stromal determinant. Nat Immunol. 2006;7:843–850. doi: 10.1038/ni1363. [DOI] [PubMed] [Google Scholar]
- 5.Mombaerts P, Arnoldi J, Russ F, Tonegawa S, Kaufmann SHE. Different roles of αβ and γδ T cells in immunity against an intracellular bacterial pathogen. Nature. 1993;365:53–56. doi: 10.1038/365053a0. [DOI] [PubMed] [Google Scholar]
- 6.Roberts SJ, et al. T-cell αβ+ and γδ+ deficient mice display abnormal but distinct phenotypes toward a natural, widespread infection of the intestinal epithelium. Proc Natl Acad Sci USA. 1996;93:11774–11779. doi: 10.1073/pnas.93.21.11774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Souza CD, et al. An anti-inflammatory role for γδ T lymphocytes in acquired immunity to Mycobacterium tuberculosis. J Immunol. 1997;158:1217–1221. [PubMed] [Google Scholar]
- 8.King DP, et al. Cutting edge: protective response to pulmonary injury requires γδ T lymphocytes. J Immunol. 1999;162:5033–5036. [PubMed] [Google Scholar]
- 9.Moore TA, Moore BB, Newstead MW, Standiford TJ. γδ-T cells are critical for survival and early proinflammatory cytokine gene expression during murine Klebsiella pneumonia. J Immunol. 2000;165:2643–2650. doi: 10.4049/jimmunol.165.5.2643. [DOI] [PubMed] [Google Scholar]
- 10.Selin LK, Santolucito PA, Pinto AK, Szomolanyi-Tsuda E, Welsh RM. Innate immunity to viruses: control of Vaccinia virus infection by γδ T cells. J Immunol. 2001;166:6784–6794. doi: 10.4049/jimmunol.166.11.6784. [DOI] [PubMed] [Google Scholar]
- 11.Zachariadis O, Cassidy JP, Brady J, Mahon BP. γδ T cells regulate the early inflammatory response to Bordetella pertussis infection in the murine respiratory tract. Infect Immun. 2006;74:1837–1845. doi: 10.1128/IAI.74.3.1837-1845.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girardi M, et al. Regulation of cutaneous malignancy by γδ T cells. Science. 2001;294:605–609. [Google Scholar]
- 13.Shiohara T, Moriya N, Hayakawa J, Itohara S, Ishikawa H. Resistance to cutaneous graft-vs.-host disease is not induced in T cell receptor d gene-mutant mice. J Exp Med. 1996;183:1483–1489. doi: 10.1084/jem.183.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng SL, Madaio MP, Hayday AC, Craft J. Propagation and regulation of systemic autoimmunity by γδ T cells. J Immunol. 1996;157:5689–5698. [PubMed] [Google Scholar]
- 15.Girardi M, et al. Resident skin-specific γδ T cells provide local, nonredundant regulation of cutaneous inflammation. J Exp Med. 2002;195:855–867. doi: 10.1084/jem.20012000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jameson J, et al. A role for skin γδ T cells in wound repair. Science. 2002;296:747–749. doi: 10.1126/science.1069639. [DOI] [PubMed] [Google Scholar]
- 17.Wang T, et al. γδ T cells facilitate adaptive immunity against West Nile virus infection in mice. J Immunol. 2006;177:1825–1832. doi: 10.4049/jimmunol.177.3.1825. [DOI] [PubMed] [Google Scholar]
- 18.Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 19.Rat Genome Sequencing Project Consortium. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 20.Kühnlein P, et al. The canonical T cell receptor of dendritic epidermal γδ T cells is highly conserved between rats and mice. Eur J Immunol. 1996;26:3092–3097. doi: 10.1002/eji.1830261240. [DOI] [PubMed] [Google Scholar]
- 21.Hein WR, Dudler L. TCR γδ+ cells are prominent in normal bovine skin and express a diverse repertoire of antigen receptors. Immunology. 1997;91:58–64. doi: 10.1046/j.1365-2567.1997.00224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Rhijn I, et al. Massive, sustained γδ T cell migration from the bovine skin in vivo. J Leukoc Biol. 2007;81:968–973. doi: 10.1189/jlb.0506331. [DOI] [PubMed] [Google Scholar]
- 23.Bos JD, et al. T-cell receptor γδ bearing cells in normal human skin. J Invest Dermatol. 1990;94:37–42. doi: 10.1111/1523-1747.ep12873333. [DOI] [PubMed] [Google Scholar]
- 24.Holtmeier W, et al. The TCR δ repertoire in normal human skin is restricted and distinct from the TCR δ repertoire in the peripheral blood. J Invest Dermatol. 2001;116:275–280. doi: 10.1046/j.1523-1747.2001.01250.x. [DOI] [PubMed] [Google Scholar]
- 25.Ebert LM, Meuter S, Moser B. Homing and function of human skin γδ T cells and NK cells: relevance for tumor surveillance. J Immunol. 2006;176:4331–4336. doi: 10.4049/jimmunol.176.7.4331. [DOI] [PubMed] [Google Scholar]
- 26.Rhodes DA, Stammers M, Malcherek G, Beck S, Trowsdale J. The cluster of BTN genes in the extended major histocompatibility complex. Genomics. 2001;71:351–362. doi: 10.1006/geno.2000.6406. [DOI] [PubMed] [Google Scholar]
- 27.Sharpe AH, Freeman JG. The B7–CD28 superfamily. Nat Rev Immunol. 2002;2:116–126. doi: 10.1038/nri727. [DOI] [PubMed] [Google Scholar]
- 28.Valentonyte R, et al. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet. 2005;37:357–364. doi: 10.1038/ng1519. [DOI] [PubMed] [Google Scholar]
- 29.Fujiwara K, Igarashi J, Irahara N, Kimura M, Nagase H. New chemically induced skin tumour susceptibility loci identified in a mouse backcross between FVB and dominant resistant PWK. BMC Genet. 2007;8:39. doi: 10.1186/1471-2156-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tigelaar RE, Lewis JM, Bergstresser PR. TCR γ/δ+ dendritic epidermal T cells as constituents of skin-associated lymphoid tissue. J Invest Dermatol. 1990;94:58S–63S. doi: 10.1111/1523-1747.ep12875138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.